Abstract
CREPT (cell-cycle related and expression-elevated protein in tumor), a novel gene also called RPRD1B and C20ORF77, was recently identified to promote tumorigenesis through up-regulation of the expression of genes related to cell cycle. The previous study demonstrated that CREPT is highly expressed in a variety of tumors and enhances the expression of Cyclin D1 by promoting the formation of a chromatin loop. To study the correlation of CREPT expression with clinical factors in different tumors, we generated a monoclonal antibody (3E10) using purified recombinant human GST-CREPT protein as an antigen. In this study, we characterized the specificity of the monoclonal antibody and cloned the gene encoding the antibody for preparation of industrial production. Our results showed that the monoclonal antibody 3E10 was sensitive and specific to recognize human endogenous CREPT protein. We have mapped the epitope of the antibody and cloned the variable region sequence of the gene encoding the antibody. We confirmed that the cloned gene produced an equivalent antibody as that produced by the original hybridoma. This study provided a basis for large-scale production of the CREPT antibody, which will be useful for the study of the role of CREPT in different tumors.
Introduction
Cell-cycle related and expression-elevated protein in tumor (CREPT) was identified by sequence alignment through the NCBI protein database with the recently reported protein p15RS, a novel p15INK4b-related gene involved in G1/S progression,(1) which was regarded as a negative regulator in cell cycle control in the G1 phase. p15RS was reported to negatively regulate cell proliferation, migration, and invasion.(1–3) In opposition, CREPT was highly expressed in a variety of tumors and promoted cell proliferation.(4) Previously Lu and colleagues found that the highly expressed CREPT was correlated with a shorter survival time of 117 stomach cancer patients.(4) Previously, Jung and colleagues reported that CREPT, together with seven other genes, was significantly elevated in SV40-immortalized cells, cancer cells, and non-small-cell lung cancer (NSCLC) tissues,(5) consistent with these observations. Recently, CREPT was reported to be highly expressed in activated T cells in the Chinese psoriatic cohort and might promote epidermal hyper-proliferation.(6) These studies suggest that CREPT might be a novel marker for diseases including cancers.
To understand the role of CREPT, we have generated a specific monoclonal antibody against CREPT. In the present study, we report the characterization of the antibody that recognizes human and mouse CREPT. We also mapped the epitope and cloned the gene of the antibody to engineer a chimeric antibody produced by HEK293T cells. Finally, we confirmed that the chimeric antibody is equivalent to that produced by the hybridoma.
Materials and Methods
Plasmids and reagents
The constructs with Flag-tagged human CREPT (pcDNA3.1/Flag-CREPT), Flag-CREPT-RPR domain (pcDNA3.1/Flag-CREPT-RPR), and Flag-CREPT-CCT domain (pcDNA3.1/Flag-CREPT-CCT) were constructed in our laboratory. GST-tagged CREPT (pGEX-5X2/CREPT) were constructed by a PCR based on the sequence of pcDNA3.1/Flag-CREPT. Anti-Flag (M2) monoclonal antibody and anti-β-actin (C-2) monoclonal antibody were purchased from Sigma-Aldrich (St. Louis, MO). Anti-Myc (9E10) and anti-p15RS (FF-8) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Generation of hybridomas and collection of mouse ascites
A female BALB/c mouse was subcutaneously injected with 0.1 mg of GST-CREPT protein in PBS and Freund's complete adjuvant. Animals were boosted three times with 0.1 mg antigen each time. The titer of antibodies in serum was measured by an ELISA assay. Mouse spleen cells were isolated and fused with an equivalent number of vital cells of mouse myeloma line SP2/0 by PEG, then suspended in HAT selection medium supplemented with 20% FCS (Gibco, Grand Island, NY) and seeded into 96-well plates. To select melted cells, aminopterin was added for 14 days (HAT supplement). Hypoxanthine and thymidine were added to the selection medium for a further period of 7 days. After selection, the remaining hybridoma cells were cultivated in RPMI1640/glutamine medium supplemented with 10% FCS. Hybridoma cells were subjected to selection with CREPT proteins purified from Escherichia coli. Three clones of hybridoma with positive activity of CREPT binding were obtained. For monoclonal antibody enrichment of antibody production, 1×106 hybridoma cells were intraperitoneally injected into a BALB/c mouse. Ascites were collected after 7 days.
ELISA
ELISA plates were coated with 50 ng/well anti-human IgG antibodies, 100 ng/well GST-CREPT, or 1 μg/well of synthesized peptides by incubation at 4°C overnight. BSA was used as a negative control. After washing three times with PBST, plates were blocked with 200 μL of blocking buffer (10% FBS in PBS) for 1 h. 100 μL/well murine CREPT monoclonal antibody and HEK293T cell lysates transfected with heavy and light chain plasmids were added and incubated for 1 h. An irrelevant plasmid was used as negative control. Unbound antibodies were removed by PBST before incubation with 100 μL/well HRP-coupled anti-mouse (1:2500) or anti-human IgG (1:3000) (Promega, Madison, WI). After 45 min of incubation with the secondary antibody, unbound antibodies were removed by PBST. Assay was developed using TMB substrate according to the manufacturer's instructions. ODs at 405 nm were measured to present the binding ability.
Mammalian cell transfection and immunoblotting
HEK293T cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum at 37°C in a 5% CO2 containing atmosphere. Lipofectamine 2000 (Invitrogen, Carlsbad, CA) was used for transient transfection of HEK293T cells according to the manufacturer's instructions. Cells were transfected with the indicated plasmids. After transfection for 24 h, cells were lysed in 200 μL of lysis buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 1% Nonidet P-40, and 1 mM sodium orthovanadate) in the presence of protease inhibitors. The cell lysates were separated by 12% SDS-PAGE and subsequently transferred to nitrocellulose membranes. Blocking of membranes was achieved by 1 h incubation in 10% skim milk in PBST. Membranes were incubated with monoclonal antibodies overnight in PBST at 4°C. After washing three times with PBST, membranes were incubated for 1 h at 37°C with anti-mouse or anti-human IgG conjugated with HRP, followed by washing four times and subsequent exposure.
Immunostaining and confocal microscopy
Cells growing on glass cover slips in 6-well plate were briefly washed with PBS and then fixed with 4% paraformaldehyde in PBS for 20 min at room temperature. After permeabilization using 0.3% Triton X-100 in PBS, cells were blocked with 10% normal goat serum for 1 h at room temperature. Cover slips were incubated with primary antibody of CREPT MAb 3E10 (diluted in 3% BSA-PBS) overnight at 4°C and bound antibody was detected with FITC- conjugated goat anti-mouse IgG. Cover slip was mounted in a glycerol-based anti-fade mounting medium and analyzed with a laser scanning confocal microscopy with a 60 X oil-immersion objective.
Human tumor specimens and immunohistochemical staining
Cancer tissues were collected in the Chinese PLA General Hospital. Tissues were kept and stained according to routine protocols.(7) The tissue collection procedure with informed consent was approved by the Clinical Ethic Committee of the Chinese PLA General Hospital. Paraffin-embedded specimens were fixed on glass slides for immunohistochemical analyses, which were performed according to an immunohistochemistry polymer double detection kit (ZhongShan Golden Bridge Bio, Beijing, China). Specifically, after blocking with 3% BSA in PBS, sections were incubated overnight with 3E10, a mouse anti-CREPT monoclonal antibody, at a dilution of 1:2000.
Epitope mapping of monoclonal antibody 3E10 using yeast surface library
A yeast surface library containing 100–500 bp fragments of CREPT coding sequence (CDS) was constructed following a previously described protocol.(8) The library was qualified through sequencing and aligning against the full-length CREPT sequence. The induced library cells were incubated with MAb 3E10 enriched from ascites (1:100) on ice for 1 h with gentle agitation and washed three times with PBS. Then the cells were incubated with fluorescent-labeled anti-Myc or anti-mouse secondary antibody (1:200) on ice for 1 h with agitation. Finally, cells were washed twice with cold PBS and analyzed by FACS using Aria II (BD Biosciences, San Jose, CA). The positive cells were sorted twice and isolated. Adequate positive clones were sequenced and aligned against the full-length CREPT sequence.
Cloning and expression of Ig genes as a recombinant chimeric antibody
Total RNA of monoclonal antibody 3E10 hybridoma cells that produce an antibody against CREPT was extracted and cDNA was synthesized using random hexamers. IgH and IgK or Igλ light chain genes were amplified in separate nested PCR reactions using cDNA as a template and water was used as a negative control. The PCR products were sequenced and subjected to IGBLAST to determine the V and J region of IgH and IgK or Igλ, respectively. Specific primers of V and J genes with restriction sites were used to amplify and clone the specific products into corresponding expression vectors. Vectors to express variable heavy chain and light chain of the antibody were transfected into HEK293T cells. Cell supernatant were collected after 48 h of transfection. Detailed methods were described in a previous report.(9)
Results
Examination of specificity and sensitivity of monoclonal antibodies against CREPT
To determine the correlation of CREPT expression and clinical characters of tumors, we generated a monoclonal antibody against CREPT. A BALB/c mouse was immunized with purified recombinant human GST-CREPT protein and B cells isolated from the spleen were extracted and fused with SP2/0, an osteosarcoma cell line. We finally obtained three hybridoma clones, which we called 3E10, 5F4, and 4H1.
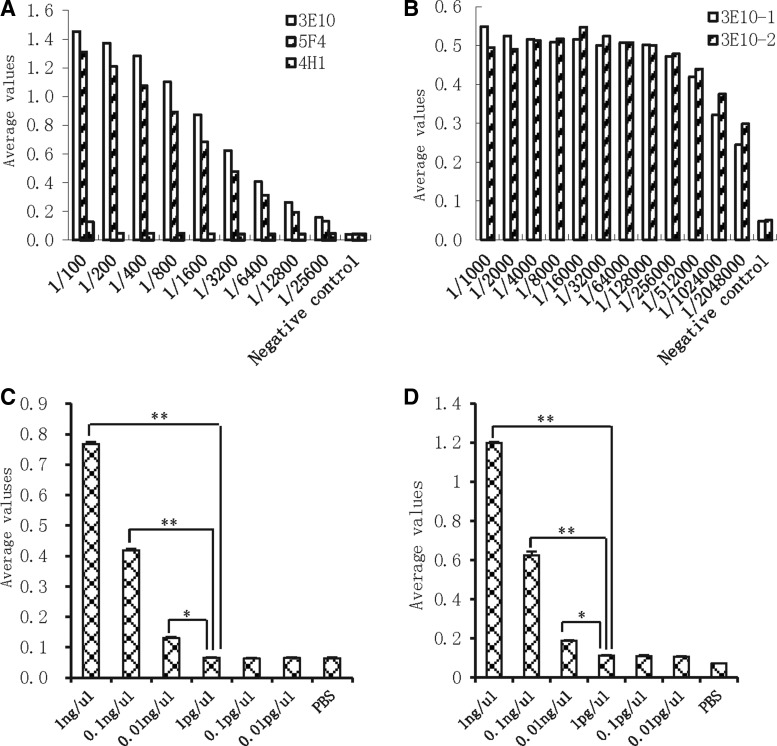
To obtain sufficient amounts of monoclonal antibodies, we injected the hybridoma cells into BALB/c mice intraperitoneally. The ascites were collected and subjected to an examination of antibody titration. ELISA assays showed that the ascites generated from hybridomas 3E10 and 5F4, but not 4H1, produced a significant amount of binding activity to CREPT up to a dilution of 1:25,600 (Fig. 1A). These results suggested that hybridoma 3E10 and 5F4 produced a high titer of antibodies. We further confirmed that hybridoma 3E10 produced equal amounts of antibodies in different mice, with a titer of as high as 1/2048000 (Fig. 1B).
FIG. 1.
Examination of specificity and sensitivity of antibodies. (A) ELISA assays were performed to detect the titer and specificity of ascites from different CREPT hybridoma clones. (B) ELISA assays were performed to detect the difference of ascites from individuals injected with the same CREPT hybridoma. (C) Sandwich ELISA assay was performed, coating with different dilutions of CREPT multiple clonal antibodies. Different concentrations of GST-CREPT proteins were added to detect the sensitivity of the antibody. Minimal concentration was determined as the concentration with no significant difference to base level of BSA. (D) Capture ELISA assay was performed, coating with CREPT MAb 3E10. Different concentrations of GST-CREPT were used to detect the minimal concentration of CREPT protein that the antibody recognized. The results were from three independent repeats and averages with standard errors are presented. *p<0.05; **p<0.01.
To examine the sensitivity of the antibody, we performed a sandwich ELISA experiment using purified GST-CREPT protein. The results showed that the antibody, with a dilution of 1:100, recognized up to 0.01 ng/μL of the GST-CREPT protein coated with CREPT multiple antibodies (produced by a rabbit) in comparison with the background of GST or PBS (Fig. 1C). In another experiment with capture ELISA analysis, we observed that 3E10 recognized up to 0.01 ng/μL of the antigen (Fig. 1D). These results showed that the antibody 3E10 recognized 0.01 ng/μL of purified CREPT protein.
MAb 3E10 recognizes endogenous CREPT by Western blot analysis, immunofluorescence, and immunohistochemistry
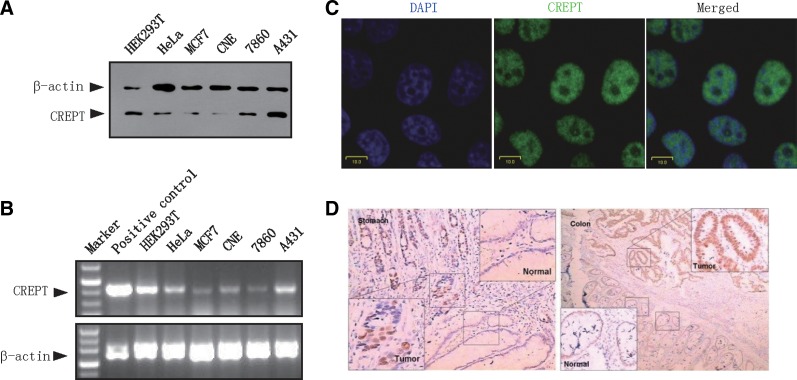
To examine the availability of 3E10 monoclonal antibody, we performed Western blot, immunofluorescence, and immunohistochemistry experiments. First, a Western blot analysis demonstrated that 3E10 antibody detected a specific band in the lysates from different cells (Fig. 2A), suggesting that the antibody recognized endogenous CREPT protein. As the levels of CREPT varied among the cells, we decided to perform an RT-PCR analysis on the CREPT expression. The results showed that the mRNA levels of CREPT among different cells were at consistent levels as examined by the Western blot (Fig. 2B). These results suggest that 3E10 recognizes endogenous protein of CREPT assayed by Western blot.
FIG. 2.
Epitope mapping of CREPT MAb 3E10. (A) HEK293T cells were transfected with Flag-tagged CREPT or control vector. Cell lysates were blotted with MAb 3E10 and then, after stripping the membrane, with an anti-Flag antibody. (B) HEK293T cells were transfected with Myc-tagged CREPT and Myc-tagged p15RS. Western blot was performed using the indicated antibody. No cross-reaction was observed. (C) Random clones from the whole library were aligned to full-length CREPT sequence. (D) Positive clones binding 3E10 were enriched by a sorting process. (E) Alignment of positive clones. Green lines represent the forward sequences and red lines represent reverse ones. The overlapping nucleotide sequences of positive yeast clones were selected and aligned to full-length CREPT sequence (marked in red box). The below sequence of 160–168 amino acids, corresponding to the red box, was shown for the comparison with the sequence of p15RS. The sequences of the epitope in different animals are shown. (F) HEK293T cells were transfected with Flag-tagged full-length RPR domain and CCT domain of CREPT. Cell lysates were detected with either an anti-Flag antibody or monoclonal antibody 3E10.
Next, we performed an immunostaining assay to examine whether 3E10 could be used to recognize the intact CREPT protein in situ. The results showed that 3E10 strongly stained in the nucleus of MCF7 cells (Fig. 2C). Since Lu and colleagues previously confirmed the nuclear localization of CREPT,(4) the strong nuclear staining confirmed recognition of CREPT by 3E10. Finally, we questioned whether 3E10 could detect CREPT protein in human tissues. For this purpose, immunohistochemical assays were performed in colon and stomach cancer samples. The results showed that 3E10 detected strong expression of CREPT in the nucleus of colon and stomach cancer cells but not in cells from normal tissues (Fig. 2D). Taken together, the data indicated that the monoclonal antibody 3E10 works in Western blot, immunofluorescence, and immunohistochemistry assays.
Identification of epitope of CREPT antibody
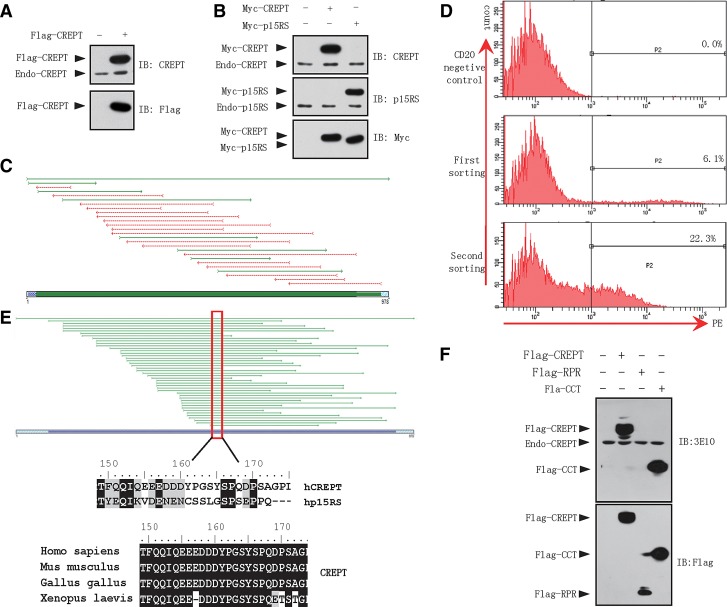
In order to map the epitope of the CREPT antibody 3E10 we first examined whether 3E10 could recognize the CREPT protein in vivo using Flag-CREPT protein expressed in HEK293T cells. Western blot analyses demonstrated that 3E10 recognized two bands in the cell lysates, an upper band specifically presented in the lysate of cells over-expressing Flag-CREPT and a lower band presented in the lysates from both control and Flag-CREPT over-expression cells (Fig. 3A, upper panel). To confirm the upper band, we blotted the membrane, after being stripped from E310 blotting, with an anti-Flag antibody. The result showed that only one band presented in the blot by the Flag-antibody at the same position as that detected by 3E10 (Fig. 3A, lower panel). These results suggested that 3E10 specifically recognized CREPT. We considered the low band as endogenous CREPT.
FIG. 3.
Application of monoclonal antibody 3E10 produced by mouse ascites. (A) Protein levels of CREPT in different cell lines. Western blot was performed with a CREPT MAb 3E10. (B) mRNA levels of human and mouse homologous sequence in different cell lines. RT-PCR assay was performed using the indicated cells. (C) MCF7 cells were immunostained with 3E10. DAPI was used for nucleus staining. Merged image is shown for the demonstration of co-localization of CREPT and DAPI. Scale, 10 μm. (D) CREPT is highly expressed in stomach (left) and colon (right) tumor tissues. Immunohistological staining assays were performed with mouse ascites CREPT MAb 3E10 (DAB staining).
Since CREPT shared a high similarity of protein sequences with p15RS, we questioned whether 3E10 contains any cross-reaction between CREPT and p15RS. We used a commercial antibody against p15RS as a control. Western blot analysis showed that 3E10 only recognized Myc-CREPT but failed to bind to Myc-p15RS (Fig. 3B, upper panel). Interestingly, the antibody against p15RS only recognized Myc-p15RS (Fig. 3B, middle panel). These results suggest that 3E10 is specific to recognize the CREPT protein without any cross-reaction to the homologue protein p15RS.
To further map the epitope of 3E10, we constructed a yeast library to display random fragments of human CREPT on the yeast surface. The random fragments of CREPT sequences in the library were widely aligned to cover the full length of CREPT with the expected length (Fig. 3C). We incubated 3E10 antibody with yeast clones from the library and selected positive clones showing interaction with 3E10. Finally, after two enrichments (Fig. 3D), we obtained positive clones and identified a common sequence of residues 160 to 168 (Fig. 2E, upper panel) using Sequencher 4.9 (Gene Codes, Ann Arbor, MI). Therefore we concluded that the epitope of 3E10 antibody is the sequence from amino acid 160 to 168 in CREPT (Fig. 3E). Interestingly, the mapped epitope in CREPT is located in the region with diversified amino acid sequences between CREPT and p15RS (Fig. 3E, middle panel). However, this epitope remains identical in CREPT proteins from human to frog (Fig. 3C, bottom panel).
To further demonstrate the epitope that 3E10 antibody recognized, Western blot was performed using Flag-tagged full-length CREPT, RPR (a domain responsible for interaction with RNA splicing factors), and CCT (coiled-coil C-terminus) domains. The results showed that 3E10 antibody recognized full length Flag-CREPT and Flag-CCT but not Flag-RPR expressed in HEK293T cells (Fig. 3F). Since the epitope that 3E10 recognized is located in the CCT domain, which covers amino acids from 136 to 326, but not in the RPR domain, which covers amino acids from 1 to 135, it is explicable that 3E10 retained strong binding ability to both the full-length and CCT domain of the CREPT protein. These results confirmed the epitope we identified.
Cloning of 3E10 variable region for engineered expression of a chimeric antibody
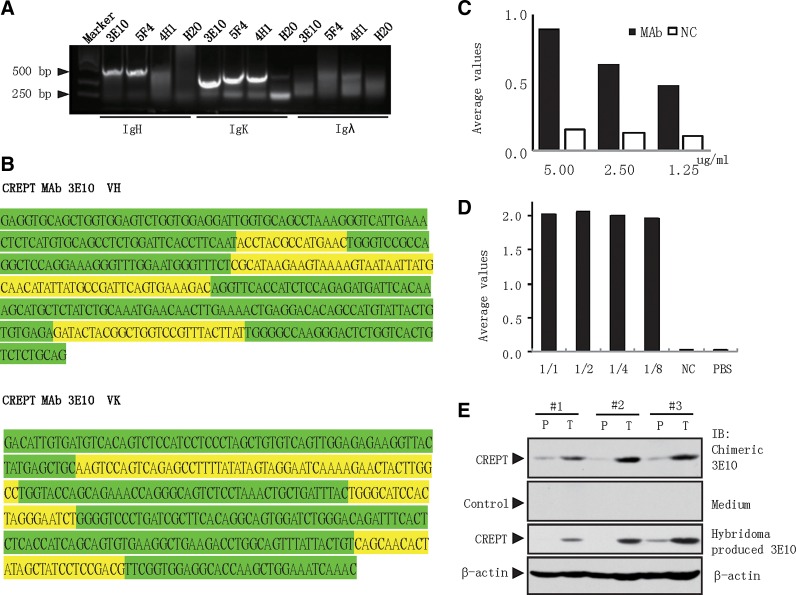
To develop large-scale production of the monoclonal antibody, we decided to clone the variable region of the 3E10 monoclonal antibody from the 3E10 hybridoma cells. A PCR experiment was performed to amplify the gene that encodes the IgH and IgK chains of the 3E10 monoclonal antibody (Fig. 4A). Based on the sequence information listed in Table 1, we designed primers according to the IgH V and IgK V sequences, with restriction enzyme sites (named 5′ AgeI P-mVH06 and 3′ SalI P-mJH03 for IgH V region primers, and 5′ AgeI P-mVK12 and 3′ BsiWI P-mJK01 for IgK V region primers). Finally, the IgH and IgK variable regions from CREPT monoclonal antibody 3E10 hybridoma cell were amplified (Fig. 4B).
FIG. 4.
Cloning of monoclonal antibody 3E10 variable regions and production of chimeric antibody. (A) IgH, IgK, and Igλ V regions of CREPT monoclonal antibody 3E10 were amplified from hybridoma cells of 3E10. Water was used as a negative control. (B) Variable sequences of CREPT monoclonal antibody Ig heavy and light chain from 3E10 clone of hybridoma cells. Framework regions are marked with green color and complementarity determining regions are marked with yellow. (C) ELISA assays were performed using different dilutions of CREPT monoclonal antibody 3E10 ascites from mouse hybridoma cells as a primary antibody. An irrelevant antibody was used as negative control. (D) ELISA assays were performed with different dilutions of CREPT monoclonal antibody 3E10 produced in supernatant from HEK293T cells expressing the chimeric antibody. The supernatant from HEK293T cells transfected vector was used as negative control. (E) Western blot analysis was performed in colon cancer samples. CREPT monoclonal antibody 3E10 produced in mouse ascites and supernatant produced by 293T cells were used as primary antibodies to detect colon tumor tissues. P, paired non-tumor tissue; T, tumor tissue from same patient.
Table 1.
Top V, D, and J Regions of 3E10 Heavy and Light Chains Matched with Ig Sequence
| Top V gene match | Top D gene match | Top J gene match | Identity (%) | |
|---|---|---|---|---|
| VH | IGHV10-1*02 | IGHD1-2*01 | IGHJ3*01 | 98.7 |
| VK | IGKV8-30*01 | – | IGKJ1*01, IGKJ1*02 | 99.0 |
After sequencing, we obtained the information of the gene sequence and performed a BLAST analysis. The results showed that the variable regions in IgH (VH) and IgK (VK) of 3E10 shared 98.7% and 99.0% of identity to the original sequences of the recombined variable light chain, suggesting that VH and HK of 3E10 remained less mutated after the maturation.
Next, we engineered the IgH and IgK variable regions of 3E10 into an expression vector to produce a chimeric CREPT monoclonal antibody. The construct was transfected into HEK293T cells for antibody production. To examine the activity of the produced chimeric antibody, an ELISA assay was performed with the supernatant and a peptide coupled to bovine serum albumin (BSA). The peptide was designed to cover the epitope of 3E10 monoclonal antibody and contained several extended amino acids to insure the binding affinity. After a test (data not shown), we synthesized a peptide covering the region from residues 158 to 172 in the CREPT protein (see Fig. 2E). This peptide was coupled to BSA at the C-terminus and was confirmed to be recognized by the 3E10 monoclonal antibody produced by the hybridoma cell (Fig. 4C). An ELISA experiment showed that the chimeric monoclonal antibody produced by 293T cells recognized the specific peptide but not BSA (Fig. 4D). These results indicated that the chimeric monoclonal antibody engineered from 3E10 can be expressed in HEK293T cells and specifically recognize the peptide of CREPT.
Finally, we examined whether the chimeric monoclonal antibody could recognize endogenous CREPT protein. For this purpose, we performed a Western blot analysis using tissues from human colon cancer patients to compare the CREPT protein levels detected by the chimeric monoclonal antibody and 3E10 produced originally by the hybridoma. The results showed that the chimeric monoclonal antibody maintained a similar sensitivity for the examination of CREPT expression in human colon cancers (Fig. 4E).
Discussion
Previously Lu and colleagues demonstrated that CREPT, a novel protein that facilitates tumor cell growth, is highly expressed in several tumors.(4) The study revealed that CREPT increased the transcription of Cyclin D1 during tumorigenesis through enhancing the recruitment of RNAPII to the promoter region as well as chromatin looping. We further raised three clones of hybridomas to produce monoclonal antibodies against CREPT protein. In the present study, we mapped the epitope of a monoclonal antibody 3E10 and identified an epitope from residues 160 to 168 in the CCT domain of CREPT protein. We further cloned the variable regions of the antibody and engineered a chimeric antibody. Our results demonstrated that the chimeric antibody worked specifically and efficiently, as well as the hybridoma-produced antibody. This study provided a basis for large-scale production of the antibody, which can be used for a broad range of examinations related to the function of CREPT in human cancer development.
In the process of epitope mapping, we adopted a yeast display strategy by constructing a library that covered deleted and truncated forms of CREPT fragments. We used the antibody 3E10 produced by hybridoma in mouse ascites to capture the CREPT fragments. We have confirmed the equivalence of the antibody collected from the ascites and supernatant of cultured hybridoma cells. Therefore we obtained an amount of antibody to enrich the positive clones from the yeast library. This was a critical process for the success of enough clones to cover the potential epitope. Indeed, we identified the epitope from over 40 independent positive clones (see Fig. 2E).
To confirm the equivalence of the chimeric antibody produced by HEK293T cells and the original 3E10 produced by the hybridoma, we used a peptide covering the epitope. Initially, we failed to detect the binding activity of the chimeric antibody with the epitope peptide of which remained nine amino acids. We reasoned that the failure might be due to the short length of the epitope peptide. Indeed, we also failed to detect the nine amino acid peptide by 3E10 produced from the hybridoma (data not shown). We speculated that the short linear epitope was unstable, making it difficult to keep the correct conformation. It could also be that this length of a nine amino acid epitope can be easily influenced by the coating process with BSA. Therefore we decided to extend the length of the peptide. Interestingly, when we extended the peptide to 15 amino acids (from 158 to 172 in CREPT), we detected a strong binding activity of the chimeric antibody with the extended peptide. These results suggested that the flank sequence may maintain a special structure for epitope recognition by the antibody. Interestingly, the flank region of the epitope is identical among CREPT proteins from humans to chickens. We speculate that the antibody could recognize CREPT proteins in mouse and chicken.
In summary, this study reports the biochemical characterization of 3E10 monoclonal antibody against CREPT. We have mapped the epitope and cloned the variable region of the antibody. Our results demonstrated an equivalence of the chimeric antibody. We expect that the CREPT monoclonal antibody 3E10, produced from the engineered antibody, will be a valuable tool for cancer diagnosis.
Acknowledgments
This work was supported by grants from 973 and 863 Project (2011CB910502, 2013ZX08011-006, 2014AA020802), NSFC (81301700, 81230044, and 81372167), Beijing Natural Science Foundation (511003), and Tsinghua Science Foundation (20121080018) in China.
Author Disclosure Statement
The authors have no financial interests to disclose.
References
- 1.Liu J, Liu H, Zhang X, Gao P, Wang J, and Hu Z: Identification and characterization of P15RS, a novel P15(INK4b) related gene on G1/S progression. Biochem Biophys Res Commun 2002;299(5):880–885 [DOI] [PubMed] [Google Scholar]
- 2.Wu Y, Zhang Y, Zhang H, Yang X, Wang Y, Ren F, Liu H, Zhai Y, Jia B, Yu J, and Chang Z: 1 p15RS attenuates Wnt/{beta}-catenin signaling by disrupting {beta}-catenin.TCF4 interaction. J Biol Chem 2010;285(45):34621–34631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang X, Cao Q, Liu X, Liu S, Wang J, Sun S, Wang O, Tian Z, Liu H, Kuang J, and Zhang W: Cellular and molecular evidence for malignancy inhibitory functions of p15RS. Cell Cycle 2011;11(10) [DOI] [PubMed] [Google Scholar]
- 4.Lu D, Wu Y, Wang Y, Ren F, Wang D, Su F, Zhang Y, Yang X, Jin G, Hao X, He D, Zhai Y, Irwin DM, Hu J, Sung JJ, Yu J, Jia B, and Chang Z: CREPT accelerates tumorigenesis by regulating the transcription of cell-cycle-related genes. Cancer Cell 2012;21(1):92–104 [DOI] [PubMed] [Google Scholar]
- 5.Jung HM, Choi SJ, and Kim JK: Expression profiles of SV40-immortalization-associated genes upregulated in various human cancers. J Cell Biochem 2009;106(4):703–713 [DOI] [PubMed] [Google Scholar]
- 6.Li X, Li J, Yang Y, Hou R, Liu R, Zhao X, Yan X, Yin G, An P, Wang Y, and Zhang K: Differential gene expression in peripheral blood T cells from patients with psoriasis, lichen planus, and atopic dermatitis. J Am Acad Dermatol 2013;69(5):e235–243 [DOI] [PubMed] [Google Scholar]
- 7.Rong Y, Cheng L, Ning H, Zou J, Zhang Y, Xu F, Lui L, Chang Z, and Fu XY: Wilms' tumor 1 and signal transducers and activators of transcription 3 synergistically promote cell proliferation: a possible mechanism in sporadic Wilms' tumor. Cancer Res 2006;66(16):8049–8057 [DOI] [PubMed] [Google Scholar]
- 8.Zuo T, Shi X, Liu Z, Guo L, Zhao Q, Guan T, Pan X, Jia N, Cao W, Zhou B, Goldin M, and Zhang L: Comprehensive analysis of pathogen-specific antibody response in vivo based on an antigen library displayed on surface of yeast. J Biol Chem 2011;286(38):33511–33519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, and Wardemann H: Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods 2008;329(1–2):112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]