Abstract
Mycobacterium avium subsp. paratuberculosis is the causative agent of Johne's Disease, an economically important intestinal ailment of ruminants. Due to the considerable genetic and serologic cross-reactivity with closely related and ubiquitous members of the M. avium complex, a species-specific method for the serological diagnosis of Johne's disease is unavailable. Computational and PCR-based analysis of the complete genome sequence of M. avium subsp. paratuberculosis led to the identification of 13 open reading frames with no identifiable homologs. One of these sequences is a putative insertion element present in six copies on the M. avium subsp. paratuberculosis genome. These novel M. avium subsp. paratuberculosis genes were cloned into Escherichia coli expression vectors, and nine were successfully expressed as recombinant fusion proteins. Five of these proteins were purified in sufficient amounts to allow immunoblot analyses of their reactivity with sera from naturally infected cattle as well as mice and rabbits exposed to M. avium subsp. paratuberculosis. Fusion proteins representing MAP0862, MAP3732c, and MAP2963c were recognized by nearly all of the sera tested, including those from cattle in the clinical stages of disease. Notably, further analysis of the protein encoded by MAP0862 showed that it reacted with sera from additional infected cattle but not with sera from uninfected control animals. The fusion product of MAP0860c did not react with any of the sera tested, while the remaining four proteins were variably recognized by sera from M. avium subsp. paratuberculosis-infected cattle. Collectively, the results of this study demonstrate the utility of genomic data to identify potential diagnostic sequences.
Mycobacterium avium subspecies paratuberculosis is a gram-positive, acid-fast bacillus that is the causative agent of Johne's disease, a chronic infection of ruminant animals characterized by inflammation of the digestive tract leading to nutrient malabsorption, emaciation, and culling of afflicted animals. It has been estimated that Johne's disease results in up to $250 million per year of lost productivity to the United States dairy industry alone (20). One of the challenges preventing the effective management of affected herds is the identification of infected animals before they present with clinical symptoms. Detection of M. avium subsp. paratuberculosis, especially during the often-lengthy subclinical phase of the disease, remains difficult due to intermittent shedding of small numbers of bacteria and a lack of effective diagnostic reagents (22).
Currently used diagnostic tests for Johne's disease need to be improved due to deficiencies in their sensitivity or specificity. Problems of specificity are due to the high degree of similarity that exists between M. avium subsp. paratuberculosis and environmental mycobacteria, especially the closely related M. avium subsp. avium. It is not surprising therefore that cell-mediated immune assays such as the intradermal (skin) test, which uses a complex, undefined M. avium subsp. paratuberculosis-secreted protein preparation to stimulate an immune reaction in the host, are prone to false-positive results due to cross-reactivity with similar proteins present in other mycobacteria. An assay used to measure gamma interferon levels in animals infected with M. avium subsp. paratuberculosis (4) was successful in identifying animals in the early stages of infection. However, even uninfected animals may intermittently test positive, perhaps suggesting a lack of specificity inherent in the assay (16). Serological assays including agar gel immunodiffusion, enzyme-linked immunosorbent assay, and complement fixation tests have been developed to detect antibodies against M. avium subsp. paratuberculosis in animal sera (6, 8). Depending on the antigens used, these assays can yield false-positive results and may not consistently detect infected animals in the early stages of disease (13). Hence, it is recognized that the identification of well-defined antigens specific to M. avium subsp. paratuberculosis will be of great utility for the development of specific, and perhaps, sensitive immunoassays for the diagnosis of Johne's disease.
Previous investigations have led to the identification of several immunogenic proteins in M. avium subsp. paratuberculosis, including heat shock proteins, reductases, proteases, cytoplasmic or cell wall proteins, and several putative proteins that have not yet been fully characterized (5, 7, 9-12, 17-19, 21, 24). Unfortunately, none of these proteins are unique to M. avium subsp. paratuberculosis, thus limiting their utility as diagnostic reagents. Ideally, immunogenic proteins unique to M. avium subsp. paratuberculosis would provide the best candidates for potential diagnostic reagents. More than 20 specific M. avium subsp. paratuberculosis sequences have already been identified by our laboratory and others using a comparative genomic approach (2) and subtractive hybridization (14). Expression and analysis of these proteins has recently identified five candidate antigens (1). In this study, we have compared the recently completed, publicly available, but unpublished genome sequences of M. avium subsp. avium and M. avium subsp. paratuberculosis to identify 13 additional putative open reading frames (ORFs) that are unique to M. avium subsp. paratuberculosis. We confirmed the specificity of these sequences relative to other mycobacterial species by PCR analysis. In addition, the cloning, expression in Escherichia coli, and characterization of these genes was undertaken in order to evaluate their utility as diagnostic antigens for the detection of M. avium subsp. paratuberculosis in infected animals.
MATERIALS AND METHODS
Identification and cloning of M. avium subsp. paratuberculosis unique genes.
At the project's inception, 4455 ORFs were identified from 223 contiguous sequences representing the unfinished genome of M. avium subsp. paratuberculosis. Consecutive BLAST searches against multiple public databases and the M. avium subsp. avium genome sequence resulted in the identification of 111 unique M. avium subsp. paratuberculosis ORFs. Once the genome sequence of M. avium subsp. paratuberculosis was completed (GenBank accession no. NC_002944), duplicated ORFs were eliminated and the number of unique ORFs was reduced to 34. Some of these ORFs have been previously characterized in our lab (1, 2). Of the remaining ORFs, 13 were successfully amplified via PCR from M. avium subsp. paratuberculosis genomic DNA but not M. avium subsp. avium DNA. These ORFs were named for their position relative to the origin of replication.
The 13 unique genes were amplified from M. avium subsp. paratuberculosis K10 genomic DNA via PCR using standard reaction conditions and the addition of 5% dimethyl sulfoxide (2). The PCR products were cloned into pBAD102/D-TOPO (Invitrogen, Carlsbad, Calif.) and pMal-c2 (New England Biolabs, Beverly, Mass.) expression vectors by following the manufacturers' instructions. Recombinant clones were verified and determined to be in-frame by DNA sequencing.
After the unique genes were initially cloned and expressed as fusion proteins, several changes were made in the final annotation of the M. avium subsp. paratuberculosis genome sequence. Four of the unique genes were reannotated as longer ORFs—MAP0855 (+123 bp), MAP0860c (+3 bp), MAP0862 (+3 bp), and MAP3732c (+84 bp)—whereas MAP2154c (−90 bp) was predicted to start 90 bp further downstream than originally thought. The sequence originally designated 177-31 was later found to be a reverse complement truncation of the ORF now annotated as MAP0858. Similarly, another unique ORF designated 210-133 was identified, cloned, and expressed, but this was subsequently determined to be a noncoding sequence from a region complementary to the N-terminal portion of ISMAP02.
Antisera.
Sera from New Zealand White rabbits that were immunized with live and heat-killed preparations of M. avium subsp. paratuberculosis were obtained in a previous study (23). Similarly, BALB/c mice were immunized with a sonicated lysate of M. avium subsp. paratuberculosis K10 as previously described (1). Sera from six well-characterized cattle in the clinical stage of Johne's disease and three control cattle were used in immunoblot analysis for detection of antibodies that bind unique M. avium subsp. paratuberculosis proteins. Five of the clinical cattle were housed on site at the National Animal Disease Center, while serum from the sixth cow (may bunny) was provided by Michael Collins at the University of Wisconsin.
Protein expression using the pBAD vector.
Overnight cultures of E. coli TOP10 containing the cloned constructs were grown and used to inoculate a 100-ml volume of Luria-Bertani broth containing ampicillin at 100 μg/ml. Cells were grown at 37°C with shaking until they reached an optical density at 600 nm of approximately 0.5. At this point, 1-ml samples of the uninduced cultures were taken, briefly centrifuged, and resuspended in 50 μl of 1× sample buffer (0.05 M Tris pH 6.8, 1% sodium dodecyl sulfate [SDS], 10% glycerol, 0.001% bromphenol blue, 0.1% β-mercaptoethanol). The remaining cultures were induced by adding arabinose to a final concentration of 0.2% and incubated at 37°C for 4 h. A 1-ml sample of the induced cultures was collected as before, except the cells were resuspended in 100 μl of 1× sample buffer. All of the collected samples were boiled for 5 min and separated on an SDS-polyacrylamide (10%, wt/vol) gel. The proteins were then transferred to nitrocellulose membranes as described below and probed with a six-histidine-tagged monoclonal antibody conjugated to horseradish peroxidase (Clontech, Palo Alto, Calif.).
Protein expression using the pMal vector.
Expression of the unique genes fused with maltose binding protein (MBP) was performed as previously described (1). Briefly, recombinant E. coli DH5α containing the unique genes cloned into pMal-c2 were induced with 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 to 3 h, harvested by centrifugation and purified by affinity chromatography using an amylose resin (New England Biolabs).
Polyhistidine-tagged protein purification.
Recombinant M. avium subsp. paratuberculosis fusion proteins that displayed high levels of expression in pBAD102 were purified using affinity columns. An overnight culture of a selected clone was used to inoculate 1 liter of Luria-Bertani broth containing ampicillin at 100 μg/ml. As before, cells were grown at 37°C with shaking until they reached an optical density at 600 nm of approximately 0.5, induced with 0.2% arabinose, and incubated at 37°C for 4 h. The cells were then harvested by centrifugation, resuspended in 30 ml of wash buffer (50 mM NaH2PO4, 300 mM NaCl [pH 7.0]), and sonicated on ice three times for 1 min intervals with 1 min of rest between each sonication. The lysate was centrifuged, and the supernatant saved on ice while the pellet of cellular material was resuspended in 30 ml of wash buffer and sonicated as before. The resulting lysates were then combined and centrifuged at 14,000 × g for 30 min at 4°C. The clarified lysate was mixed with 6 ml of Talon resin (Clontech) that had been washed twice in 30 ml of wash buffer. This mixture was then incubated at room temperature for one hour with gentle rocking to prevent settling of the resin. The solution was centrifuged and the resin was washed with 30 ml of wash buffer, resuspended in 20 ml of wash buffer, applied to a 20-ml column, and allowed to settle. The column was drained and then washed with 150 ml wash buffer and 400 ml of wash buffer containing 7.5 mM imidazole. The protein was eluted from the column in wash buffer containing 150 mM imidazole. Fractions containing the protein of interest were identified by SDS-polyacrylamide gel electrophoresis, pooled, and dialyzed using Slide-A-Lyzer cassettes (Pierce, Rockford, Ill.) overnight in 1 liter of phosphate-buffered saline (PBS) (150 mM NaCl, 10 mM NaPO4, pH 7.4) with three exchanges.
Immunoblotting.
Separation of protein samples was performed on an SDS-10% polyacrylamide gel. For immunoblotting, proteins within the gel were transferred to a Protran nitrocellulose membrane (Schleicher & Schuell, Keene, N.H.) using a Bio-Rad (Hercules, Calif.) Trans Blot Cell in sodium phosphate buffer (25 mM, pH 7.8) at 0.9 A for 90 min. The membranes were then incubated overnight in a blocking solution (PBS containing 2% bovine serum albumin and 0.1% Tween 20). Membranes were exposed to either rabbit, mouse, or cattle sera for 2 h, washed three times in PBS containing 0.1% Tween 20, incubated for 1.5 h with an appropriate secondary antibody conjugated to peroxidase and diluted 1:20,000 in blocking solution, washed as before, and developed for 5 min in SuperSignal (Pierce). Secondary antibodies used were protein A (Pierce), anti-mouse (Pierce), and anti-goat (Vector Labs, Burlingame, Calif.) for rabbit, mouse, and bovine sera, respectively.
RESULTS
Identification of unique genes.
Through a comparative genomic approach with mycobacterial genome sequence data, we have identified a collection of 13 genes that are present only in M. avium subsp. paratuberculosis. Furthermore, these genes lack homology to any sequences currently found in publicly available sequence databases and are not present in the closely related M. avium subsp. avium 104 genome (http://tigrblast.tigr.org/ufmg/). In addition, they are different from the 21 predicted coding sequences identified previously in our laboratory (1, 2). PCR amplification using genomic DNA from 39 different isolates of M. avium subsp. paratuberculosis shows that 8 of 13 sequences are 100% conserved in this subspecies (Table 1). An additional four sequences (MAP0862, MAP3732c, MAP3736c, and MAP2149c) were present in 38 of the 39 isolates tested. All four of these genes were absent in only one of the human isolates of M. avium subsp. paratuberculosis (Table 1). Importantly, we were unable to detect any of the unique genes in five M. avium subsp. avium strains as well as 16 other mycobacterial species, including other members of the M. avium complex (Table 1). Of these sequences, the putative insertion sequence, ISMAP02, is present in 6 copies on the M. avium subsp. paratuberculosis genome (genes are identified in Table 2). This fact, combined with its presence only in M. avium subsp. paratuberculosis, makes this sequence an attractive alternative for IS900 in PCR-based diagnosis. Overall, these data suggest novel M. avium subsp. paratuberculosis sequences that may be important candidates for future diagnostic tests.
TABLE 1.
PCR amplification of M. avium subsp. paratuberculosis unique genes from genomic DNA of mycobacteria
| Organisma | Amplificationb of:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAP3817c | ISMAP02 | MAP0855 | MAP0860c | MAP0862 | MAP1345 | MAP3732c | MAP2963c | MAP0858 | MAP2154c | MAP2756c | MAP3736c | MAP2149c | |
| M. avium subsp. paratuberculosis K10 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| M. avium subsp. paratuberculosis 20 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| M. avium subsp. paratuberculosis 109 | + | + | + | + | + | + | + | + | + | + | + | + | + |
| M. avium subsp. paratuberculosis of origin | |||||||||||||
| Bovine (n = 24) | 24 | 24 | 24 | 24 | 24 | 24 | 24 | 24 | 24 | 24 | 24 | 24 | 24 |
| Caprine (n = 5) | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| Human (n = 2) | 2 | 2 | 2 | 2 | 1 | 2 | 1 | − | 2 | 2 | 2 | 1 | 1 |
| Murine | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Ovine (n = 3) | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 2 | 3 | 3 | 3 | 2 | 2 |
| Unknown | + | + | + | + | + | + | + | + | + | + | + | + | + |
| M. avium subsp. avium 1015 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| M. avium subsp. avium 104 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| M. avium subsp. avium1282 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| M. avium subsp. avium 1285 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| M. avium subsp. avium 1611 | − | − | − | − | − | − | − | − | − | − | − | − | − |
| M. silvaticum | − | − | − | − | − | − | − | − | − | − | − | − | − |
| M. smegmatis | − | − | − | − | − | − | − | − | − | − | − | − | − |
| M. phlei | − | − | − | − | − | − | − | − | − | − | − | − | − |
| M. abscessus | − | − | − | − | − | − | − | − | − | − | − | − | − |
| M. fortuitum | − | − | − | − | − | − | − | − | − | − | − | − | − |
| M. celatum | − | − | − | − | − | − | − | − | − | − | − | − | − |
| M. cheloneae | − | − | − | − | − | − | − | − | − | − | − | − | − |
| M. xenopi | − | − | − | − | − | − | − | − | − | − | − | − | − |
| MAC | − | − | − | − | − | − | − | − | − | − | − | − | − |
| MAC human (n = 5) | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Salmonella (n = 2) | − | − | − | − | − | − | − | − | − | − | − | − | − |
The number of isolates evaluated within each category is indicated in parentheses.
The number of positive reactions is represented numerically; −, negative reaction for all isolates tested.
TABLE 2.
M. avium subsp. paratuberculosis-specific genes in this study
| ORF | Gene identification no. | Size (bp) | Functional domain(s) | Cloneda | Expressedb |
|---|---|---|---|---|---|
| ISMAP02 | 2717932 | 1,674 | None | pBAD, pMal | |
| 2717466 | |||||
| 2719200 | |||||
| 2718191 | |||||
| 2718683 | |||||
| 2720300 | |||||
| MAP0855 | 2720245 | 945 | ATP binding, ATPase | pBAD, pMal | pBAD, pMal |
| 177-31c | 357 | None | pBAD | ||
| MAP0858 | 2718672 | 549 | None | ||
| MAP0860c | 2719015 | 891 | None | pBAD, pMal | pBAD, pMal |
| MAP0862 | 2718315 | 1,083 | None | pBAD | pBAD |
| MAP3732c | 2718816 | 714 | None | pBAD, pMal | pBAD |
| MAP3736c | 2719217 | 960 | Aldolase | pBAD, pMal | |
| MAP1345 | 2718344 | 600 | Methylase | pBAD, pMal | pMal |
| MAP2149c | 2717464 | 645 | None | pMal | |
| MAP2154c | 2717876 | 576 | Protein kinase | pBAD, pMal | pBAD |
| MAP2963c | 2720464 | 2,625 | Heme binding | pMal | pMal |
| MAP3817c | 2718568 | 939 | Thiamine biosynthesis | pBAD, pMal | pMal |
| MAP2756c | 2718333 | 4,911 | None | pBAD, pMal | pBAD, pMal |
Indicates which expression vector the sequence was successfully cloned into.
Absence of data means that no protein was produced despite successful cloning in an expression vector.
This ORF was later found to be a reverse complement truncation of MAP0858
Several of these M. avium subsp. paratuberculosis-specific equences contain regions that resemble the functional domains of known proteins (Table 2). Genes designated MAP3736c and MAP3817c appear to encode proteins that have similarities to those involved in amino acid and thiamine biosynthesis, respectively, while the proteins produced by MAP0855, MAP1345, and MAP2154c possess similarities to regulatory proteins. It is anticipated that genes encoding biosynthetic or regulatory proteins would not be highly immunogenic, as they are often located in the cytoplasm and therefore may remain undetected by the host immune system. Conversely, the large (875-amino-acid) protein encoded by MAP2963c contains two domains that resemble heme-binding proteins, suggesting an association with the cell surface. The remaining genes have no recognizable functional domains present in the proteins they encode. While homology searches provide us with a limited insight into the potential functions of the proteins encoded by these unique genes, the true measure of their utility as diagnostic reagents will be determined by their reactivity with clinical samples.
Expression of unique genes.
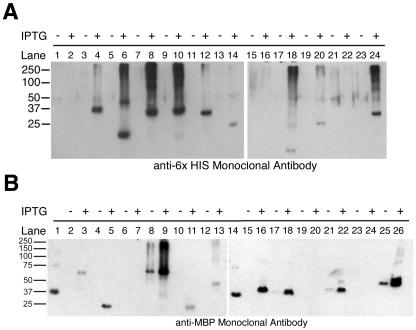
All 13 of the novel genes identified in M. avium subsp. paratuberculosis were amplified from strain K10 genomic DNA, and 11 were cloned into at least one of the two expression vectors used in this study (Table 2). Nine of these were successfully expressed in E. coli as fusion proteins with hexahistidine, MBP, or both (Table 2; Fig. 1). Most of the proteins successfully expressed with a six-His tag migrated near their expected molecular weight (Fig. 1A). However, the product of MAP1345 appeared to migrate as a smaller protein than was predicted by the amino acid sequence. Six of the pBAD102 constructs were not successfully expressed in E. coli and will require additional optimization of the expression conditions used for this study.
FIG. 1.
Immunoblot analysis of M. avium subsp. paratuberculosis proteins expressed in E. coli. Genes were cloned into pBAD (A) or pMal (B) and induced by arabinose or IPTG, respectively. A 25-μl sample of each E. coli culture was taken, mixed with sample buffer, and run on an SDS-10% polyacrylamide gel. Proteins were transferred to nitrocellusose, blocked, and probed with antibodies against hexahistidine (A) or MBP (B). (A) Lane 1, ISMAP02 (C-terminal) uninduced (U); lane 2, ISMAP02 (C-terminal half) induced (I); lane 3, MAP0855 U; lane 4, MAP0855 I; lane 5, 177-31 U; lane 6, 177-31 I; lane 7, MAP0860c U; lane 8, MAP0860c I; lane 9, MAP0862 U; lane 10, MAP0862 I; lane 11, 210-133 U; lane 12, 210-133 I; lane 13, MAP3732c U; lane 14, MAP3732c I; lane 15, MAP3736c U; lane 16, MAP3736c I; lane 17, MAP1345 U; lane 18, MAP1345 I; lane 19, MAP2154c U; lane 20, MAP2154c I; lane 21, MAP3817c U; lane 22, MAP3817c I; lane 23, MAP2756c U; lane 24, MAP2756c I. (B) Lane 1, MBP affinity tag; lane 2, MAP0855 uninduced (U); lane 3, MAP0855 induced (I); lane 4, ISMAP02 (C-terminal half) U; lane 5, ISMAP02 (C-terminal half) I; lane 6, MAP3736c U; lane 7, MAP3736c I; lane 8, MAP2963c (C-terminal half) U; lane 9, MAP2963c (C-terminal half) I; lane 10, MAP3732c U; lane 11, MAP3732c I; lane 12, MAP3817c U; lane 13, MAP3817c I; lane 14, MBP affinity tag; lane 15, MAP2756c U; lane 16, MAP2756c I; lane 17, MAP2963c (N-terminal) U; lane 18, MAP2963c (N-terminal) I; lane 19, MAP2149c U; lane 20, MAP2149c I; lane 21, MAP1345 U; lane 22, MAP1345 I; lane 23, MAP3736c U; lane 24, MAP3736c I; lane 25, MAP0860c U; lane 26, MAP0860c I.
Six of the M. avium subsp. paratuberculosis-specific genes (MAP0855, MAP0860c, MAP1345, MAP3817c, MAP2756c, and the C-terminal half of MAP2963c) were successfully expressed as fusion proteins with MBP (Fig. 1B). Of these, two recombinants expressed fusion proteins regardless of the presence of IPTG inducer (MAP0860c and the C-terminal half of MAP2963c). Three of the cloned genes expressed proteins that were significantly smaller than expected (MAP2963c (N-terminal half), MAP3732c, and ISMAP02 (C-terminal half)). Sequencing of these clones revealed that frame-shift mutations had occurred, resulting in truncated or misassembled proteins. No proteins were produced by recombinants encoding MAP2149c and MAP3736c. Two independent MAP3736c recombinants were analyzed in this experiment and neither recombinant produced protein despite a correct, in-frame sequence for both clones. Additional protein expression techniques will need to be evaluated in order to purify M. avium subsp. paratuberculosis proteins that were not successfully expressed as fusion proteins with hexahistidine or MBP.
Reactivity of fusion proteins with sera from animals exposed to M. avium subsp. paratuberculosis.
The 6x his-tagged fusion proteins representing the products of sequences MAP0862, MAP3732c, and MAP2154c were selected for purification due to their abundant expression in our preliminary analysis (data not shown). Likewise, the MBP-tagged fusion proteins representing MAP0860c, MAP0855, MAP1345, and the C-terminal half of MAP2963c were also expressed at high levels and selected for purification. Samples of sufficient purity and concentration to probe sera from immunized or infected animals were obtained for MAP0860c, MAP0862, MAP3732c, MAP2154c, and MAP2963c (C-terminal half).
These five fusion proteins were screened against a panel of sera from experimentally immunized and naturally infected animals in order to examine the ability of the unique gene products to elicit an immune response. The MAP0862, MAP3732c, and MAP2963c fusion proteins reacted with sera from two rabbits immunized with heat-killed M. avium subsp. paratuberculosis (273 and 275), while the MAP2154c protein reacted only with sera from rabbit 273 (Table 3). A previously described major membrane protein (MMP) (3) that was included on some of the blots also reacted with sera from both of the rabbits (data not shown). Only the fusion proteins representing genes MAP0862, MAP3732c, MAP2154c, and MAP2963c (C-terminal half) were detected by serum from a mouse immunized with a sonicate lysate of M. avium subsp. paratuberculosis (Table 3 and Fig. 2), while none of the fusion proteins reacted with serum from a mouse immunized with the major membrane protein fused to MBP (data not shown).
TABLE 3.
Summary of unique gene product reactivity with antisera
| Sample | Product reactivity
|
||||
|---|---|---|---|---|---|
| MAP0862 | MAP3732c | MAP2154c | MAP0860ca | MAP2963c (C terminal half)a | |
| Mouse 48 | + | + | + | − | + |
| Rabbit 273 | + | + | + | − | + |
| Rabbit 275 | + | + | − | − | + |
| Cow 47 | + | + | − | − | + |
| Cow May Bunny | + | + | − | − | + |
MBP fusion protein.
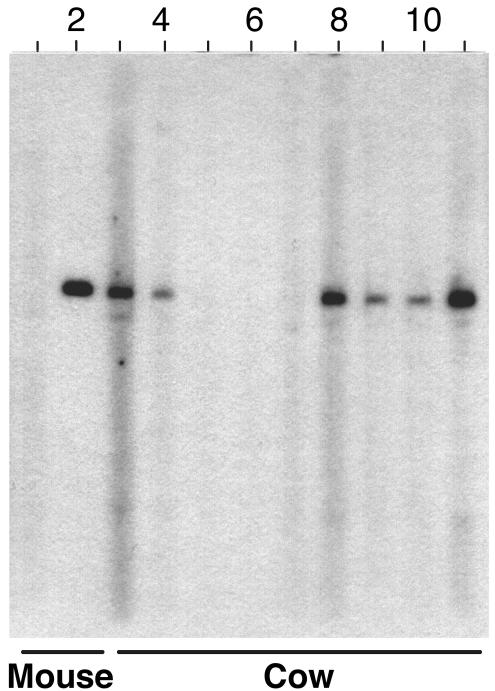
FIG. 2.
Preparative immunoblot analysis of affinity purified M. avium subsp. paratuberculosis unique gene MAP0862. The purified six-histidine-tagged fusion protein representing gene MAP0862 was separated on an SDS-10% polyacrylamide gel, transferred to nitrocellulose, blocked, and probed with sera from mouse 48 and infected and healthy cattle. Lane 1, mouse anti-major membrane protein antibody; lane 2, serum from mouse 48; lane 3, clinical serum from cow May Bunny; lane 4, clinical cow 47; lane 5, control serum from cow 108; lane 6, control serum from cow 181; lane 7, control serum from cow 111; lane 8, clinical serum from cow 27; lane 9, clinical serum from cow 85; lane 10, clinical serum from cow 110; lane 11, clinical serum from cow 135.
Sera from two naturally infected cows were also examined for the presence of antibodies against the recombinant proteins. The five fusion proteins reacted variably to sera from different cattle (Table 3). Only MAP0862, MAP3732c, and MAP2963c (C-terminal half) reacted with both of the serum samples from Johne's disease cows. The fusion protein representing MAP0862 was used to probe additional cattle sera (Fig. 2). The protein was not recognized by sera from three uninfected cattle but was readily detected by sera from four naturally infected animals. These preliminary data indicate that the protein encoded by gene MAP0862 may be the most useful as a diagnostic marker of M. avium subsp. paratuberculosis infection.
These results suggest that the products of genes MAP0862, MAP3732c, and MAP2963c (C-terminal) are antigenic and readily recognized by the immune systems of multiple host species. The fusion protein representing MAP0860c did not react with any of the sera tested, while MAP2154c was detected by only two of the serum samples. The variability of reactions between M. avium subsp. paratuberculosis purified proteins and animal sera illustrate the necessity of using multiple antigenic targets for the development of diagnostic tests in order to account for potential temporal changes in both bacterial gene expression and the host immune response.
DISCUSSION
Thanks to the enormous efforts of several investigators, the field of mycobacterial genomics has moved forward rapidly. A recent study has used a comparative genomic approach and hybridization techniques to identify the core set of genes present among all mycobacteria tested (15). In the present study, a comparative genomic approach as well as PCR analysis was used to identify a set of sequences present only in M. avium subsp. paratuberculosis. The identification of these specific sequences represent an initial step in defining more specific diagnostic reagents. An urgent need in the control of Johne's Disease is a reliable, easy to perform diagnostic test capable of distinguishing between infected and healthy animals. This would enable animal producers to more successfully manage their herds or flocks and prevent the spread of disease. The development of diagnostic tests around antigenic proteins specific to M. avium subsp. paratuberculosis holds great promise, as they should not be susceptible to many of the problems that plague currently available tests developed using complex protein preparations such as purified protein derivative. Together with previously identified sequences unique to M. avium subsp. paratuberculosis (1, 2, 14), the 13 genes identified in our present study have yielded a valuable pool of potential diagnostic reagents.
A PCR-based survey of 60 mycobacterial strains and isolates using M. avium subsp. paratuberculosis-specific sequences identified by a comparative genomic approach revealed that eight of the genes were detected in all of the M. avium subsp. paratuberculosis isolates tested, while none of the genes were detected in other mycobacteria. These data show 100% conservation among all 39 tested M. avium subsp. paratuberculosis isolates from at least five host species. Furthermore, these genes are not conserved among members of the avium complex despite their high overall genomic similarity. Of particular interest is the ISMAP02 sequence present in six copies on the M. avium subsp. paratuberculosis genome. This sequence was only identified through the M. avium subsp. paratuberculosis genome project and may serve as an alternative to IS900-based PCR diagnostics or to confirm IS900 positive reactions.
Eleven of the unique genes were cloned into at least one of two expression vectors, and 9 were successfully expressed as fusion proteins. Three of the six-histidine-tagged fusion proteins (MAP0862, MAP3732c, and MAP2154c) and two of the MBP fusions (MAP0860c and MAP2963c [C-terminal half]) were purified in sufficient amounts to be used for immunoblot analysis. Unfortunately, this leaves seven to eight proteins that remain to be tested for antigenicity with sera from Johne's disease cattle, owing to the difficulty in heterologously producing M. avium subsp. paratuberculosis proteins. Further optimization of protein expression conditions and the use of additional expression systems are ongoing and will be needed to obtain usable amounts of purified protein for the remaining unique genes. Nonetheless, these 13 sequences remain solid diagnostic targets in nucleic acid-based tests to be developed in the future.
Immunoblot analysis revealed that most of the fusion proteins were recognized by at least one serum sample from three species of animals that had been infected or immunized with M. avium subsp. paratuberculosis. The products of genes MAP0862, MAP3732c, and MAP2963c (C-terminal half) were strongly reactive with all of the sera tested, indicating that they are immunogenic. Further evaluation of the MAP0862 fusion protein against sera from naturally infected and uninfected cattle indicated that antibodies against the protein are present in sera from the infected animals but not uninfected controls, suggesting that this protein may provide an ideal target for the development of an antigen-based diagnostic test.
MAP0860c was not detected by any of the sera, while the remaining purified proteins were variably reactive. One of two possibilities may account for this lack of reactivity seen for MAP0860c. First, it may simply not be antigenic; and second, the gene prediction could be inaccurate and the protein might not even be produced by M. avium subsp. paratuberculosis. Proteomic approaches should distinguish between these two possibilities.
The results of this study suggest that novel and specific M. avium subsp. paratuberculosis genes will be useful as diagnostic reagents. Among the sequences analyzed in this study, ISMAP02 appears to be the best nucleic acid target sequence and MAP0862 appears to be the best candidate antigen. Notably, none of the unique sequences were homologous to previously characterized genes, and only 6 of the proteins encoded by these genes possessed recognizable functional domains. Characterizing proteins of unknown function is one of the major challenges that must be addressed by postgenome research and will be a vital part of future research on the pathogenesis of M. avium subsp. paratuberculosis now that the complete genome sequence has been elucidated. It is possible that some of the genes identified in this study will not only prove useful as diagnostic tools but also give us new insights into the biology of M. avium subsp. paratuberculosis.
Acknowledgments
The expert technical assistance of Janis K. Hansen is greatly appreciated.
This work was supported by the USDA's Agricultural Research Service and USDA-NRI grants to J.P.B. and V.K.
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA.
REFERENCES
- 1.Bannantine, J. P., J. K. Hansen, M. L. Paustian, J. R. Stabel, and V. Kapur. 2004. Expression and immunogenicity of proteins encoded by sequences specific to Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 42:106-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannantine, J. P., E. Baechler, Q. Zhang, L. Li, and V. Kapur. 2002. Genome scale comparison of Mycobacterium avium subsp. paratuberculosis with Mycobacterium avium subsp. avium reveals potential diagnostic sequences. J. Clin. Microbiol. 40:1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannantine, J. P., and J. R. Stabel. 2001. Identification of two Mycobacterium avium subspecies paratuberculosis gene products differentially recognised by sera from rabbits immunised with live mycobacteria but not heat-killed mycobacteria. J. Med. Microbiol. 50:795-804. [DOI] [PubMed] [Google Scholar]
- 4.Billman-Jacobe, H., M. Carrigan, F. Cockram, L. A. Corner, I. J. Gill, J. F. Hill, T. Jessep, A. R. Milner, and P. R. Wood. 1992. A comparison of the interferon gamma assay with the absorbed ELISA for the diagnosis of Johne's disease in cattle. Aust. Vet. J. 69:25-28. [DOI] [PubMed] [Google Scholar]
- 5.Cameron, R. M., K. Stevenson, N. F. Inglis, J. Klausen, and J. M. Sharp. 1994. Identification and characterization of a putative serine protease expressed in vivo by Mycobacterium avium subsp. paratuberculosis. Microbiology 140:1977-1982. [DOI] [PubMed] [Google Scholar]
- 6.Clarke, C. J., I. A. Patterson, K. E. Armstrong, and J. C. Low. 1996. Comparison of the absorbed ELISA and agar gel immunodiffusion test with clinicopathological findings in ovine clinical paratuberculosis. Vet. Rec. 139:618-621. [PubMed] [Google Scholar]
- 7.Cobb, A. J., and R. Frothingham. 1999. The GroES antigens of Mycobacterium avium and Mycobacterium paratuberculosis. Vet. Microbiol. 67:31-35. [DOI] [PubMed] [Google Scholar]
- 8.Colgrove, G. S., C. O. Thoen, B. O. Blackburn, and C. D. Murphy. 1989. Paratuberculosis in cattle: a comparison of three serologic tests with results of fecal culture. Vet. Microbiol. 19:183-187. [DOI] [PubMed] [Google Scholar]
- 9.De Kesel, M., P. Gilot, M. Coene, and C. Cocito. 1992. Composition and immunological properties of the protein fraction of A36, a major antigen complex of Mycobacterium paratuberculosis. Scand. J. Immunol. 36:201-212. [DOI] [PubMed] [Google Scholar]
- 10.El-Zaatari, F. A., S. A. Naser, L. Engstrand, C. Y. Hachem, and D. Y. Graham. 1994. Identification and characterization of Mycobacterium paratuberculosis recombinant proteins expressed in E. coli. Curr. Microbiol. 29:177-184. [DOI] [PubMed] [Google Scholar]
- 11.El-Zaatari, F. A., S. A. Naser, and D. Y. Graham. 1997. Characterization of a specific Mycobacterium paratuberculosis recombinant clone expressing 35,000-molecular-weight antigen and reactivity with sera from animals with clinical and subclinical Johne's disease. J. Clin. Microbiol. 35:1794-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilot, P., M. De Kesel, L. Machtelinckx, M. Coene, and C. Cocito. 1993. Isolation and sequencing of the gene coding for an antigenic 34-kilodalton protein of Mycobacterium paratuberculosis. J. Bacteriol. 175:4930-4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hilbink, F., D. M. West, G. W. de Lisle, R. Kittelberger, B. D. Hosie, J. Hutton, M. M. Cooke, and M. Penrose. 1994. Comparison of a complement fixation test, a gel diffusion test and two absorbed and unabsorbed ELISAs for the diagnosis of paratuberculosis in sheep. Vet. Microbiol. 41:107-116. [DOI] [PubMed] [Google Scholar]
- 14.Klitgaard Nielsen, K., and P. Ahrens. 2002. Putative in vitro expressed gene fragments unique to Mycobacterium avium subspecies paratuberculosis. FEMS Microbiol. Lett. 214:199-203. [DOI] [PubMed] [Google Scholar]
- 15.Marmiesse, M., P. Brodin, C. Buchrieser, C. Gutierrez, N. Simoes, V. Vincent, P. Glasser, S. T. Cole, and R. Brosch. 2004. Macro-array and bioinformatics analysis reveal mycobacterial ′core' genes, variation in ESAT-6 gene family and new phylogenetic markers for the Mycobacterium tuberculosis complex. Microbiology 150:483-496. [DOI] [PubMed] [Google Scholar]
- 16.McDonald, W. L., S. E. Ridge, A. F. Hope, and R. J. Condron. 1999. Evaluation of diagnostic tests for Johne's disease in young cattle. Aust. Vet. J. 77:113-119. [DOI] [PubMed] [Google Scholar]
- 17.Mullerad, J., A. H. Hovav, R. Nahary, Y. Fishman, and H. Bercovier. 2003. Immunogenicity of a 16.7kDa Mycobacterium paratuberculosis antigen. Microb. Pathog. 34:81-90. [DOI] [PubMed] [Google Scholar]
- 18.Mutharia, L. M., W. Moreno, and M. Raymond. 1997. Analysis of culture filtrate and cell wall-associated antigens of Mycobacterium paratuberculosis with monoclonal antibodies. Infect. Immun. 65:387-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olsen, I., L. J. Reitan, G. Holstad, and H. G. Wiker. 2000. Alkyl hydroperoxide reductases C and D are major antigens constitutively expressed by Mycobacterium avium subsp. paratuberculosis. Infect. Immun. 68:801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ott, S. L., S. J. Wells, and B. A. Wagner. 1999. Herd-level economic losses associated with Johne's disease on US dairy operations. Prev. Vet. Med. 40:179-192. [DOI] [PubMed] [Google Scholar]
- 21.Silbaq, F. S., S. N. Cho, S. T. Cole, and P. J. Brennan. 1998. Characterization of a 34-kilodalton protein of Mycobacterium leprae that is isologous to the immunodominant 34-kilodalton antigen of Mycobacterium paratuberculosis. Infect. Immun. 66:5576-5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stabel, J. R. 1998. Johne's disease: a hidden threat. J. Dairy Sci. 81:283-288. [DOI] [PubMed] [Google Scholar]
- 23.Stabel, J. R., M. R. Ackermann, and J. P. Goff. 1996. Comparison of polyclonal antibodies to three different preparations of Mycobacterium paratuberculosis in immunohistochemical diagnosis of Johne's disease in cattle. J. Vet. Diagn. Invest. 8:469-473. [DOI] [PubMed] [Google Scholar]
- 24.White, W. B., D. L. Whipple, J. R. Stabel, and C. A. Bolin. 1994. Comparison of cellular and extracellular proteins expressed by various isolates of Mycobacterium paratuberculosis and other mycobacterial species. Am. J. Vet. Res. 55:1399-1405. [PubMed] [Google Scholar]