Abstract
We compared Streptococcus pneumoniae serotype 1 isolates causing disease among children in six geographic regions of the United States to determine genetic relatedness. Genomic fingerprints were determined by repetitive element polymorphism PCR (Rep-PCR). Multilocus sequence type characterization was performed on selected isolates. Four different genomic banding patterns were identified by Rep-PCR. One profile (clone 1) was predominant and matched sequence type 227.
The use of the 7-valent pneumococcal conjugate vaccine since early 2000 has reduced the rate of invasive pneumococcal disease in young children by approximately 70% in the United States (10, 15). However, one concern is that disease caused by nonvaccine serotypes will become more prevalent. Streptococcus pneumoniae serotype 1 is not a commonly recovered pneumococcal serotype among children with invasive or local respiratory disease in the United States and is not included in the 7-valent pneumococcal conjugate vaccine (6, 7). Several investigators have documented an increase in invasive disease caused by S. pneumoniae capsular serotype 1 in selected populations in North America (3, 5, 14).
Compared with other pneumococcal serotypes, S. pneumoniae serotype 1 is less often isolated from carriers but appears to have a tendency to spread epidemically (2, 8). Recently, this serotype has been associated with an increase in the incidence of parapneumonic empyemas complicating pneumococcal pneumonia in children (3, 4, 14).
The U.S. Pediatric Multicenter Pneumococcal Surveillance Study Group has been prospectively identifying children with systemic pneumococcal infections in both inpatients and outpatients in eight centers since 1993 with approval of each institution's institutional review board. The methodology of data and isolate collection is described elsewhere (9, 10, 14). Isolates are serotyped and serogrouped by the Quellung reaction with antisera obtained from the Statens Seruminstitut, Copenhagen, Denmark. The purpose of this study is to compare S. pneumoniae serotype 1 isolates causing disease among children in eight diverse geographic regions and determine whether the strains are genetically related.
Between 1993 and 2002, a total of 73 patients with pneumococcal disease caused by serotype 1 were identified. Fifty-five isolates were available for testing and included in the study. The distribution of isolates according to centers and spectrum of infections is depicted in Table 1. Template DNA from each strain was isolated using the UltraClean microbial DNA kit as recommended by the manufacturer (MO Bio Laboratories, Inc., Solana Beach, Calif.).
TABLE 1.
Distribution of serotype 1 S. pneumoniae isolates by study center
| Center | No. of isolates | Mean age (yr) (SD) | No. of isolates in disease category:
|
|||
|---|---|---|---|---|---|---|
| Bacteremia | Meningitis | Otitis | Pneumonia | |||
| Pittsburgh, Pa. | 17 | 8.56 (±4) | 1 | 0 | 3 | 13 |
| Houston, Tex. | 4 | 9.12 (±3) | 1 | 0 | 0 | 3 |
| Columbus, Ohio | 12 | 7.78 (±4) | 1 | 0 | 1 | 10 |
| Little Rock, Ark. | 12 | 7.02 (±5) | 2 | 1 | 1 | 8 |
| San Diego, Calif. | 5 | 5.88 (±4) | 1 | 0 | 1 | 3 |
| Winston-Salem, N.C. | 5 | 12.36 (±5) | 0 | 0 | 0 | 5 |
| Total | 55 | 8.2 (±4) | 6 | 1 | 6 | 42 |
All isolates were typed by repetitive element polymorphism PCR (Rep-PCR) with a commercial Rep-PCR fingerprinting kit according to the instructions of the manufacturer (Bacterial Barcodes, Houston, Tex.). A PTC-200 Peltier thermocycler PCR system (MJ Research, Reno, Nev.) was used for the PCR, and the amplicons were separated by electrophoresis on a 1.5% agarose gel in 1× TAE (0.04 M Tris-HCl, 0.001 M EDTA). The fingerprints were visualized by UV after ethidium bromide staining. The fingerprints were compared digitally by Pearson correlation analysis with GelComparII computer software (Applied Maths, Kortrijk, Belgium).
Multilocus sequence type (MLST) determination was performed according to the instructions posted at the MLST website (www.mlst.net), and the sequence type (ST) was determined using the pneumococcal MLST database located at Imperial College, London, United Kingdom, and funded by the Wellcome Trust.
Fifty-one percent of the patients were male, and the majority of the patients were Caucasian (80%), followed by black (9%) and Hispanic (7%). The mean age of these patients was 8.2 years (standard deviation, ±4.0) with the youngest patient being 7 days old and the oldest being 16 years of age. The number of serotype 1 isolates increased with age (5% under 1 year of age, 5% between 1 and 2 years, 4% between 2 and 3 years, 16% between 3 and 4 years, and 69% over the age of 5 years). Eighty-one percent of these children had no underlying conditions that would predispose them to invasive pneumococcal disease. Of the 55 infections caused by S. pneumoniae serotype 1, pneumonia accounted for 76%. Other infections included bacteremia (11%), otitis media (11%), and meningitis (2%).
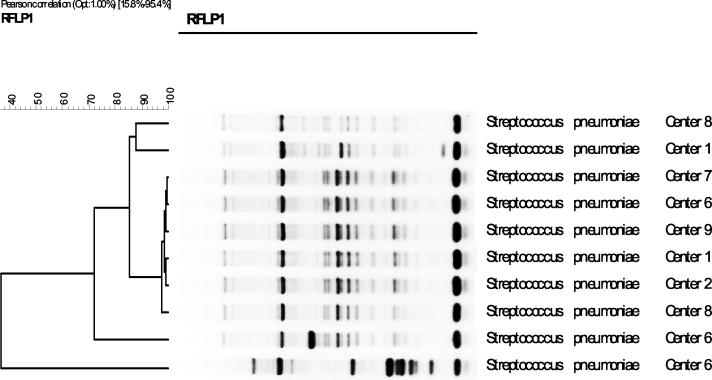
The majority of the serotype 1 strains collected from children over the study period from eight geographically separated regions in the United States appeared to be clonally related; four banding patterns for Rep-PCR genomic profiles were identified among the strains (Fig. 1). One banding profile was predominant, accounting for 49 strains (89%) (designated as clone SP1 and identified as MLST 227). Four strains (clone SP2) differed from clone SP1 by four bands (16% by Pearson correlation) and from each other by 12%. Two additional strains differed both from each other and from clone 1 by more than 70% (clones SP3 and SP4).
FIG. 1.
GelComparII analysis of Rep-PCR results from strains representing all centers and all clones. Center 1, Pittsburgh, Pa.; center 2, Houston, Tex.; center 6, Columbus, Ohio; center 7, Little Rock, Ark.; center 8, San Diego, Calif.; center 9, Winston-Salem, N.C.
Brueggemann et al. (1) recently reported the clonal diversity of serotype 1 isolates and their geographical distribution worldwide. Eighteen isolates from the United States were analyzed in the study, including eight from the Navajo Indian population. They found that the predominant ST in the United States isolates was ST227. This was also found to be the most common clone in England and Canada. A select three isolates within our clone SP1 were characterized by MLST and matched ST227 (Table 2). Since 89% of our strains had the same Rep-PCR profile, we assume that ST227 is the predominant clone in children in the United States, corroborating the findings of Brueggemann et al.
TABLE 2.
MLST of pneumococcal isolates from three medical centers
| Isolate no. | Clone | Allelic profile
|
ST | Centera | Yr | Site from which isolated | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aroE | gdh | gki | recP | spi | xpt | ddl | ||||||
| 7122 | 1 | 12 | 5 | 13 | 5 | 17 | 4 | 20 | 227 | NC | 2002 | Blood |
| 7095 | 1 | 12 | 5 | 13 | 5 | 17 | 4 | 20 | 227 | AR | 2002 | Pleural fluid |
| 4384 | 1 | 12 | 5 | 13 | 5 | 17 | 4 | 20 | 227 | PA | 1998 | Pleural fluid |
NC, Winston-Salem, N.C.; AR, Little Rock, Ark.; PA, Pittsburgh, Pa.
Susceptibility to penicillin and macrolides was determined by broth microdilution and agar disk diffusion, respectively, and categorized according to the 2003 National Committee for Clinical Laboratory Standards interpretative guidelines (11). Our SP1 clone had the same antimicrobial susceptibility pattern as the serotype 1 clone described by Porat et al. in Israel (12), that is, the majority of isolates (91%) were susceptible to penicillin, erythromycin, and clindamycin. It is thought that this is most likely related to the very low carriage rates of this serotype, resulting in less antibiotic exposure and hence less resistance (12). Only one of our patients had an isolate resistant to all three antibiotics. This child had a history of recurrent otitis media and had received antibiotics prior to the isolation of the pneumococcus from the middle ear. The Rep-PCR profile of this isolate (SP3) was very different from that of the predominant clone (SP1).
We conclude that there is a predominant clone of S. pneumoniae serotype 1 in the United States belonging to ST227, which is susceptible to penicillin, erythromycin, and clindamycin and is an important cause of invasive disease, especially in children over the age of 5 years. Since this capsular type is not included in the current 7-valent vaccine, close surveillance of this serotype is needed, as it may become more commonly isolated in young children in the United States, as it already is in other areas of the world such as Latin America and South Africa (6, 13).
Acknowledgments
This work was supported in part by a grant from Roche Laboratories.
We thank Linda Lamberth for technical support and the members of the U.S. Pediatric Multicenter Pneumococcal Surveillance Group.
The U.S. Pediatric Multicenter Pneumococcal Surveillance Study Group consists of Edward O. Mason, Jr., and Sheldon L. Kaplan, Texas Children's Hospital and Baylor College of Medicine, Houston, Tex.; Ellen R. Wald, Children's Hospital of Pittsburgh and University of Pittsburgh School of Medicine, Pittsburgh, Pa.; John S. Bradley, Children's Hospital San Diego and the University of California—San Diego School of Medicine, San Diego, Calif.; William J. Barson, Columbus Children's Hospital and Ohio State University College of Medicine, Columbus, Ohio; Tina Q. Tan and Ram Yogev, Children's Memorial Hospital and Northwestern University School of Medicine, Chicago, Ill.; Gordon E. Schutze, Children's Hospital and University of Arkansas for Medical Sciences, Little Rock, Ark.; Laurence B. Givner, Brenner Children's Hospital and Bowman Gray School of Medicine, Wake Forest University, Winston-Salem, N.C.; and Jill Hoffman, Children's Hospital of Los Angeles and the University of Southern California School of Medicine, Los Angeles, Calif.
REFERENCES
- 1.Brueggemann, A. B., and B. G. Spratt. 2003. Geographic distribution and clonal diversity of Streptococcus pneumoniae serotype 1 isolates. J. Clin. Microbiol. 41:4966-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruyn, G. A., B. J. Zegers, and R. van Furth. 1992. Mechanisms of host defense against infection with Streptococcus pneumoniae. Clin. Infect. Dis. 14:251-262. [DOI] [PubMed] [Google Scholar]
- 3.Byington, C. L., L. Y. Spencer, T. A. Johnson, A. T. Pavia, D. Allen, E. O. Mason, S. L. Kaplan, K. C. Carroll, J. A. Daly, J. C. Christenson, and M. H. Samore. 2002. An epidemiological investigation of a sustained high rate of pediatric parapneumonic empyema: risk factors and microbiological associations. Clin. Infect. Dis. 34:434-440. [DOI] [PubMed] [Google Scholar]
- 4.Eltringham, G., A. Kearns, R. Freeman, J. Clark, D. Spencer, K. Eastham, J. Harwood, and J. Leeming. 2003. Culture-negative childhood empyema is usually due to penicillin-sensitive Streptococcus pneumoniae capsular serotype 1. J. Clin. Microbiol. 41:521-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardie, W., R. Bokulic, V. F. Garcia, S. F. Reising, and C. D. Christie. 1996. Pneumococcal pleural empyemas in children. Clin. Infect. Dis. 22:1057-1063. [DOI] [PubMed] [Google Scholar]
- 6.Hausdorff, W. P., J. Bryant, P. R. Paradiso, and G. R. Siber. 2000. Which pneumococcal serogroups cause the most invasive disease: implications for conjugate vaccine formulation and use, part I. Clin. Infect. Dis. 30:100-121. [DOI] [PubMed] [Google Scholar]
- 7.Hausdorff, W. P., J. Bryant, C. Kloek, P. R. Paradiso, and G. R. Siber. 2000. The contribution of specific pneumococcal serogroups to different disease manifestations: implications for conjugate vaccine formulation and use, part II. Clin. Infect. Dis. 30:122-140. [DOI] [PubMed] [Google Scholar]
- 8.Kalin, M. 1998. Pneumococcal serotypes and their clinical relevance. Thorax 53:159-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaplan, S. L., E. O. Mason, Jr., E. R. Wald, T. Q. Tan, G. E. Schutze, J. S. Bradley, L. B. Givner, K. S. Kim, R. Yogev, and W. J. Barson,. 2002. Six-year multicenter surveillance of invasive pneumococcal infections in children. Pediatr. Infect. Dis. J. 21:141-147. [DOI] [PubMed] [Google Scholar]
- 10.Kaplan, S. L., E. O. Mason, Jr., E. R. Wald, G. E. Schutze, J. S. Bradley, T. Q. Tan, J. A. Hoffman, L. B. Givner, R. Yogev, and W. J. Barson. 2004. Decrease of invasive pneumococcal infections in children among 8 children's hospitals in the United States after the introduction of the 7-valent pneumococcal conjugate vaccine. Pediatrics 113:443-449. [DOI] [PubMed] [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial susceptibility testing; 13th informational supplement (aerobic dilution). M100-S13 (M7). National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 12.Porat, N., R. Trefler, and R. Dagan. 2001. Persistence of two invasive Streptococcus pneumoniae clones of serotypes 1 and 5 in comparison to that of multiple clones of serotypes 6B and 23F among children in southern Israel. J. Clin. Microbiol. 39:1827-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott, J. A. G., A. J. Hall, R. Dagan, J. M. Dixon, S. J. Eykyn, A. Fenoll, M. Hortal, L. P. Jette, J. H. Jorgensen, F. Lamothe, C. Latorre, J. T Macfarlane, D. M. Shlaes, L. E. Smart, and A. Taunay. 1996. Serogroup-specific epidemiology of Streptococcus pneumoniae: associations with age, sex, and geography in 7,000 episodes of invasive disease. Clin. Infect. Dis. 22:973-981. [DOI] [PubMed] [Google Scholar]
- 14.Tan, T. Q., E. O. Mason, Jr., E. R. Wald, W. J. Barson, G. E. Schutze, J. S. Bradley, L. B. Givner, R. Yogev, K. S. Kim, and S. L. Kaplan. 2002. Clinical characteristics of children with complicated pneumonia caused by Streptococcus pneumoniae. Pediatrics 110:1-6. [DOI] [PubMed] [Google Scholar]
- 15.Whitney, C. G., M. M. Farley, J. Hadler, L. H. Harrison, N. M. Bennett, R. Lynfield, A. Reingold, P. R. Cieslak, T. Pilishvili, D. Jackson, R. R. Facklam, J. H. Jorgensen, and A. Schuchat. 2003. Active bacterial core surveillance of the emerging infections program network. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N. Engl. J. Med. 348:1737-1746. [DOI] [PubMed] [Google Scholar]