Abstract
Biogeographic partitioning of the genome is typical of the gastric pathogen Helicobacter pylori. Such population-specific evolution could serve as a model for understanding host-pathogen interaction and the impact of genetic drift and recombination on insular populations. With a total of 320 isolates from six geographic regions (Japan, India, England, Spain, Ireland, Africa, and Peru) analyzed by enterobacterial repetitive intergenic consensus (ERIC)-based genotyping, we examined genetic affinities among various H. pylori populations in the world. Several strain-specific and region-specific differences were observed by ERIC-based typing. Polymorphic ERIC patterns indicated that the ERIC sequences are in fact dispersed in the H. pylori chromosome at different locations separated by various distances. Phylogenetic analysis of 61 representative isolates revealed three distinct genetic clusters populated by isolates with shared ERIC types independent of the cag right-junction motif type and vacA allele status. Among the notable genetic relationships were the genotypic similarities between Irish and Japanese and between Peruvian and Japanese isolates. Insular genotypic characteristics of Irish isolates amid genetic similarity to East Asian, as well as North European, strains have been once again proved in this study. Peruvian genotypes were more similar to those of Japanese isolates than to those of Iberian or European isolates. Given the current debate on the origin and age of present-day H. pylori, this is a significant finding that supports the possibility of ancient colonization of Amerindians with East Asian strains. Genotypic data presented here will be additionally helpful in realizing the importance of H. pylori geographical genomics in the development of gastroduodenal pathology.
Stomach infection with Helicobacter pylori is the second most common infectious disease of humans. The severe pathological consequences of this infection include gastric and duodenal ulcer disease, the development of gastric mucosal atrophy, gastric carcinoma, and, more rarely, malignant tumors of the lymphoma (12, 14, 15, 17, 18). H. pylori infections cause very high morbidity and mortality and are of particular concern in developing countries, where H. pylori prevalences as high as 90% have been reported (6, 20, 31).
H. pylori isolates obtained from different individuals and ethnic groups in the world exhibit substantial genomic diversity due to synonymous substitutions, insertion-deletion (indel) polymorphisms, and mobility of repetitive elements. This diversity could be further enhanced by chromosomal rearrangements due to a high level of interstrain recombination. Geographical partitioning of the gene pool exists within H. pylori, and sequences are less related between isolates from different populations than between isolates from families (1). Several molecular typing tools were tried for strain typing and identification of H. pylori isolates. These include pulsed-field gel electrophoresis (32), random fragment length polymorphism (28), randomly amplified polymorphic DNA (4, 24), amplified fragment length polymorphism (2, 11), and PCR-based genotyping of repetitive sequences, namely, repetitive extragenic palindromes (21, 22) and enterobacterial repetitive intergenic consensus elements (5). All these techniques indicate that the H. pylori population genetic structure is panmictic, and a high level of DNA diversity is found within strains. Despite this, reliable molecular methods are needed to generate data on isolate divergence and relatedness on a global scale to help us better understand the nature and consequences of host pathogen interaction in H. pylori. The importance of genome sequence-based fingerprinting techniques in explaining virulence characteristics, such as identification of tubercle bacilli linked to human immunodeficiency virus-positive patients (3) and those related to virulent Beijing genotypes (16), has been appreciated in a post-genome sequencing scenario.
The coevolution of H. pylori with the human host has been a subject of intense debate. Possible symbiotic relationships have been debated since the discovery of H. pylori.
The debate has been further intensified as recent studies have posed the intriguing possibility that H. pylori infection may be advantageous in some cases, like infant diarrhea and esophageal disease, where it confers protection on the individual (34). It is believed that H. pylori infects >50% of the world's population. However, only a small subset of infected people experience H. pylori-associated illnesses. The cause of this mystery can be known if we analyze the genetic diversity of the pathogen juxtaposed with the host diversity in the world. The available typing methods, therefore, must be robust enough with respect to reproducibility, specificity, resolving power, stability, and the molecular clocks of the markers used. Unfortunately, for H. pylori, no single method has been deemed a “gold standard” for strain typing and phylogeny.
Enterobacterial repetitive intergenic consensus (ERIC)-PCR was originally developed for typing Mycobacterium tuberculosis (29) and has been useful for differentiating strains of Vibrio cholerae, Salmonella enterica serovar Typhimurium, S. enterica serovar Typhi, Escherichia coli, and Klebsiella pneumoniae (13, 23). These sequences are a novel class of highly conserved DNA elements in gram-negative bacteria and other enterobacteria. The ERIC sequence is 126 bp long and includes a central inverted-repeat sequence which could form a stem-loop structure when transcribed into RNA. ERIC sequences are highly conserved at the nucleotide sequence level, but their chromosomal locations differ among species and strains (19). The elements have been successfully used for molecular typing purposes. By use of ERIC-PCR, differences in band sizes representing polymorphism due to repetitive elements in different genomes can be analyzed for developing phylogeographic relationships. This also allows the identification of interstrain genotypic diversity (35). ERIC sequences have been of special interest to us, as they have been reported to be conserved in enteric bacteria that have very long evolutionary histories. This contrasts with the evolutionary history of H. pylori, whose phylogenetic descent has been a subject of intense argument. It is believed that H. pylori might have recently diverged from its ancestor after agriculture began in Europe (17). These concerns and our continued interest in understanding the nature and extent of H. pylori diversity in a global scenario (2, 7) has led to the present study to establish the global phylogeny of H. pylori based on ERIC profiling.
MATERIALS AND METHODS
Patients, bacterial isolates, and DNA samples.
A total of 320 different isolates recovered from unrelated patients from seven different countries were selected irrespective of the disease severity. All of the isolates were recovered from patients undergoing upper gastrointestinal endoscopy after informed consent. All the protocols related to the collection and analysis of patient samples were approved by the institutional biosafety committees of the concerned institutes and hospitals participating in the study. Bacterial isolates were cultured from antral biopsy specimens as described previously (2). Almost all of the isolates were positive for the presence of cagA and vacA genes. Only two English isolates were cagA negative. Genomic DNA samples were prepared, quantified, and preserved according to methods described elsewhere (2). Genomic-DNA preparations belonging to patients from Peru, Spain, Japan, England, and South Africa were received from Douglas E. Berg, Washington University, St. Louis, Mo. DNA isolates from Ireland were provided by Ian M. Carroll (Department of Microbiology, Moyne Institute of Preventive Medicine, Trinity College, Dublin, Ireland).
ERIC-PCR.
PCRs were carried out using ERIC primers (forward primer-1R, 5′ ATGTAAGCTCCTGGGGATTCAC 3′, and reverse primer-2R, 5′ AAGTAAGTGACTGGGGTGAGCG 3′) (5). This primer pair had been extensively used previously (8, 25, 26, 29, 35). All the PCRs were performed in a total volume of 20 μl containing 1.0 μl of DNA (100 ng/μl), 10× PCR buffer (Applied Biosystems), 2 mM MgCl2 (MBI), 80 μM deoxynucleoside triphosphates (Amersham Biosciences), 10 pM (each) forward and reverse primers, and 2 U of Taq DNA polymerase (Applied Biosystems). The total volume was made up with autoclaved Milli-Q water. Furthermore, appropriate positive controls containing purified genomic DNA that gave the maximum number of distinct bands and negative controls lacking either DNA or primers or both were included along with each test run. Dedicated pipettes and gloves and plugged pipette tips were used to prevent template carryover. Different laboratory spaces were dedicated to template preparation and PCR setup to avoid contamination. PCR was performed in a 9700 thermal cycler (Applied Biosystems). The amplification conditions were optimized as follows: initial denaturation for 5 min at 94°C, followed by 35 cycles of denaturation at 90°C for 30 s, annealing at 49°C for 1 min, and extension at 70°C for 5 min. After a final extension at 70°C for 10 min, the amplified products were separated on a 1% agarose gel in 1× TAE buffer (40 mM Tris-acetate, 1 mM EDTA, pH 8.0) containing ethidium bromide (0.5 μg/ml). The DNA was then visualized under UV light. Interlaboratory reproducibility of the method was established after repeating the experiments with sample replicates in two different laboratories (at the Centre for DNA Fingerprinting and Diagnostics [CDFD] and Deccan College of Medical Sciences, Hyderabad, India).
Gel analysis and phylogeny construction.
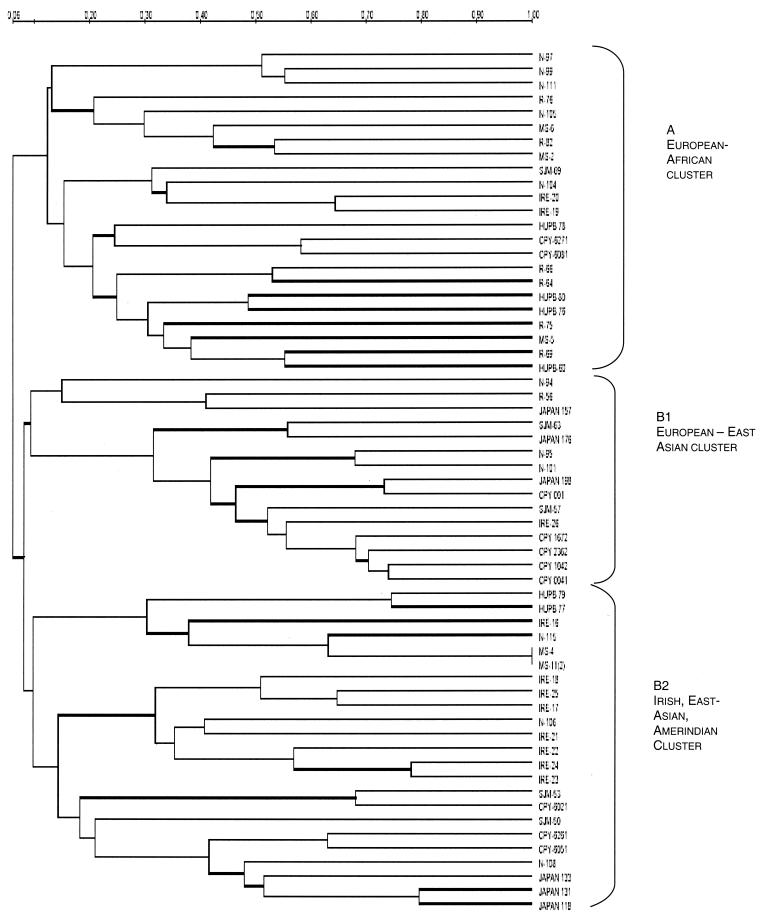
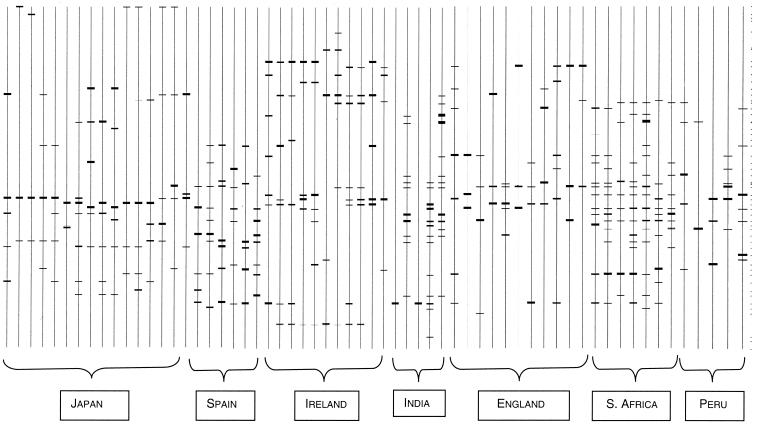
All of the gel images were analyzed using Quantity version 1.0 software in a gel documentation system (Bio-Rad). The images were then uploaded into the Diversity version 2.2.0 database (Bio-Rad). Band sizes, band attributes, and standard molecular weights were assigned according to the molecular weight markers. Cluster analysis of all 61 isolates was conducted on the basis of the fingerprint characteristics generated. Based on the data for the presence or absence of 3 to 15 different DNA fragments in the fingerprints of the 61 strains of H. pylori, a binary data matrix was created. Overall similarity between the pairs of strains was calculated from the binary data matrix using the simple matching-dice coefficient. The resulting similarity matrix was used as the input data for cluster analysis by the unweighted-pair-group method with arithmetic averages (27) to generate a dendrogram (Fig. 1 and 2).
FIG. 1.
Phylogenetic tree based on ERIC-PCR profiles of representative isolates from different geographical regions. The scale bar at the top indicates the extent of genetic relatedness among the isolates, with 1.00 corresponding to 100% identity.
FIG. 2.
Symbolic banding patterns for ERIC fingerprints generated with representative isolates using Quantity version 1.0 software. Short horizontal lines denote ERIC amplicon traces observed on the agarose gel. Each vertical line signifies a single lane on the gel corresponding to one genotype identifier for a single isolate.
RESULTS
Amplification of H. pylori genomic DNA with ERIC oligonucleotides.
All of the strains were found to be analyzable by the ERIC-PCR method and yielded significant PCR products. This indicates that the ERIC sequences are in fact dispersed in the H. pylori chromosome at different locations separated by various distances. These widely spread repetitive sequences could be used as primer binding sites, and PCR amplification between them might yield divergent fingerprints when separated by agarose gel electrophoresis. Depending upon the abundance and availability of ERIC sequences, fingerprints of the isolates representing different geographic regions comprised 3 to 15 bands of various sizes. The maximum number (up to 15) of amplified fragments was observed for strains from South Africa and India, whereas a few polymorphic fragments were observed for East Asian, European, and South American isolates. The sequenced strain J99 produced four amplified fragments. All the isolates were found to have ERIC amplicons sufficient to score for similarity indices. However, a couple of Indian isolates failed to generate many fragments and could amplify only a single locus. We believe that this might be due to degradation of the genomic DNA.
Specificity of ERIC amplification.
All the profiles generated by ERIC PCR were found to be highly specific. This was confirmed by homologies for DNA sequence similarity to the two sequenced genomes (http://genolist.pasteur.fr/PyloriGene/index.html). Both the primer sequences and short stretches of amplified fragments were mapped to several important genes, particularly those coding for GuaB, NhaH, Ppic, Fba, Tbr, PpsA, and UvrA and rRNA. It appears that the sequences of primers we used were fairly abundant in the genome, flanking the sequences of variable length, thus generating robust, polymorphic profiles.
Reproducibility and resolution power.
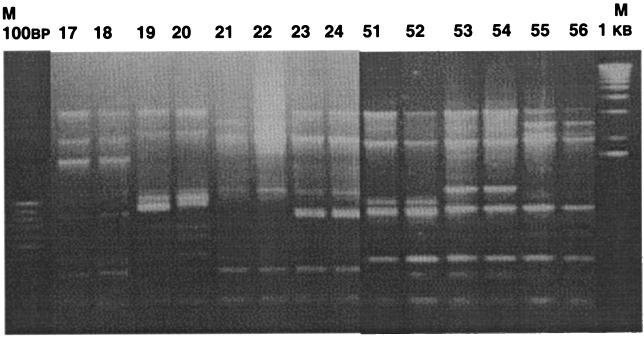
In our experiments, the ERIC profiling results were 100% reproducible on all occasions and were not found to be discrepant within and between laboratories. In our analysis, no two isolates were found to be 100% similar. All the isolates produced highly specific and individualized fingerprints. This reflects the resolution power of this technique, by which isolates from the same population or geographic region can be distinguished and assigned a unique genotype identifier for tracing the movement and descent of pathogenic clones in communities. This contrasts with earlier observations of Vibrio parahaemolyticus and other pathogens in which the amplification of a large number of monomorphic fragments was recorded for unrelated isolates (35). In our case, ERIC-PCR was able to resolve even the microevolution of paired isolates (Fig. 3) obtained from some patients months apart. These isolates were equally resolvable with other typing techniques, such as fluorescent amplified fragment length polymorphism (FAFLP) analysis (data not shown). FAFLP is based on selective PCR amplification of restriction fragments of genomic DNA (11), while ERIC-PCR involves amplification of the genomic DNA flanked by the ERIC sequences. Overall, the ERIC-PCR technique was found to be robust, reproducible, portable, easy to perform, and highly discriminating once standardization was accomplished.
FIG. 3.
ERIC profiles generated with paired Irish isolates. Numbers identifying isolates are given at the top. M, molecular mass markers with 100-bp (left) and 1-kb (right) ladders.
Genetic affinities and identification of population-specific clusters.
Genetic relationships among H. pylori strains were examined through cluster analysis of the PCR-generated patterns from representative isolates (n = 61) and were presented in the form of a dendrogram (Fig. 1). The most important relationships obtained were as follows. Cluster A (European-African) included 23 of 61 isolates (37.7%), mainly from England, Spain, South Africa, and India. Clustering, however, was independent of the disease type or severity, as clusters contained isolates irrespective of cagA gene status and vac allele type. Also, the genetic affinities for clustering were independent of the motif types based on indel and substitution patterns found at the right end of the H. pylori cag pathogenecity island, as described previously (17). Isolates belonging mainly to two different indel categories, such as type III and type I, were found to be genetically similar based on ERIC profiles. A few English strains belonging to indel type III and vac allele type s1m2, however, formed a separate branch within the cluster. Subcluster B1 (European-East Asian) comprised 15 isolates (24.5%) representing mainly Japanese and European isolates. These again included mixed indel profiles but had mainly s1m2 alleles of the vacA gene. The cluster comprised independent isolates from Japan and Peru. One Irish isolate with rare type IV indel motifs was also found to cluster with Japanese isolates in this subcluster. Subcluster B2 (Irish-East Asian-Amerindian) included equal number of isolates from Japan and Ireland, highlighting the possibility of some genetic relatedness between the two insular human populations. It is also composed of a dedicated lineage leading to an exclusively Irish subsubcluster. The Irish aggregate belonged to vac allele type s1am2, although the indel motif type varied. However, none of the Irish isolates were found to be clonal, although a few weak clonal associations were discerned. A third branch in subcluster B2 described genetic relationships among Japanese and native Peruvian isolates. None of the isolates in this class showed similar indel motifs as well as vac allele types. The cag motif types and vacA allele statuses for isolates included in the study are listed in Table 1.
TABLE 1.
cag right-junction motif types and vac allele statuses of representative isolates typed by ERIC-PCR
| Cluster | Isolate | cag motif type | vac status |
|---|---|---|---|
| A | N 97 | 3a | s1m2 |
| N 99 | 3a | s1m2 | |
| N 111 | 3a | s1m2 | |
| R 76 | 1a/1b | NDa | |
| N 105 | 1a | s1m1a | |
| MS 6 | 3b | s2m2 | |
| R 82 | 1a/1b | ND | |
| MS 2 | ND | s2m2 | |
| SJM 69 | 1a/1b | ||
| N 104 | CagA minus strain | s1m2 | |
| Ire 20 | ND | ND | |
| Ire 19 | ND | ND | |
| HupB 78 | 1a/1b | ND | |
| CPY 6271 | 2 | ND | |
| CPY 6081 | 2 | ND | |
| R 66 | 1a/1b | ND | |
| R 64 | 1a/1b | ND | |
| HUPB 80 | 1a/1b | ND | |
| HUBP 76 | 1a/1b | ND | |
| R 75 | 1a/1b | ND | |
| MS 5 | 3a | s2m1b | |
| R 69 | 1a/1b | ND | |
| HUPB 60 | 1a/1b | ND | |
| B1 | N 94 | CagA minus strain | s1m2 |
| R 56 | 1a/1b | ND | |
| JAPAN 157 | 2 | ND | |
| SJM 63 | 1a/1b | ND | |
| JAPAN 176 | 2 | ND | |
| N 95 | 1a | s1m1a | |
| N 101 | 1b | s1m2 | |
| JAPAN 198 | 2 | ND | |
| CPY 001 | 2 | ND | |
| SJM 57 | 1a/1b | ND | |
| IRE 26 | 4 | s1am2 | |
| CPY 1672 | 2 | ND | |
| CPY 2362 | 2 | ND | |
| CPY 1042 | 2 | ND | |
| CPY 0041 | 2 | ND | |
| B2 | HUPB 79 | 1a/1b | ND |
| HUPB 77 | 1a/1b | ND | |
| IRE 16 | ND | ND | |
| N 115 | 2 | s1 | |
| MS4 | 3a | s1m1b | |
| MS11 (2) | - | s1m2 | |
| IRE 18 | 2 | s1am2 | |
| IRE 25 | 4 | s1am2 | |
| IRE 17 | 2 | s1am2 | |
| N 106 | 1b | s1m2 | |
| IRE 21 | 1a | s1am2 | |
| IRE 22 | 1a | s1am2 | |
| IRE 24 | ND | ND | |
| IRE 23 | ND | ND | |
| SJM 53 | 1a/1b | ND | |
| CPY 6021 | 2 | ND | |
| SJM 50 | 1a/1b | ND | |
| CPY 6261 | 2 | ND | |
| CPY 6051 | 2 | ND | |
| N 108 | 2 | s1m1a | |
| JAPAN 133 | 2 | ND | |
| JAPAN 131 | 2 | ND | |
| JAPAN 118 | 2 | ND |
ND, data not available.
The main message from phenetic analysis of the isolates in our collection revealed no impact of indel motif type and vac allele status on the phylogeographic clustering based on ERIC patterns. Highly shared ERIC types among Irish isolates are a striking observation. Our analysis revealed homogeneous fingerprinting profiles among Irish isolates (compared to isolates from other geographical regions) that have major genetic relatedness with European and East Asian isolates. These findings are in agreement with a recent report (9) highlighting an Asian or oriental influence on certain European populations of H. pylori. Japanese and Peruvian isolates may be genetically linked due to ancient Asian ancestry of the present-day Amerindians in the Americas. Interestingly, these findings are perfectly in agreement with recent (9, 10) studies highlighting the possibility that Amerindian strains may carry certain alleles from their ancient predecessors of Japanese origin.
DISCUSSION
In this study, we analyzed the nature of prevalent genetic diversity among various H. pylori isolates in the world. The isolates were carefully selected to fulfill the requirement for a truly representative international collection of strains. ERIC sequence-based genotyping was identified as a novel method for phylogenetic analysis of H. pylori. These sequences are examples of bacterial interspersed mosaic elements that appear to be transcribed, although they may be present in the intergenic regions or in the 3′ or 5′ untranslated regions of the protein coding regions. The presence of intergenic regions in the bacterial chromosome helps in controlling potential signals for the processes of transcription and translation. These sequences include transcriptional promoters and terminators. However, there is no correlation between the presence of ERICs and the function of an operon. Furthermore, ERICs can be found in either orientation with respect to the direction of transcription, implying that any function is at a secondary structural level rather than being determined by the primary sequence (13). An earlier study by Veralovic and colleagues (33) indicated that these sequences are fairly conserved across gram-negative enteric bacteria and related species for at least hundreds of millions of years. This suggests that their presence precedes even the formation of the gram-negative enteric bacterial lineage. Since these sequences represent sites of interaction of DNA with essential proteins (such as DNA gyrase and polymerase I in E. coli), substitutions, deletions, and genetic rearrangements may be constraints. Also, they may propagate themselves as RNA intermediates, accounting for the widespread presence and conservation of these prokaryotic repetitive elements (33).
ERIC-PCR has been employed to investigate the rate of human intestinal infections with more than a single Campylobacter strain and to study the genetic variabilities of these strains throughout an infection episode (30).
Recently, fingerprints generated by ERIC-PCR suggested the presence of such repeating sequences in H. pylori (5). It was also shown that a mixed population of different H. pylori strains with marked variation, both genotypically and phenotypically, may colonize the same patient (5). Our study, however, was aimed at testing the utility of such a typing system to analyze H. pylori genotypic partitioning on a global scale and to establish its merit in identification of isolates for epidemiology and phylogeography. In our experiments, the exact positions and potential functionalities of the genomic landmarks of these sequences were located in the H. pylori chromosome. However, there were only partial matches with the total 126-bp ERIC sequence in the H. pylori genome. It appears that the primer-annealing regions were found scattered throughout the chromosome, separated by regions of various lengths. Furthermore, no transposons were found in the flanking genes of the open reading frames in which ERIC sequences have been located.
Of all 61 isolates typed by ERIC-PCR, no two isolates generated identical profiles, except for a case of paired isolates and two Indian isolates that failed to generate multiple bands for unknown reasons. However, prominent bands were common in strains from individual countries. For instance, an amplified product of 800 bp was present in all Japanese isolates, fragments of 200 bp and 2.0 and 3.5 kb were specific to Irish isolates, and fragments of 700 and 270 bp demarcated South African isolates. Phylogenetic analysis revealed interesting links between isolates from Japan and Peru, contrasting with earlier observations established with the same set of strains (17). Our results based on ERIC typing were also not in agreement with recent observations of Japanese and Peruvian isolates based on FAFLP analysis, in which Peruvian isolates were found to be phylogenetically more closely related to Spanish isolates than to Japanese isolates (7). However, the number of isolates analyzed in that study was insufficient to establish conclusive data (only one Japanese isolate was studied) (7). Compared to FAFLPs, the reliability of ERIC-based phylogenies may be argued on the basis of the fact that these markers are reported to be evolutionarily conserved and stable across the enterobacterial branch in evolution (13, 26, 33). Our results, on the other hand, support the findings based on sequence analysis of some evolutionarily important gene loci and housekeeping genes (9, 10) for which a possible evolutionary link between South American and Japanese isolates has been argued.
Another interesting observation based on ERIC typing was the distinctiveness of Irish isolates. This is yet more evidence to confirm previous findings, which argued that Irish isolates have certain distinctive features compared to isolates from other European countries (7). These features include weakly clonal population structure, homogeneous fingerprinting profiles from unrelated individual isolates, and evolutionary links with both East Asian and Northern European isolates. We suggest that such features might have evolved as a result of a complex interplay between the host and pathogen in which recombination has not been sufficiently frequent to destroy the evidence for clonality in this insular European population. This is in accordance with a recent landmark study based on the sequences of seven housekeeping genes (atpA, efp, mutY, ppa, trpC, ureI, and yphC) and one virulence gene (vacA) (9) that demonstrated that Indian and European H. pylori isolates grouped in the same subpopulation and that East Asian and a subset of European isolates share an ancestral relationship and diverged from each other recently.
During this study, we found the ERIC-PCR technique to be easy to perform compared to restriction fragment length polymorphism or FAFLP, with reproducibility of >95%. We agree that any molecular marker to be used as a discriminative tool in molecular epidemiological studies needs to be inherited stably over the years. This has been proven efficiently with ERIC sequences, as ERIC-PCR carried out on paired isolates from Ireland that were obtained from the same patient over a period of time produced identical profiles (data not shown). These results might also give an idea of the conserved nature of these sequences over time, considering that the rest of the H. pylori genome is quickly evolving. However, further studies of the molecular clocks of these sequences in H. pylori are clearly required.
We do not believe that shared genotypes observed in our phylogeny were the result of artifacts or contamination. The inclusion of appropriate controls and the use of dedicated instruments and laboratory spaces made DNA carryover very unlikely. However, since the technique was used in isolation, its discriminative potential could not be assessed in comparison with techniques based on other repetitive sequences, namely, repetitive extragenic palindromes, etc. The ERIC profiles generated in this study are suitable for and amenable to electronic transmission and deposit in various national and international databases. It may be possible to develop a web-enabled archive of such fingerprint profiles that allows archiving and comparison of ERIC profiles with fingerprint profiles obtained with various other typing systems on a populationwide interface.
Finally, the present study of H. pylori genotypes from different human populations may be useful to our understanding of bacterium-host interactions and the origin and evolution of the bacterium in humans. Furthermore, it will also be helpful to realize the importance of H. pylori geographical genomics in gastroduodenal pathology.
Acknowledgments
We are very grateful to Seyed E. Hasnain for his patronage, encouragement, guidance, and support. Special thanks are due to Douglas E. Berg (Washington University at St. Louis) for the supply of genomic DNA from H. pylori isolates from non-Irish and non-Indian origins, originally obtained by Robert H. Gilman, Teresa Alarcon, Manuel Lopez Brea, and John Atherton. We also thank Ian M. Carroll and Cyril J. Smyth (Trinity College, Dublin, Ireland) for providing genomic DNA samples of Irish isolates. Thanks are also due to C. S. Jyothirmayee and K. Rajender Rao for their support and to Surya K. Mishra for help with phylogeny construction.
This study was funded in part by a grant from the Department of Biotechnology, Government of India (DBT Task Force on Human Genetics and Genomics-Helicobacter Program 2001) to N.A. and C.M.H. N.A. is a staff scientist and group head at CDFD and assistant professor of molecular biology (adjunct) at the Faculty of Veterinary Sciences, ANG Ranga Agricultural University, Hyderabad, India.
REFERENCES
- 1.Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459-470. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed, N., A. A. Khan, A. Alvi, S. Tiwari, C. S. Jyothirmayee, F. Kauser, M. Ali, and C. M. Habibullah. 2003. Genomic analysis of Helicobacter pylori from Andhra Pradesh, South India: molecular evidence for three major genetic clusters. Curr. Sci. 85:101-108. [Google Scholar]
- 3.Ahmed, N., L. Caviedes, M. Alam, K. R. Rao, V. Sangal, P. Sheen, R. H. Gilman, and S. E. Hasnain. 2003. Distinctiveness of Mycobacterium tuberculosis genotypes from human immunodeficiency virus type 1- seropositive and -seronegative patients in Lima, Peru. J. Clin. Microbiol. 41:1712-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akopyantz, N., N. O. Bukanov, T. U. Westblom, S. Kresovich, and D. E. Berg. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 20:5137-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ann-Catrin, E. T., N. Hosseini, A. M. Svennerholm, and I. Bölin. 2000. Different Helicobacter pylori strains colonize the antral and duodenal mucosa of duodenal ulcer patients. Helicobacter 5:69-78. [DOI] [PubMed] [Google Scholar]
- 6.Atherton, J. C. 1997. The clinical relevance of strain types of Helicobacter pylori. Gut 40:701-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll, I. M., N. Ahmed, S. M. Beesley, A. A. Khan, S. Ghousunnissa, C. A. O'Morain, and C. J. Smyth. 2003. Fine-structure molecular typing of Irish Helicobacter pylori isolates and their genetic relatedness to strains from four different continents. J. Clin. Microbiol. 41:5755-5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eric, M., E. Jumas-Bilak, A. Allardet-Servent, D. O'Callaghan, and M. Ramuz. 1996. Polymorphism in Brucella strains detected by studying distribution of two short repetitive DNA elements. J. Clin. Microbiol. 34:1299-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falush, D., T. Wirth, B. Linz, J. K. Pritchard, M. Stephens, M. Kidd, M. J. Blaser, D. Y. Graham, S. Vacher, G. I. Perez-Perez, Y. Yamaoka, F. Mégraud, K. Otto, U. Reichard, E. Katzowitsch, X. Wang, M. Achtman, and S. Suerbaum. 2003. Traces of human migrations in Helicobacter pylori populations. Science 299:1582-1585. [DOI] [PubMed] [Google Scholar]
- 10.Ghose, C., G. I. Perez-Perez, M. G. Dominguez-Bello, D. T. Pride, C. M. Bravi, and M. J. Blaser. 2002. East Asian genotypes of Helicobacter pylori strains in Amerindians provide evidence for its ancient human carriage. Proc. Natl. Acad. Sci. USA 99:15107-15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibson, J. R., E. Slater, J. Xerry, D. S. Tompkins, and R. J. Owen. 1998. Use of an amplified-fragment length polymorphism technique to fingerprint and differentiate isolates of Helicobacter pylori. J. Clin. Microbiol. 36:2580-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, J. Q., S. Sridhar, Y. Chen, and R. H. Hunt. 1998. Meta-analysis of the relationship between Helicobacter pylori seropositivity and gastric cancer. Gastroenterology 114:1169-1179. [DOI] [PubMed] [Google Scholar]
- 13.Hulton, C. S. J., C. F. Higgins, and P. M. Sharp. 1991. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium and other enterobacteria. Mol. Microbiol. 5:825-834. [DOI] [PubMed] [Google Scholar]
- 14.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. 1994. Schistosomes, liver flukes and Helicobacter pylori: views and expert opinions of an IARC working group on the evaluation of carcinogenic risks to humans. IARC Monographs, p. 177-240. International Association for Research on Cancer, Lyon, France. [PMC free article] [PubMed]
- 15.Ikeno, T., H. Ota, A. Sugiyama, K. Ishida, T. Katsuyama, R. M. Genta, and S. Kawasaki. 1999. Helicobacter pylori-induced chronic active gastritis, intestinal metaplasia, and gastric ulcer in Mongolian gerbils. Am. J. Pathol. 154:951-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Judith, R. G., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kersulyte, D., A. K. Mukhopadhyay, B. Velapatiño, W. W. Su, Z. J. Pan, C. Garcia, V. Hernandez, Y. Valdez, R. S. Mistry, R. H. Gilman, Y. Yuan, H. Gao, T. Alarcón, M. López-Brea, G. B. Nair, A. Chowdhury, S. Datta, M. Shirai, T. Nakazawa, R. Ally, I. Segal, B. C. Y. Wong, S. K. Lam, F. Olfat, T. Borén, L. Engstrand, O. Torres, R. Schneider, J. E. Thomas, S. Czinn, and D. E. Berg. 2000. Differences in genotypes of Helicobacter pylori from different human populations. J. Bacteriol. 182:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuipers, E. J. 1999. Exploring the link between Helicobacter pylori and gastric cancer. Aliment. Pharmacol. Ther. 13:3-11. [DOI] [PubMed] [Google Scholar]
- 19.Lupski, J. R., and G. M. Weinstock. 1992. Short, interspersed repetitive DNA sequences in prokaryotic genomes. J. Bacteriol. 174:4525-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matysiak-Budnik, T., and F. Megraud. 1997. Epidemiology of Helicobacter pylori with special reference to professional risk. J. Physiol. Pharmocol. 48:3-17. [PubMed] [Google Scholar]
- 21.Miehlke, S., R. Thomas, O. Gutierrez, D. Y. Graham, and M. F. Go. 1999. DNA fingerprinting of single colonies of Helicobacter pylori from gastric cancer patients suggests infection with a single predominant strain. J. Clin. Microbiol. 37:245-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nathalie, E., M. van Doorn, F. Namavar, L. J. van Doorn, Z. Durrani, E. J. Kuipers, and C. M. J. E. Vandenbroucke-Grauls. 1999. Analysis of vacA, cagA, and IS605 genotypes and those determined by PCR amplification of DNA between repetitive sequences of Helicobacter pylori strains isolated from patients with nonulcer dyspepsia or mucosa-associated lymphoid tissue lymphoma. J. Clin. Microbiol. 37:2348-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olive, D. M., and P. Bean. 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Owen, R. J., J. Bickley, M. Costas, and D. R. Morgan. 1991. Genomic variation in Helicobacter pylori: application for identification of strains. J. Gastroenterol. 181:43-50. [DOI] [PubMed] [Google Scholar]
- 25.Pettigrew, M. M., B. Foxman, Z. Ecevit, C. F. Marrs, and J. Gilsdorf. 2002. Use of pulsed-field gel electrophoresis, enterobacterial repetitive intergenic consensus typing, and automated ribotyping to assess genomic variability among strains of nontypeable Haemophilus influenzae. J. Clin. Microbiol. 40:660-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rivera, G., M. A. R. Chowdhury, A. Huq, D. Jacobs, M. T. Martins, and R. R. Colwell. 1995. Enterobacterial repetitive intergenic consensus sequences and the PCR to generate fingerprints of genomic DNAs from Vibrio cholerae O1, O139, and non-O1 strains. Appl. Environ. Microbiol. 61:2898-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romesbrug, H. C. 1984. Cluster analysis for researchers. Lifetime Learning Publications, Belmont, Calif.
- 28.Salaun, L., C. Audibert, G. Le Ley, C. Burucoa, J. L. Fauchere, and B. Picard. 1998. Panmictic structure of Helicobacter pylori demonstrated by the comparative study of six genetic markers. FEMS Microbiol. Lett. 161:231-239. [DOI] [PubMed] [Google Scholar]
- 29.Sechi, L. A., S. Zanetti, I. Dupré, G. Delogu, and G. Fadda. 1998. Enterobacterial repetitive intergenic consensus sequences as molecular targets for typing of Mycobacterium tuberculosis strains. J. Clin. Microbiol. 36:128-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinbrueckner, B., F. Ruberg, and M. Kist. 2001. Bacterial genetic fingerprint: a reliable factor in the study of the epidemiology of human Campylobacter enteritis? J. Clin. Microbiol. 39:4155-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suerbaum, S., and P. Michetti. 2002. Helicobacter pylori infection. N. Engl. J. Med. 347:1175-1186. [DOI] [PubMed] [Google Scholar]
- 32.Takami, S., T. Hayashi, H. Akashi, T. Shimoyama, and T. Tamura. 1994. Genetic heterogeneity of Helicobacter pylori by pulse-field gel electrophoresis and re-evaluation of DNA homology. Eur. J. Gastroenterol. Hepatol. Suppl. 1:S53-S60. [PubMed] [Google Scholar]
- 33.Veralovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in Eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitfield, J. 2003. Gut reaction. Nature 423:583-584. [DOI] [PubMed] [Google Scholar]
- 35.Wong, H. C., and C. H. Lin. 2001. Evaluation of typing of Vibrio parahaemolyticus by three PCR methods using specific primers. J. Clin. Microbiol. 39:4233-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]