Over the past few years, clinical microbiologists have assumed increasing responsibility for the rapid, accurate detection of a diverse number of emerging (new) and reemerging pathogens. Ironically, these responsibilities come at a time when resources are limited and budgets are significantly constrained. Indeed, in the past decade, some institutions, especially those housing smaller clinical microbiology laboratories, have outsourced microbiology testing. Such a practice, often in response to budgetary cuts, had, in effect, relegated the importance of microbiology services to the bottom of the clinical pathology services “food chain” (25).
One tragic event in U.S. history, though terrible as it was, had a positive effect on the relevance of on-site microbiology services that test for reemerging infectious diseases 24 h/day, 7 days/week. The intentional release of anthrax spores in the U.S. mail in late 2001 was a somber but emphatic message to health care providers and health care leadership (including hospital administrators, public health officials, and politicians), as well as the general public, that comprehensive, state-of-the art, on-site microbiology services are essential, if not expected. The 2002-2003 outbreak of another highly fatal but emerging disease, severe acute respiratory syndrome (SARS), further emphasized the need for on-site diagnostic testing, in this case, for the rapid detection of respiratory pathogens (6). Because a specific diagnostic test was not available to detect SARS coronavirus (SARS CoV) early in the outbreak, it was important to rule out infections caused by much more common pathogens, like influenza viruses, the clinical presentations of which could mimic those of SARS.
Fortunately, the biotechnology boom of the late 1990s and early 2000s fueled the development of highly automated nucleic acid-based testing methods, which had important implications for the identification of infectious pathogens in human specimens (29). One of these technologies, commonly referred to as real-time PCR, has gained considerable popularity. This method combines nucleic acid amplification and fluorescent detection of the amplified product in the same closed system (1, 8, 9, 28). The promulgation of real-time PCR as an important testing platform in clinical microbiology was catapulted by U.S. homeland security efforts to produce rapid reliable testing methods for identifying potential agents of bioterrorism. The Laboratory Response Network (LRN), an integrated group of public health, armed forces, and private referral laboratories, was created by the Centers for Disease Control and Prevention (CDC) to serve as a reference laboratory network for identifying and confirming agents of bioterrorism. In a very short period of time, scientists at CDC successfully developed a number of real-time PCR assays for detection of agents of bioterrorism, and these assays are now available at many of the LRN laboratories.
Numerous reports have described the utility of this user-friendly technology for the rapid (same-day) and accurate detection of many emerging (new) and reemerging pathogens as well as pathogens commonly encountered in medical practice. A search for all articles published in the Journal of Clinical Microbiology from 2000 through 2003 which evaluated real-time PCR as a test method for pathogen detection and/or identification of genes or mutations associated with antimicrobial resistance in pathogens revealed a total of 109 articles. Among these articles, 84 described assays with the LightCycler instrument (Roche Diagnostics Corporation, Indianapolis, Ind.); 21 described assays with the ABI PRISM 7000, 7700, or 7900H instrument (Applied Biosystems, Foster City, Calif.); 2 described assays with the SmartCycler instrument (Cepheid, Sunnyvale, Calif.); and 2 described assays with the iCycler instrument (Bio-Rad Laboratories, Hercules, Calif.). The availability of nucleic acid-based technology, such as real-time PCR, along with conventional staining and culture methods and immunoassays, can provide laboratories of many sizes with a comprehensive and responsible approach to the detection of both commonly encountered and emerging or reemerging pathogens.
The objectives of the present minireview are twofold. First, a short description of emerging and reemerging pathogens is provided. Second, a review of state-of-the art testing methods for the rapid and accurate identification of selected organisms is presented.
WHAT ARE EMERGING OR REEMERGING INFECTIOUS DISEASES?
The World Health Organization (WHO; www.who.int/inf-fs/en/fact097.html) defines “emerging infectious diseases” as those “resulting from newly identified and previously unknown infections, which cause public health problems either locally or internationally.” SARS is an example of an emerging (new) infectious disease. WHO defines “reemerging infectious diseases” as those that are “due to the reappearance of, and an increase in, the number of infections from a disease, which is known, but which had formerly caused so few infections that it had no longer been considered a public health problem.” Anthrax is an example of a reemerging infectious disease.
Table 1 displays the emerging and reemerging human pathogens and the corresponding infectious diseases which are of primary concern in developed countries. This list was compiled from recent publications by WHO (www.who.int/inf-fs/en/fact097.html) and CDC (www.cdc.gov/ncidod/diseases/eid/disease_sites.htm). For the purposes of the present discussion, only organisms that are in boldface in Table 1 are covered. The authors have made this arbitrary decision for several reasons. First, until the recent availability of rapid nucleic acid-based testing methods, like real-time PCR, no rapid testing method was available for the detection of these pathogens. Second, rapid detection of the organism or other organisms that may cause similar clinical presentations is important, because any of these organisms may produce significant morbidity or mortality if treatment is not provided expeditiously. Third, rapid identification of the organism in either symptomatic individuals or carriers is essential to prevent the spread of the disease to others. Relevant to these three points, laboratories located in close proximity to the ambulatory care clinics and hospitals that they serve can best accomplish the rapid detection of this subset of organisms in boldface in Table 1. If specimens or isolates require shipping to distant regional or referral laboratories, a significant time delay may occur before the pathogen is detected. An untoward outcome could then result for the patient, or the disease could be spread to others if isolation procedures or other preventative measures are delayed.
TABLE 1.
Emerging (new) or reemerging infectious diseases recognized by CDC and WHO and of primary concern in developed countriesa
| Agent(s) | Infectious disease |
|---|---|
| Bacteria | |
| Bacillus anthracis | Anthrax |
| Borrelia burgdorferi sensu lato complex (B. burgdorferi | |
| senso stricto, Borrelia garinii, and Borellia afzelii) | Lyme disease |
| Bartonella henselae | Cat scratch disease, bacillary angiomatosis, bacillary peliosis, endocarditis |
| Bordetella pertussis | Whooping cough |
| Ehrlichia spp. and Anaplasma phagocytophilum | Ehrlichiosis |
| Mycobacterium tuberculosis | Tuberculosis |
| MRSA | Nosocomial and community infections, including soft tissue and bone and joint infections and bacteremias |
| VRE | Nosocomial infections, including urinary tract and wound infections and bacteremias |
| Viruses | |
| Influenza virus | Influenza |
| Rotavirus | Diarrhea |
| Variola virus | Variola major and variola minor (smallpox) |
| West Nile virus | Central nervous system infection |
| SARS CoV | SARS |
Organisms that are in boldface are specifically addressed in this minireview.
Variola virus (which causes smallpox) is included in Table 1; however, at present it is not classified as either an emerging or a reemerging pathogen. Because this highly virulent virus could be used as an agent of bioterrorism and the clinical presentation caused by other common viruses can mimic that caused by variola virus, the authors have included it in this minireview.
Although the reason that some of the other pathogens in Table 1 have emerged is unknown (natural emergence), it is clear that others have emerged as the result of human intervention. Examples of the latter are methicillin (oxacillin)-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus spp. (VRE), which likely emerged as the result of antibiotic pressure. An example of the former (natural emergence) may be SARS CoV. However, even this virus may also have arisen as the result of human intervention, as contact with exotic animals, including Himalayan palm civets, which carried the virus or a precursor virus, may have played a role (16). Clearly, the most direct example of a human intervention responsible for a reemergent disease is the recent intentional release of Bacillus anthracis in the United States.
TESTING METHODS FOR SELECTED BACTERIAL AGENTS OF EMERGING AND REEMERGING INFECTIOUS DISEASES
B. pertussis.
Bordetella pertussis is a fastidious minute coccobacillary gram-negative bacterium that can cause serious morbidity, including central nervous system abnormalities, and occasionally death, especially in infants. The incidence of pertussis has increased substantially in some developed countries due to decreased pertussis vaccine use and waning postvaccination immunity in the elderly population (17). In one study (26) it was estimated that as many as 20 to 30% of adults with prolonged cough may have pertussis. The laboratory diagnosis of pertussis in adults, even those who have only mild symptoms, may be important, as they may transmit the disease to infants, who are more susceptible to serious complications.
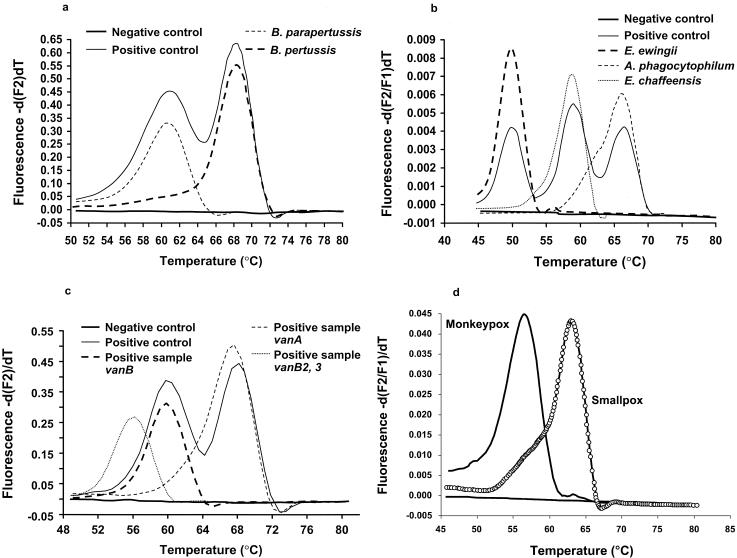
Due to its remarkably enhanced sensitivity, PCR amplification and detection of IS481 insertion sequences in the B. pertussis genome has replaced direct fluorescent-antibody (DFA) methods and culture as the “gold standard” method for detection of this upper respiratory pathogen from nasopharyngeal secretions. A comprehensive, seminal study by Loeffelholz and colleagues (19) demonstrated that the sensitivities of conventional PCR, culture, and DFA for the detection of B. pertussis in nasopharyngeal swab specimens were 93.5, 52.2, and 15.2%, respectively. Recently, our group at Mayo Clinic showed that a rapid-cycle real-time PCR method with dual fluorescent energy transfer (FRET) probes and the LightCycler instrument was over 200% more sensitive than culture (27, 28). Due to their complexity, conventional “home-brewed” PCR assays (those developed in-house) can be used only in highly specialized laboratories, such as institutional molecular core testing laboratories or referral laboratories. The recent availability in the United States of analyte-specific reagents (ASRs) by one manufacturer for use on the LightCycler real-time PCR instrument (LightCycler Bordetella IS481/1001detection assay; Roche Diagnostics Corporation) makes this an easily adaptable method for many clinical microbiology laboratories. The LightCycler platform, like the other real-time PCR testing platforms listed above, is a walk-away system that automatically performs PCR and detects PCR products in real time. The system, like most other real-time PCR instrument systems, is closed, so the chances for carryover of amplified nucleic acid (amplified product or amplicon contamination) are considerably less than those with conventional open PCR systems. Representative melting curves are shown for the Roche LightCycler Bordetella IS481/1001 ASR in Fig. 1a. It is likely that other rapid nucleic acid amplification assays for the detection of B. pertussis from other vendors will be commercially available in the near future and will also be used in many clinical microbiology laboratories.
FIG. 1.
Representative melting curves for selected real-time PCR assays with FRET probes and the LightCycler instrument. (a) Assays for B. pertussis and B. parapertussis (LightCycler Bordetella IS481/1001 detection assay; Roche Diagnostics Corporation); (b) assays for E. chafeensis, E. ewingii, and A. phagocytophilum (developed in-house; Mayo Clinic); (c) assay for VRE (LightCycler vanA/vanB detection assay; Roche Diagnostics Corporation); (d) assays for monkey poxvirus and variola virus (developed in-house; Mayo Clinic).
Agents of ehrlichiosis.
Ehrlichia chaffeensis, Ehrlichia ewingii, and Anaplasma phagocytophilum are the primary agents associated with ehrlichiosis in the United States and Europe; infections caused by Ehrlichia sennetsu are limited to the Far East. All of these agents can result in serious and sometimes fatal disease. These organisms are small gram-negative organisms that cannot be cultured by routine laboratory techniques. Shock, respiratory failure, and death are more frequent in elderly individuals, individuals infected with human immunodeficiency virus, or patients receiving immunosuppressive drugs (12). The diagnosis can be supported by observing the mulberry-like inclusions (morulae) of the organisms in infected leukocytes on Giemsa-stained thin blood films of smeared peripheral blood, but a definitive diagnosis requires isolation of the organism by culture; detection of serum antibodies, usually by immunofluorescence techniques; or PCR of blood. Culture requires specialized techniques with mammalian cell lines, and organisms may not be identified by this method for more than 1 month. Antibody detection is generally available only in referral laboratories, and a positive result may not occur during acute infection. In one study (10) only 22% of serum samples tested in the first week of illness were positive. PCR techniques, especially real-time PCR, offer the best approach for the rapid and sensitive identification of Ehrlichia spp. and Anaplasma phagocytophilum in blood samples, as demonstrated in a recent study in which the iCyler instrument and TaqMan fluorescent probes were used (20). No commercial nucleic acid-based detection ASRs or kits are available at present. However, should ASRs or kits that use real-time PCR platforms become available, this type of testing should be adaptable for many laboratories. Our research group has developed a real-time PCR assay which uses dual FRET probes and melting-curve analysis to detect and differentiate E. chaffensis, E. ewingii, and A. phagocytophilum (Fig. 1b). This real-time PCR assay is easy to perform and provides same-day results, and initial experience shows that it is as sensitive as and much easier to perform than the conventional PCR method that we used previously. (J. J. Germer, J. R. Uhl, F. R. Cockerill III, C. A. Bell, R. Patel, and J. D. C. Yao, Abstr. 103rd Gen. Meet. Am. Soc. Microbiol., abstr. C-306, 2003).
Emerging nosocomial bacterial pathogens: MRSA and VRE.
Rates of MRSA and VRE infections continue to increase in U.S. hospitals (11). What makes this of particular concern is that bacteremias caused by these gram-positive bacteria are associated with rates of mortality higher than those associated with their susceptible counterparts, methicillin-susceptible S. aureus and vancomycin-susceptible Enterococcus spp. (8).
In May 2003, the Society for Healthcare Epidemiologists of America (SHEA) published a guideline for preventing the nosocomial transmission of MRSA and VRE (22). Essential to the prevention of MRSA and VRE transmission are active surveillance programs that can identify colonized patients and then use the contact precautions recommended by CDC. Numerous studies have demonstrated that such a “search and containment” approach and/or a “search and destroy” approach (in which an attempt is made to eliminate carriage of the organism from the carrier [e.g., MRSA can be eliminated by nasal application of mupuricin]) can reduce the incidence of nosocomial infections caused by these organisms and be cost saving (22).
The conventional detection of MRSA and VRE carriers is achieved by culture. In our bacteriology laboratory at the Mayo Clinic, which operates 24 h/day, 7 days/week, we recently determined that the mean time for detection of MRSA from nasal swab specimens was ≅48 h and that the mean time for detection of VRE from perianal swab specimens was ≅72 h (8). If comprehensive surveillance programs are undertaken by health care facilities in accordance with the SHEA guidelines, large-scale culture evaluation may be particularly demanding, if not impossible. Moreover, the lack of sensitivity of culture, especially for detection of VRE in stool specimens, and the time required to generate a final result by culture may affect the ability to rapidly and consistently reduce or eliminate nosocomial outbreaks. DNA-based amplification techniques, in contrast to culture, have been shown to have improved sensitivity, especially for the detection of VRE from rectal or perianal swab specimens (23), and to dramatically decrease the time required for a result for both MRSA from nasal swab specimens and VRE from rectal or perianal swab specimens (8).
At least two manufacturers now or soon will have ASRs or kits available for use with real-time PCR instrumentation for detection of VRE and MRSA. Roche Diagnostics Corporation provides separate ASRs for VRE (LightCycler vanA/vanB detection assay; Fig. 1) and MRSA (LightCycler mecA detection assay) detection with the LightCycler instrument. Infectio Diagnostics (IDI; Quebec, Quebec, Canada) has recently received Food and Drug Administration approval for a kit that can directly screen nasal swab specimens for MRSA (IDI-MRSA with the SmartCycler instrument [Cepheid]). At the Mayo Clinic, we have used the Roche VRE detection assay in combination with an automated nucleic acid extraction instrument, the MagNA Pure instrument (Roche Diagnostics Corporation), which is designed to be used in tandem with the LightCycler instrument (Fig. 1c). It has been determined that the Roche VRE detection assay is over 120% more sensitive than standard VRE culture screening plates; and final results are available within 3.5 h, whereas culture requires ≥3 days (27a). In a study with the IDI-MRSA and the SmartCycler instrument, the sensitivity of the assay for the detection of MRSA directly from nasal swab specimens equaled that of culture, and the results were available considerably faster (within 2 h, whereas culture requires 48 to 72 h) (R. S. Liao, D. K. Warren, L. R. Merz, and W. M. Dunne, Jr., Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. K-1748, 2003).
In order to make large-scale surveillance programs for MRSA and VRE feasible from the perspective of the workload in the laboratory and effective from the perspective of the prevention of nosocomial outbreaks, rapid, sensitive, easy-to-perform tests like the LightCycler and SmartCyler nucleic acid-based tests will be essential.
B. anthracis.
B. anthracis should be suspected if large spore-forming gram-positive bacilli are observed on Gram stains performed directly on clinical specimens or from nonhemolytic, nonmotile, catalase-positive colonies that grow on blood agar. Due to national security concerns, forensic requirements in potential criminal situations, safety concerns at the local laboratory level, and the necessity to confirm true-positive results, it is essential that any suspected case of anthrax be immediately reported to public health officials.
As mentioned previously, LRN, sponsored by CDC in the United States, is primarily focused on providing confirmatory clinical microbiology diagnostic testing for agents of bioterrorism. Confirmation of the isolation of these agents from human specimens or environmental samples (e.g., envelopes containing a powdery substance) is vitally important. Many clinical microbiology laboratories were deluged with requests to test environmental samples during the hysteria that followed the intentional release of anthrax spores in the United States in late 2001. False-positive results could result in undo anxiety and unnecessary medical interventions, including the provision of prophylactic antibiotics or vaccines to exposed individuals. In its role as a confirmatory testing laboratory, LRN serves an important complementary function to local laboratories, which provide lower-level testing for institutions directly involved in patient care.
B. anthracis test kits or ASRs are commercially available from manufacturers for use with several real-time PCR testing platforms. The LightCycler Bacillus anthracis Detection kit (Roche Applied Science, Indianapolis, Ind.) and the Bacillus anthracis Biothreat Screening kit (Idaho Technology, Salt Lake City, Utah) are designed for use on the LightCycler instrument. Artus (Hamburg, Germany) provides RealArt B. anthracis PCR kits, which can be used on the LightCycler instrument, the ABI Prism instruments (7000, 7700, and 7900H), or the Rotor-Gene instrument (Corbett Research, Sydney, Australia). When such PCR methods are coupled with autoclaving of specimens, they may provide a rapid, reliable, user-friendly, and safe detection method for local laboratories that are nearest the site of a bioterrorism event but that do not have biosafety level 3 capabilities. Two recent studies have demonstrated that autoclaving of B. anthracis or vaccinia virus (a surrogate for smallpox virus) does not affect the sensitivities of either conventional or real-time PCR assays (14, 15). Therefore, autoclaving should obviate concerns that individuals transporting or testing the specimens may be exposed to the agent. Even if real-time PCR tests are performed, LRN laboratories will still need to confirm the results. Culture of specimens may still be performed by LRN facilities, which generally have higher-level biosafety facilities. Culture may be necessary for susceptibility testing and strain identification, if required for treatment or forensic evaluation.
It is the authors' opinion that the high rates of mortality associated with B. anthracis infections and infections caused by other potential bacterial agents of bioterrorism, like Francisella tularensis and Yersinia pestis, and both the high rates of mortality and the significant chance for the communal spread of variola virus (the agent of smallpox) necessitate the availability of rapid detection methods in close proximity to patient care, i.e., on-site in microbiology laboratories at health care institutions. Several papers (2, 18, 21) have described the utility of real-time PCR assays for the rapid identification of B. anthracis. To ensure that virulent strains can be differentiated from avirulent strains (which may be used in hoaxes), assays should include primers and probes for the detection of virulence genes on both plasmid pX0 and plasmid pX02.
METHODS FOR TESTING FOR SELECTED VIRAL AGENTS OF EMERGING AND REEMERGING INFECTIOUS DISEASES
Variola virus: the importance of ruling out the presence of common viral pathogens that can cause cutaneous vesicular disease.
CDC has developed an algorithm for the clinical and laboratory evaluation of potential cases of smallpox (www.cdc.gov). Depending on the risk for the disease, the clinical presentations of patients infected with common viruses that cause cutaneous vesicular lesions (i.e., herpes simplex virus [HSV], varicella-zoster virus [VZV], enterovirus, or disseminated vaccinia virus following smallpox vaccination) may mimic those of patients with smallpox. Another complicating feature is that some recipients of the smallpox vaccine may develop erythema multiforme, which can also present as vesicular lesions. It has been our experience that by using real-time PCR assays, one can rapidly discriminate among these possibilities. ASRs or kits for the detection of HSV or VZV with the LightCycler instrument are available from at least two vendors (RealArt HSV 1/2 kit and RealArtVZV PCR kit [Artus]; LightCycler Herpes Simplex Virus 1/2 and LightCycler VZV ORF29 [Roche Diagnostics Corporation]). Kits are also available for testing for VZV (RealArt VZV PCR kit; Artus) with the ABI PRISM 7000, 7700, and 7900H instruments. We have used the assays with the Roche LightCycler instrument to routinely detect HSV and VZV and have developed an in-house real-time PCR assay for poxviruses, including variola virus, that uses the LightCycler instrument. These assays have been invaluable for providing a rapid result, especially for military personnel who have developed cutaneous vesicular lesions as a complication of receiving the smallpox vaccine and who have been on assignment in areas of the world at significant risk for bioterrorism events. Importantly, the home-brewed real-time assay that we have developed can discriminate among several poxviruses and was useful in the identification of viremia in a recent case of monkey pox virus disease in a patient from the upper Midwest (13). An ASR for the detection of variola virus with the LightCycler instrument is also available from Artus (RealArt Orthopox PCR kit).
West Nile virus: the importance of ruling out treatable causes of viral central nervous system disease.
West Nile virus, a RNA virus of the family Flaviviridae, has a predilection for the central nervous system and can be associated with significant morbidity and mortality. The first human cases of West Nile virus infection occurred in the northeastern United States in the summer of 1999; since then the disease has progressed relentlessly from east to west across the continental United States. As yet, no effective therapy has been defined (24).
Traditionally, during the summer and early fall in the United States, viral central nervous system disease is most frequently caused by enterovirus. In most regions of the United States, West Nile virus infection must now also be considered during this time of the year. HSV can cause encephalitis at any time of the year, and antiviral therapy is available and effective. Therefore, ruling out HSV infection should be a priority, especially when encephalitis is encountered. Real-time PCR has replaced viral culture as the gold standard for the rapid and accurate detection of HSV in cerebrospinal fluid. As mentioned previously, ASRs or kits for the detection of HSV are available from Artus and Roche. Artus also has a kit that can be used to test for enterovirus (RealArt Enterovirus RT PCR kit) with the LightCycler instrument.
Limited studies have shown that PCR detection of West Nile virus in cerebrospinal fluid is less sensitive than immunoassay for immunoglobulin M antibodies (24). At present, only a few referral and public health laboratories have the capability to perform immunoassays. At least two companies offer ASRs or kits for real-time PCR (RealArt WNV RT PCR kit [Artus]; LightCycler WNV Detection Kit [Roche Applied Science]) with the LightCycler platform. If effective antiviral therapy becomes available, the rapid on-site diagnosis of West Nile virus disease in areas of endemicity may be desirable.
SARS CoV: the importance of ruling out influenza.
One important lesson learned from the 2002-2003 winter outbreak of SARS was that the early identification and quarantine of individuals with suspected cases of SARS were essential for controlling the disease, especially in institutional settings (7, 16). This effective approach toward the control of a communicable infectious disease adds credence to the concept that similar measures can be effective for controlling and preventing nosocomial VRE and MRSA outbreaks. No laboratory tests were available for the detection of SARS CoV during much of the outbreak, as the etiological agent was not confirmed until early March 2003. Eventually, real-time PCR tests were developed and were available commercially from at least two manufacturers for use with several real-time PCR testing platforms (RealArt HPA-Coronavirus RT PCR Kits [Artus] for use with the LightCyler instrument, the ABI PRISM 7000, 7700, and 7900H instruments, and the Rotor-Gene instrument; and LightCycler SARS-CoV [Roche Diagnostics Corporation] for use with the LightCycler instrument). During the outbreak it was important to rule out treatable influenza virus type A or B infections, whose clinical presentations can mimic those of SARS CoV.
Rapid antigen tests for the detection of influenza virus (both type A and type B) are relatively easy to perform and may be useful in the local setting for the detection of cases of influenza; however, these tests lack sensitivity. As infections due to both influenza virus type A and influenza virus type B are now treatable, rapid on-site diagnostic capabilities are important. Recently, a real-time PCR assay that uses the LightCycler platform was demonstrated to have much greater sensitivity than antigen detection (100 and 44%, respectively) for the detection of influenza virus type A infections (3).
Following the 2002-2003 SARS outbreak, many LRN member laboratories developed the capability to detect SARS CoV. Should another outbreak occur, this public health laboratory network should facilitate the laboratory diagnosis of cases, especially when testing at the local level is not available.
CONCLUSIONS
Clinical microbiology laboratories at the local level have an increasing responsibility to provide rapid and accurate diagnostic services for emerging (new) and reemerging infectious diseases, especially those diseases for which significant mortality or morbidity may occur as the result of a delay in diagnosis. Rapid, accurate diagnosis of emerging and reemerging infectious diseases may also be critical at the local level to ensure optimal infection control. Detection of these pathogens has often required esoteric procedures like conventional PCR, which could be performed only at referral laboratories or, recently, at public health laboratories.
Recent technical advances in molecular diagnostics have resulted in the development of user-friendly automated testing platforms, such as real-time PCR. These novel testing methods can be used to detect emerging and reemerging pathogens as well as common pathogens and have the potential for broad-scale use in smaller laboratories in close proximity to the delivery of care.
During the writing of this minireview, a large outbreak of influenza virus type A (H3N2) was peaking in the United States, and new influenza virus type A strains (H5N1, H9N2) have been associated with both avian and human influenza in regions of the Far East (6). The apparent significant morbidity and mortality associated with these new influenza virus strains emphasize the need for rapid, accurate laboratory diagnostic capabilities at the local level (4, 5). As is the case for SARS, agents of bioterrorism, and the other pathogens discussed in this minireview, rapid diagnostic methods, such as real-time PCR, will likely play a major role in the early and sensitive detection of emerging and reemerging infectious diseases encountered in the future.
Acknowledgments
JoAnn Brunette is thanked for her assistance in preparing the manuscript.
REFERENCES
- 1.Bankowski, M. J., and S. M. Anderson. 2004. Real-time nucleic acid amplification in clinical microbiology. Clin. Microbiol. Newsl. 26:9-15. [Google Scholar]
- 2.Bell, C. A., J. R. Uhl, T. L. Hadfield, J. C. David, R. F. Meyer, T. F. Smith, and F. R. Cockerill III. 2002. Detection of Bacillus anthracis DNA by LightCycler PCR. J. Clin. Microbiol. 40:2897-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boivin, G., S. Cote, P. Dery, F. DeSerres, and M. G. Bergeron. 2004. Multiplex real-time PCR assay for detection of influenza and human respiratory syncytial viruses. J. Clin. Microbiol. 42:45-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campitelli, L., C. Fabiani, S. Puzelli, A. Fioretti, E. Foni, A. De Marco, S. Krauss, R. G. Webster, and I. Donatelli. 2002. H3N2 influenza viruses from domestic chickens in Italy: an increasing role for chickens in the ecology of influenza? J. Gen. Virol. 83(Pt 2):413-420. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2003. Prevention and control of influenza; recommendations of the Advisory Committee on Immunization Practices (ACIP). Morb. Mortal. Wkly. Rep. 52(RR-08):1-36. [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2004. Update: influenza activity—United States, January 18-24, 2004. Morb. Mortal. Wkly. Rep. 53(RR-03):63-65. [PubMed] [Google Scholar]
- 7.Choi, K. W., T. N. Chau, O. Tsang, E. Tso, M. C. Chiu, W. L. Tong, P. O. Lee, T. K. Ng, W. F. Ng, K. C. Lee, W. Lam, W. C. Yu, J. Y. Lai, and S. T. Lai. 2003. Outcomes and prognostic factors in 267 patients with severe acute respiratory syndrome in Hong Kong. Ann. Intern. Med. 139:715-723. [DOI] [PubMed] [Google Scholar]
- 8.Cockerill, F. R., III 2003. Rapid detection of pathogens and antimicrobial resistance in intensive care patients using nucleic acid-based techniques. Scand. J. Clin. Lab. Investig. 63:34-46. [Google Scholar]
- 9.Cockerill, F. R., III, and J. R. Uhl. 2002. Applications and challenges of real-time PCR for the clinical microbiology laboratory, p. 3-27. In U. Reischl, C. Wittwer, and F. Cockerill (ed.), Rapid cycle real-time PCR methods and applications. Springer-Verlag, Berlin, Germany.
- 10.Dawson, J. E., D. B. Fishbein, T. R. Eng, M. A. Redus, and N. R. Green. 1990. Diagnosis of human ehrlichiosis with the indirect fluorescent antibody test: kinetics and specificity. J. Infect. Dis. 162:91-95. [DOI] [PubMed] [Google Scholar]
- 11.Diekema, D. J., B. J. Boots Miller, T. E. Vaughn, R. F. Woolson, J. W. Yankey, E. J. Ernst, S. D. Flach, M. M. Ward, C. L. Franciscus, M. A. Pfaller, and B. N. Doebbeling. 2004. Antimicrobial resistance trends and outbreak frequency in United States hospitals. Clin. Infect. Dis. 38:78-85. [DOI] [PubMed] [Google Scholar]
- 12.Dumler, J. S., and J. S. Bakken. 1998. Human ehrlichioses: newly recognized infections transmitted by ticks. Annu. Rev. Med. 49:201-213. [DOI] [PubMed] [Google Scholar]
- 13.Espy, M. J., F. R. Cockerill III, R. F. Meyer, M. D. Bowen, G. A. Poland, T. L. Hadfield, and T. F. Smith. 2002. Detection of smallpox virus DNA by LightCycler PCR. J. Clin. Microbiol. 40:1985-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espy, M. J., J. R. Uhl, L. M. Sloan, J. E. Rosenblatt, F. R. Cockerill III, and T. F. Smith. 2002. Detection of vaccinia virus, herpes simplex virus, varicella-zoster virus, and Bacillus anthracis DNA by LightCycler polymerase chain reaction after autoclaving: implications for biosafety of bioterrorism agents. Mayo Clin. Proc. 77:624-628. [DOI] [PubMed] [Google Scholar]
- 15.Fasanella, A., S. Losito, R. Adone, F. Ciuchini, T. Trotta, S. A. Altamura, D. Chiocco, and G. Ippolito. 2003. PCR assay to detect Bacillus anthracis spores in heat-treated specimens. J. Clin. Microbiol. 41:896-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276-278. [DOI] [PubMed] [Google Scholar]
- 17.Hewlett, E. L. 2000. Bordetella species, p. 2414-2422. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, vol. 2, 5th ed. Churchill Livingstone, Philadelphia, Pa.
- 18.Lee, M. A., G. Brightwell, D. Leslie, H. Bird, and A. Hamilton. 1999. Fluorescent detection techniques for real-time multiplex strand specific detection of Bacillus anthracis using rapid PCR. J. Appl. Microbiol. 87:218-223. [DOI] [PubMed] [Google Scholar]
- 19.Loeffelholz, M. J., C. J. Thompson, K. S. Long, and M. J. Gilchrist. 1999. Comparison of PCR, culture, and direct fluorescent-antibody testing for detection of Bordetella pertussis. J. Clin. Microbiol. 37:2872-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loftis, A. D., R. F. Massung, and M. L. Levin. 2003. Quantitative real-time PCR assay for detection of Ehrlichia chaffeensis. J. Clin. Microbiol. 41:3870-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makino, S., and H. I. Cheun. 2003. Application of the real-time PCR for the detection of airborne microbial pathogens in reference to the anthrax spores. J. Microbiol. Methods 53:141-147. [DOI] [PubMed] [Google Scholar]
- 22.Muto, C. A., J. A. Jernigan, B. E. Ostrowsky, H. M. Richet, W. R. Jarvis, J. M. Boyce, and B. M. Farr. 2003. SHEA guideline for preventing nosocomial transmission of multidrug-resistant strains of Staphylococcus aureus and enterococcus. Infect. Control Hosp. Epidemiol. 24:362-386. [DOI] [PubMed] [Google Scholar]
- 23.Paule, S. M., W. E. Trick, F. C. Tenover, M. Lankford, S. Cunningham, V. Stosor, R. L. Cordell, and L. R. Peterson. 2003. Comparison of PCR assay to culture for surveillance detection of vancomycin-resistant enterococci. J. Clin. Microbiol. 41:4805-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petersen, L. R., and A. A. Marfin. 2002. West Nile virus: a primer for the clinician. Ann. Intern. Med. 137:173-179. [DOI] [PubMed] [Google Scholar]
- 25.Peterson, L. R., J. D. Hamilton, E. J. Baron, L. S. Tompkins, J. M. Miller, C. M. Wilfert, F. C. Tenover, and R. B. Thomson, Jr. 2001. Role of clinical microbiology laboratories in the management and control of infectious diseases and the delivery of health care. Clin. Infect. Dis. 32:605-611. [DOI] [PubMed] [Google Scholar]
- 26.Robbins, J. B. 1999. Pertussis in adults: introduction. Clin. Infect. Dis. 28(Suppl. 2):S91-S93. [DOI] [PubMed] [Google Scholar]
- 27.Sloan, L. M., M. K. Hopkins, P. S. Mitchell, E. A. Vetter, J. E. Rosenblatt, W. S. Harmsen, F. R. Cockerill, and R. Patel. 2002. Multiplex LightCycler PCR assay for detection and differentiation of Bordetella pertussis and Bordetella parapertussis in nasopharyngeal specimens. J. Clin. Microbiol. 40:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Sloan, L. M., J. R. Uhl, E. A. Vetter, C. D. Schleck, W. S. Harmsen, J. Manahan, R. L. Thompson, J. E. Rosenblatt, and F. R. Cockerill III. 2004. Comparison of the Roche Lightcycler vanA/vanB detection assay and culture for detection of vancomycin-resistant enterococci from perianal swabs. 42:2636-2643. [DOI] [PMC free article] [PubMed]
- 28.Uhl, J. R., and F. R. Cockerill III. 2004. The fluorescence resonance energy transfer system, p. 295-306. In D. Persing, F. C. Tenover, J. Versalovic, Y. W. Tang, E. Unger, D. Relman, and T. White (ed.), Molecular microbiology diagnostic principles and practice. ASM Press, Washington, D.C.
- 29.Wolk, D., P. S. Mitchell, and R. Patel. 2001. Principles of molecular microbiology testing methods. Infect. Dis. Clin. N. Am. 15:1157-1204. [DOI] [PubMed]