Legionella species are gram-negative environmental bacteria that replicate within amoebae and other protozoa. After transmission to humans, pneumonia, influenza-like illness, or asymptomatic infection may occur (1). So far, 50 Legionella spp. have been described (www.bacterio.cict.fr). About half of them are associated with illness in humans. The most important species is L. pneumophila. The remaining Legionella spp. account for about 5 to 10% of clinical cases, mostly in immunosuppressed patients (1).
Several Legionella spp. exhibit blue-white fluorescence under long-wavelength UV light. Some investigators have proposed placing these species in the genus Fluoribacter within the family Legionellaceae. However, recent studies using 16S rRNA gene analysis confirm that only one genus in this family, Legionella, exists. Nevertheless, the blue-white species formed clusters within this genus when 16S rRNA, mip (macrophage infectivity potentiator), and rpoB (RNA polymerase subunit beta) DNA sequences were analyzed (2, 5). The species within this cluster are L. anisa, L. bozemanii, L. cherrii, L. dumoffii, L. gormanii, L. parisiensis, L. gratiana, L. steigerwaltii, and L. tucsoniensis.
Legionella parisiensis was first isolated from environmental samples. In 1997, Lo Presti et al. (3) obtained the first strain of this species from a liver transplant patient with pneumonia. Here, we report on a nonfluorescent L. parisiensis strain that was isolated from an immunosuppressed patient with pneumonia in Germany.
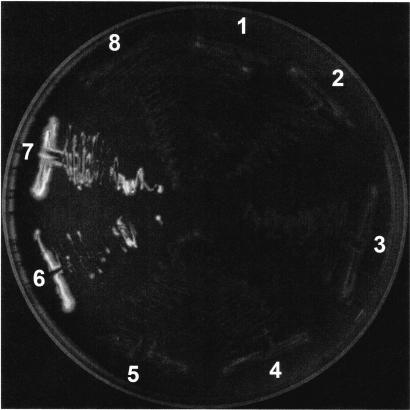
This strain showed the typical ground glass colony appearance on buffered charcoal yeast extract (BCYE) agar, did not grow on sheep blood agar, and reacted with the non-L. pneumophila reagent in a commercially available latex agglutination kit (Oxoid, Wesel, Germany). According to the manufacturer, this reagent contains rabbit antisera against L. anisa, L. bozemanii, L. dumoffii, L. gormanii, L. jordanis, L. longbeachae, and L. micdadei. By using rabbit antisera against these species prepared in our laboratory, the strain reacted strongly to rabbit anti-L. bozemanii antiserum and weakly to anti-L. anisa antiserum. In contrast, none of the colonies isolated exhibited the blue-white fluorescence under UV light (Fig. 1) typical for strains of the blue-white fluorescent group.
FIG. 1.
Growth of L. parisiensis (colonies 1 to 6) and L. bozemanii (colonies 6 and 7) on BCYE agar after 3 days at 37°C in 5% CO2 when examined under long-wavelength UV light.
To define the species of this strain exactly, we determined the DNA sequences of the 16S rRNA, mip, and rpoB genes (2, 5). Comparison of the nucleotide sequences with those in the National Center for Biotechnology Information database by using the standard nucleotide Basic Local Alignment Search Tool (BLASTn) revealed nearly complete homology to L. parisiensis: 16S rRNA, 100%; mip, 99%; and rpoB, 100%. Based on mip sequences, the next related species were L. tucsoniensis (95%), L. bozemanii (95%), and L. anisa (94%). The rpoB sequences revealed L. bozemanii (94%) and L. anisa (90%) as the closest related species. The BLAST analysis of the 16S rRNA sequence shows agreement with L. anisa (97%), L. bozemanii (97%), and L. dumoffii (97%).
Furthermore, analysis of bacterial cell wall fatty acids was performed by using the Microbial Identification System (MIDI, Newark, Del.) with the Hewlett-Packard 9890 chromatograph and the MIDI software package Aerobe Clin40 database, version 4.0. For this analysis, three colonies were grown and analyzed in separate runs. Good library comparisons—i.e., a similarity index (SI) of >0.6 and separation from the second choice of >0.1—were obtained for L. parisiensis (SI = 0.791 ± 0.04). Similarities were indicated for L. gormanii (SI = 0.588), L. anisa (SI = 0.587), and L. bozemanii (SI = 0.326). Thus, the strain could be unambiguously assigned to the species L. parisiensis.
There is no information available describing which genes are involved in the blue-white fluorescent phenotype. Furthermore, the relevance of these genes for virulence is unknown. Originally L. parisiensis was not known to cause pneumonia in humans. In 1996, O′Connell et al. (4) showed that the type strain of L. parisiensis is able to multiply in macrophage-like cells. We infected Acanthamoeba castellanii with our strain of L. parisiensis. Within 24 h, our strain multiplied approximately 1,000-fold. This result showed that the genes responsible for blue-white fluorescence seem to be not essential for intracellular multiplication. The fact that we isolated this nonfluorescent L. parisiensis strain from a patient argues for some virulence properties of this strain.
In general, the identification of Legionella spp. in the clinical laboratory is based on phenotypic properties: i.e., colony morphology, cysteine dependence, and serotyping. But, as shown here, this might be misleading. Due to the serological cross-reactivity among Legionella spp., definitive species identification must be based on sequence analysis of Legionella genes. Thus, it might be possible that in different laboratories, strains exist that have been misclassified on the basis of serotyping results. If our strain had shown the blue-white phenotype, we would not have determined the exact species by sequencing the mip gene. Finally, it must be pointed out that all Legionella spp. are susceptible to macrolides, quinolones, or rifampin. Therefore, the choices of antibiotic therapy are identical for all Legionella spp.
Nucleotide sequence accession number.
The nucleotide sequences have been submitted toGenBank EMBL under accession no. AJ601373, AJ601374, and AJ601375 for the mip, rpoB, and 16S rRNA genes, respectively.
Acknowledgments
We are grateful to Jutta Paasche for technical assistance.
This study was supported by the Federal Ministry of Education and Research of Germany, Network of Competence in Medicine CAPNetz.
REFERENCES
- 1.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ko, K. S., H. K. Lee, M.-Y. Park, K.-H. Lee, Y.-J. Yun, S.-Y. Woo, H. Miyamoto, and Y.-H. Kook. 2002. Application of RNA polymerase β-subunit gene (rpoB) sequences for the molecular differentiation of Legionella species. J. Clin. Microbiol. 40:2653-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo Presti, F., S. Riffard, F. Vandenesch, M. Reyrolle, E. Ronco, P. Ichai, and J. Etienne. 1997. The first clinical isolate of Legionella parisiensis, from a liver transplant patient with pneumonia. J. Clin. Microbiol. 35:1706-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O′Connell, W. A., L. Dhand, and N. P. Cianciotto. 1996. Infection of macrophage-like cells by Legionella species that have not been associated with disease. Infect. Immun. 64:4381-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ratcliff, R. M., J. A. Lanser, P. A. Manning, and M. W. Heuzenroeder. 1998. Sequence-based classification scheme for the genus Legionella targeting the mip gene. J. Clin. Microbiol. 36:1560-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]