Abstract
Bmi1 (B-cell-specific Moloney murine leukemia virus insertion site 1) had been found to involve in self -renewal of stem cells and tumorigenesis in various malignancies. The purpose of this study is to evaluate the role of Bmi1 in the development of tongue squamous cell carcinoma (TSCC) and its functional effect on the migration and invasion of TSCC. Initially, immunohistochemistry revealed that Bmi1 overexpression was a common event in premalignant dysplasia, primary TSCC, and lymph node metastases and was associated with a poor prognosis. A significant correlation between Bmi1 and SOD2 (manganese superoxide dismutase) expression was observed. Side population (SP) cells were used as cancer stem-like cells and further assessed by sphere and colony formation assays, and the expression of stem cell markers. TSCC cells with higher migration and invasion ability (UM1 cell lines) showed a higher proportion of SP cells and Bmi1 expression than TSCC cells with lower migration and invasion ability (UM2 cell lines). Knockdown of Bmi1 in UM1 or SP cells inhibited migration and invasion and decreased the sphere and colony formation, and the expression of stem cell markers and SOD2. Direct binding of C-myc to the Bmi1 promoter was demonstrated by chromatin immunoprecipitation and luciferase assays. Moreover, C-myc knockdown in SP cells inhibited their migration and invasion and decreased the expression of Bmi1 and SOD2. Our results indicate that the deregulation of Bmi1 expression is a frequent event during the progression of TSCC and may have a prognostic value for patients with this disease. The Bmi1-mediated migration and invasion of TSCC is related to cancer stem-like cells and involves the C-myc-Bmi1-SOD2 pathway.
Keywords: Tongue squamous cell carcinoma, Cancer stem-like cell, migration, invasion, prognosis, Bmi1.
Introduction
As a type of common malignant tumor in the oral cavity, tongue squamous cell carcinoma (TSCC) has a high mortality rate due to early metastasis and recurrence. Although the level of healthcare has greatly improved, the cure rate of TSCC remains unsatisfactory, and the 5-year survival rate is 50% 1. One of the reasons for treatment failure is thought to be related to the presence of a subpopulation of cells within the tumor called cancer stem cells (CSCs). In head and neck squamous cell carcinomas (HNSCCs), including TSCC 2, CSCs have been shown to have an integral role in tumor initiation, disease progression, metastasis, and treatment resistance 3-6.
B-cell-specific Moloney murine leukemia virus insertion site 1 (Bmi1) is a member of the Polycomb group of chromatin-modifier proteins. As a stem cell marker, Bmi1 plays a key role in the functioning of endogenous stem cells and CSCs 7,8. Moreover, Bmi1 had been found to be related with many other cancer stem cell markers 9-12, such as ALDH1, CD44, Oct4, SOX2, Nanog and ABCG2. Tsai et al found that Sphere-forming/self-renewal capability was increased in cisplatin-resistant OC2 cells (oral squamous carcinoma cells). Cisplatin-resistant OC2 cells highly expressed the stemness markers (Nanog, Oct4, Bmi1, CD117, CD133, and ABCG2) and increased migration/invasion/clonogenicity ability 9. Seo et al also found a connection between BMI-1 and Sox2 in maintaining self-renewal and identified BMI-1 as a key mediator of Sox2 function 10. In human cancers, Bmi1 overexpression had been found to drive stem-like properties associated with the induction of the epithelial-mesenchymal transition that promotes invasion, metastasis, and poor prognosis 13. Indeed, many studies have reported that Bmi1 is overexpressed in HNSCC cells when compared with normal epithelium and is thought to influence cell proliferation and survival in HNSCC 13-16. In contrast, Hayry`s study found that Bmi-1-negative tumors showed a correlation with a poor prognosis in TSCC 17. Thus, more evidence is needed to confirm the relationship between Bmi1 and the development of TSCC.
Although Bmi1 has been found to be associated with HNSCC 10, 11, the mechanistic rationale for an increased metastatic capacity of tumor cells overexpressing Bmi1 is still ambiguous and requires further investigation. As an oncogenic transcription factor, the C-myc protein recognizes an E-box recognition site in the promoter of its target genes and then exerts a wide array of biological functions in different cellular models, including cell cycle control, cell differentiation, and metastasis 18-20. C-myc has been documented to regulate the expression of an unusually large number of target genes, including Bmi1 in nasopharyngeal carcinoma 21. In our previous studies 22-24, we identified that the manganese superoxide dismutase (SOD2)-dependent production of H2O2 contributes to the migration and invasion of TSCC via the Snail (Snai1 and Slug) signaling pathway. However, the relationship between Bmi1 and SOD2 has not been uncovered to date.
The goal of this study was to investigate the role of Bmi1 in the development of TSCC and its mechanism in the migration and invasion of TSCC. First, we investigated the role of Bmi1 in the development and progress of TSCC. We then investigated whether the migration and invasion of TSCC mediated by Bmi1 were related to CSCs and whether the C-myc-Bmi1-SOD2 pathway mediates the metastasis of TSCC. Our findings suggest that Bmi1 is an important factor in the development and prognosis of TSCC. In addition, the Bmi1-mediated migration and invasion of TSCC is related to cancer stem-like cells and involves the C-myc-Bmi1-SOD2 pathway.
Methods and materials
Patients and tissues
The clinical samples used in this study were obtained from 77 cases of TSCC, 22 cases of premalignant tongue (leukoplakia, LP), and 12 normal tongue biopsies, which had been used in our previous study 23,25. Clinical characterizations of these samples are summarized in Supplementary Material: Table S1. Among the 77 cases of TSCC that we examined, follow-up results were available for 52 cases; the median duration of follow-up was 77 months (range: 8-116 months). Survival was calculated based on the date of surgery and the date of latest follow-up (or death). This study was approved by the ethical committee of the First Affiliated Hospital, Sun Yat-Sen University.
Immunohistochemistry analysis
The immunohistochemistry analysis was performed as previously described 25,26 using an anti-Bmi1 antibody (Cell Signaling Technology, Beverly, MA, USA). Sample evaluation via immunostaining was performed by 3 independent pathologists blinded for the clinical data according to criteria of staining intensity described previously by Li et al 27. All areas of tumor cells within each section were analyzed. All tumor cells in ten random high-power fields were counted. A scale of 0 to 3 was used to score the relative intensity, with 0 corresponding to no detectable immunoreactivity, and 1, 2, and 3 equivalent to low, moderate, and high staining, respectively. The representative images of each score (0-3) were supplied in Supplementary Material: Figure S1.
Cell culture and transfection
Human TSCC cell lines (UM1 and UM2) 24, with UM1 having a higher migration and invasion ability than UM2, were maintained in DMEM/F12 (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum (Hyclone, Logan, Utah, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin in a 37 °C incubator with 5% CO2. For further study, a Bmi1 siRNA, C-myc siRNA, and control non-targeting siRNA (Genepharma, Shanghai, China) were transfected into the TSCC cells using Lipofectamine Transfection Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions, as previously described 28. Three sequences of Bmi1 or C-myc siRNA were used, and then the sequence that had the best knockdown effect was chosen. The sequences of siRNA used for transfection are shown in Supplementary Material: Table S2.
Cell migration and invasion assay
The cell migration and invasion assay were performed according to our previous study 24,28. Briefly, for the migration assays, cells were seeded in the upper chamber of Transwell devices (Corning, Steuben, NY, USA) with a membrane containing 8-μm-diameter pores in serum-free medium. After 24 h, the cells on the lower surface of the membrane were fixed and stained with DAPI solution in the dark. Three random fields were captured at 20 × magnification. For the invasion assays, Biocoat Matrigel invasion chamber inserts (BD Biosciences, San Diego, CA, USA) were equilibrated for 2 h at 37 °C in serum-free medium. The cells were seeded in the upper chamber and allowed to invade through the Matrigel to the lower chamber for 24 h. After 24 h, the number of invaded cells (lower side of the membrane) was counted as described above.
Western blot analysis
Western blots were performed as described previously 24 using antibodies specific for Bmi1, SOD2, Slug (Cell Signaling Technology, Beverly, MA, USA), Nanog (Aviva Systems Biology, San Diego, CA, USA), C-myc, ABCG2 (ATP-binding cassette transporters) (Santa Cruz, Dallas, Texas, USA) and GAPDH (Sigma, San Louis, MO, USA). The results obtained from western blot were quantified by Quantity One software (Bio-Rad) and shown in Supplementary Material: Figure S2.
Side population sorting
The TSCC side population was sorted as previously described 29. Briefly, TSCC cells were incubated at 37 °C for 90 minutes in the dark with 5 μg/ml Hoechst 33342 (Sigma, San Louis, MO, USA) and intermittent mixing either alone or in the presence of 50 μM verapamil (Sigma, San Louis, MO, USA), an inhibitor of ABC transporters. The cells were then centrifuged and resuspended in ice-cold HBSS. The samples were sorted using a FACSAria II high-speed cell sorter (BD Biosciences, San Diego, CA, USA). Propidium iodide (2 μg/ml) was added to exclude dead cells. Hoechst 33342 was excited with the UV laser at 350 nm, and the fluorescence emission was measured with 405 / BP30 (Hoechst blue) and 570 / BP20 (Hoechst red) optical filters. After sorting, the SP and non-SP cells were used for further experiments.
Sphere and colony formation assays
For the spheroid-forming assay, cells were plated in 24-well ultralow attachment plates (Corning, Steuben, NY, USA) at a density of 1000 cells/ml in 100 μl of stem cell medium containing DMEM/F12 plus 1% N2, 10 ng/ml human recombinant bFGF, 10 ng/ml human recombinant EGF, and 1% antibiotic-antimycotic (Life Technologies, Carlsbad, CA, USA). Sphere formation (20 mm in diameter) in each well was calculated at days 14 after seeding.
For the colony formation assay, cells were plated at approximately 200 cells per well in 6-well coated plates in DMEM/F-12 supplemented with 10% FBS. The medium was changed twice a week. After 14 days, the cells were fixed in 4% formaldehyde and stained with Giemsa. Colonies larger than 1 mm (>50 cells/clone) in diameter were counted.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as described previously 30 following the manufacturer's recommendations (Upstate Biotechnology, Buffalo, NY, USA). A total of 1×107 UM1 or UM2 cells were crosslinked with formaldehyde, and the cell pellet was lysed and sonicated to shear the DNA into fragments of 200-500 bp. Following precleaning, 1% of each chromatin supernatant was used as the input loading control; the remaining chromatin supernatant was incubated with C-myc antibodies or preimmune IgG (PI) for immunoprecipitation. Protein-chromatin complexes were eluted and reverse-cross-linked to recover free DNA. Purified DNA was analyzed by a quantitative polymerase chain reaction (PCR) analysis with SYBR Green qPCR SuperMix Kit (Invitrogen, Carlsbad, CA, USA), as previously described 28. The primers for Bmi1 (NM_005180) were as follows: F, 5' taattcccaggccgccctta 3'; R, 5' caccggctccaaaatggctc 3'.
Dual luciferase reporter assay
A dual luciferase assay was performed as described previously 28. Briefly, the dual luciferase reporter gene construct for Bmi1 (pGL-Bmi1) was created by cloning a 925-bp fragment from the promoter of Bmi1 (NM_005180, containing the C-myc binding site (gagcacgtgac)) into the KpnI and HindIII sites of the pGL3-Control firefly luciferase reporter vector (Promega, Madison, WI, USA). The corresponding mutant constructs (pGL-Bmi1mt) were created by replacing the C-myc binding site in the promoter of Bmi1 with 5'-ctcgagcactg-3'. A plasmid containing C-myc was also constructed by cloning a 1365-bp fragment of C-myc cDNA (NM_002467.4) into the BamHI and XhoI sites of pcDNA3.1. The constructs were then verified by sequencing. The cells were transiently cotransfected with pGL-Bmi1 and pcDNA3-C-myc using Lipofectamine (Life Technologies, Carlsbad, CA, USA). The luciferase activity was measured with a luminometer (Tecan, San Jose, CA, USA), and the activities were normalized based on Renilla activity and the protein concentration (cotansfected with pRL-SV40).
Cell proliferation assays
Proliferation was measured using a Cell Counting Kit-8 (CCK-8) assay. Briefly, cells were seeded in 96-well plates at a density of 2×103 cells per well. Cell proliferation was analyzed by incubating the cells with CCK-8 (Dojindo, Kumamoto, Japan) according to the manufacturer's instructions at 72 h after transfection. Absorbance (A) was measured at 450 nm, and the cell inhibition rate was calculated as (1-Atreated/Acontrol) × 100%.
Statistical analysis
The results are representative of at least three independent experiments; data are presented as the mean ± standard error. The data were analyzed using the Statistical Package for the Social Science (SPSS, Chicago, IL, USA), Version 17.0. Pearson and Spearman Correlation Coefficients were used to assess correlations among the gene expression and clinical and histopathological parameters. A one-way ANOVA and Student's t-test were used to compare differences between groups. Survival curves were plotted using the Kaplan-Meier method and compared with the log-rank test. For all statistical analyses, P < 0.05 was considered statistically significant.
Results
Bmi1 plays an important role in the development of TSCC and correlates with SOD2
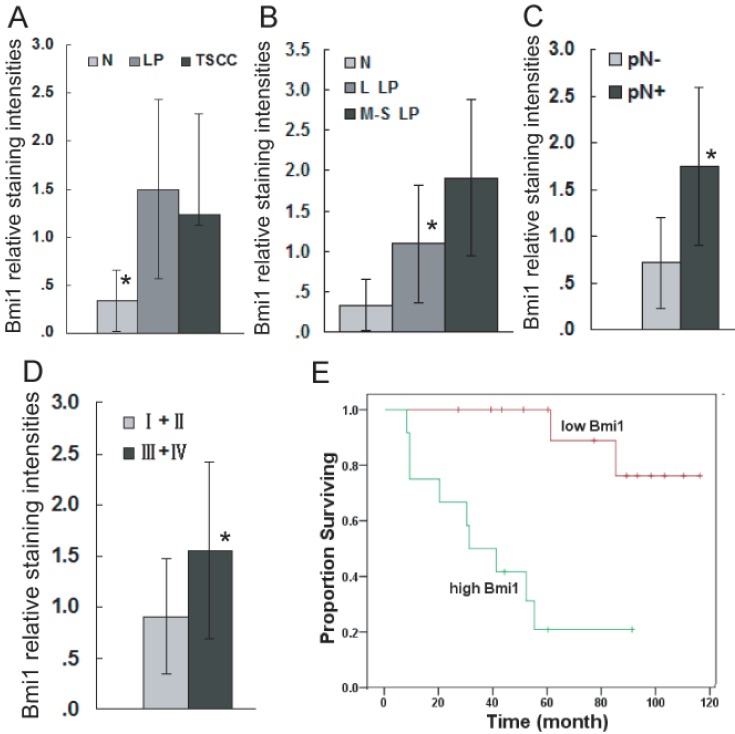
To confirm the role of Bmi1 in the development of TSCC, the expression of Bmi1 was examined by immunohistochemistry (IHC). As illustrated in Figure 1, Bmi1 IHC staining was observed in the cell nucleus. The percentage of positive cells and the staining intensity in LP and TSCC tissues were increased compared to normal tongue mucosa. Bmi1 expression was significantly higher in LP and TSCC than in normal tongue tissue (Figure 2A) and in moderate and severe LP than in light LP (Figure 2B). Among the TSCC cases, the expression level of Bmi1 was significantly higher in the patients with a positive node metastasis status (pN+) than those with a negative status (pN-) (Figure 2C) and in late-stage (stages III and IV) than in early-stage disease (stages I and II) (Figure 2D). No difference in the Bmi1 expression level was observed in TSCC cases with regard to different age, gender, pT stage, and pathological differentiation. As illustrated in Figure 2E, there was a significantly higher 5-year survival rate in the low Bmi1 expression group (mean survival = 105 months) than in the high Bmi1 expression group (mean survival = 42 months). The 5-year survival rate, as assessed by the Kaplan-Meier method, was 85.7% in the low Bmi1 expression group, whereas it was only 25% in the high Bmi1 expression group.
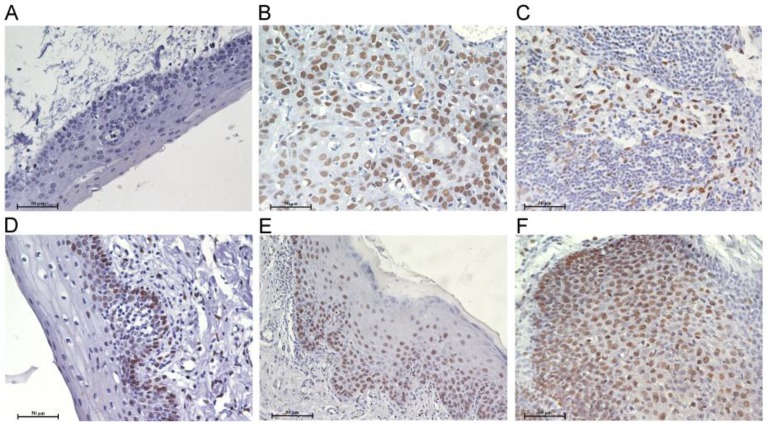
Figure 1.
Bmi1 deregulation in the development of TSCC. Immunohistochemistry analyses for Bmi1 were performed as described in Material and Methods. A: normal tongue mucosa, B: TSCC, C: lymph node metastasis tissue of TSCC, D: light LP, E: moderate LP, F: severe LP. (Scale bar: 50 μm).
Figure 2.
Bmi1 deregulation is associated with the development and poor prognosis of TSCC. Bmi1 staining intensities were presented for comparison among TSCC, LP, and normal tongue tissue (A: Bmi1 expression was significantly higher in LP and TSCC than in normal tongue tissue. No difference in the Bmi1 expression level was observed between TSCC and LP, TSCC and L LP, TSCC and M-S LP.), LP cases of different grades (B: Bmi1 expression was significant difference between normal and L LP, normal and M-S LP, L LP and M-S LP.), TSCC cases with or without lymph node metastasis (C), and TSCC cases of different clinical stages (D). Kaplan-Meier plots of overall survival in patient groups (n= 52) defined by Bmi1 immunohistochemistry was analyzed (E). The differences in survival rates were statistically significant (P <0.05). L LP: light LP, M-S LP: moderate and severe LP, High Bmi1: Bmi1 expression higher than the mean staining in TSCC (1.26), Low Bmi1: Bmi1 expressions lower than mean staining in TSCC (1.26). *P < 0.05. The actual P value for each of them was supplied in Supplementary Material: Table S3.
We further analyzed correlations among gene expression (e.g., Bmi1, SOD2, and Ki67) and clinical and pathological characteristics in the TSCC patient cohort (Table 1). The expression pattern of SOD2 and Ki67 had been published in our previous research 23. There was no significant correlation between the expression of Bmi1 and age, gender, and pT stage in the present study. However, strong positive correlations were observed between pN and the clinical stage. A significant correlation was also observed between Bmi1 expression and SOD2 expression and the Ki67 proliferation index.
Table 1.
Correlation of Bmi1 level with expression of its related genes in tumor tissues and clinical characteristics of TCSS patients.
| TSCC | Pearson | Spearman | |||
|---|---|---|---|---|---|
| r | P value | r | P value | ||
| Bmi1 expression | Age | 0.127 | 0.246 | 0.148 | 0.175 |
| Gender | -0.127 | 0.273 | -0.129 | 0.263 | |
| pT stage | -0.051 | 0.645 | -0.037 | 0.738 | |
| pN stage | 0.446 | 0.01 | 0.407 | 0.021 | |
| C stage | 0.356 | 0.004 | 0.343 | 0.006 | |
| SOD2 | 0.632 | 0.000 | 0.747 | 0.000 | |
| Ki67 | 0.453 | 0.000 | 0.468 | 0.000 | |
Bmi1-mediated TSCC migration and invasion is related to cancer stem-like cells and SOD2
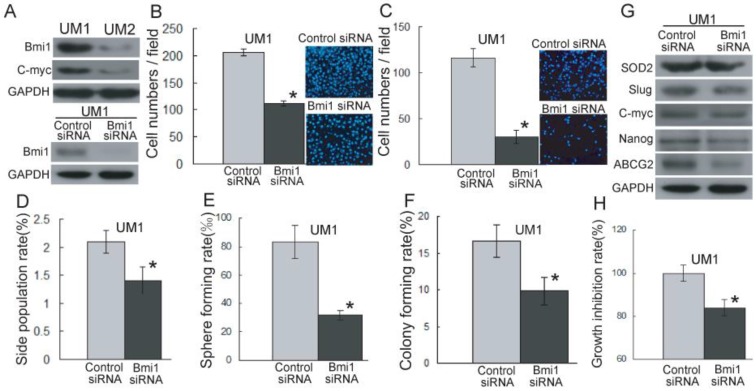
As shown in Figure 3A and S2A, Bmi1 has a higher expression level in UM1 cells than in UM2 cells. However, after the knockdown of Bmi1 in UM1 cells (Figure 3A), the migration (Figure 3B) and invasion (Figure 3C) abilities were significantly inhibited compared to control siRNA transfection.
Figure 3.
Bmi1-mediated TSCC migration and invasion are related to cancer stem cells. (A) The expression of Bmi1 and C-myc was detected by western blotting using GAPDH as loading control. (B, C) The migration (B) and invasion (C) abilities in UM1 cells after transfection with Bmi1 siRNA were significantly inhibited compared to control siRNA transfection. (D-F) Significantly higher SP subpopulation (D), sphere formation (E), and colony formation (F) were found in UM1 cells transfected control siRNA compared to UM1 cells transfected with Bmi1 siRNA. (G) The expression of SOD2, Slug, and stem cell markers (ABCG2, Nanog) was decreased in UM1 cells transfected with Bmi1 siRNA. (H) UM1 cells transfected with control siRNA showed a significantly higher proliferation rate than UM1 cells transfected with Bmi1 siRNA. *P < 0.05.
To investigate whether TSCC migration and invasion mediated by Bmi1 is related to cancer stem-like cells, we sorted and characterized SP cells (cancer stem-like cells) from TSCC cell lines. As shown in Supplementary Material: Figure S3, the proportion of SP cells, sorted by flow cytometry, was decreased in the presence of verapamil. The proportion of UM1 and UM2 SP cells was 3.2% and 2.5%, respectively. The SP cells expressed a high level of Bmi1 and migration and invasion abilities in comparison to non-SP cells (Supplementary Material: Figure S4). As shown in Figure 3D, the proportion of SP cells was significantly decreased in UM1 cells after Bmi1 knockdown, and after 14 days of culture, the UM1 cells transfected with Bmi1 siRNA displayed significantly decreased sphere formation (Figure 3E) and colony formation (Figure 3F) compared to the cells transfected with control siRNA. The expression of SOD2, Slug, and stem cell markers (ABCG2, Nanog) in UM1 cells were significantly decreased after transfection with Bmi1 siRNA (Figure 3G and S2B). Moreover, the proliferation of UM1 cells was significantly inhibited after transfection with Bmi1 siRNA compared to the cells transfected with control siRNA (Figure 3H).
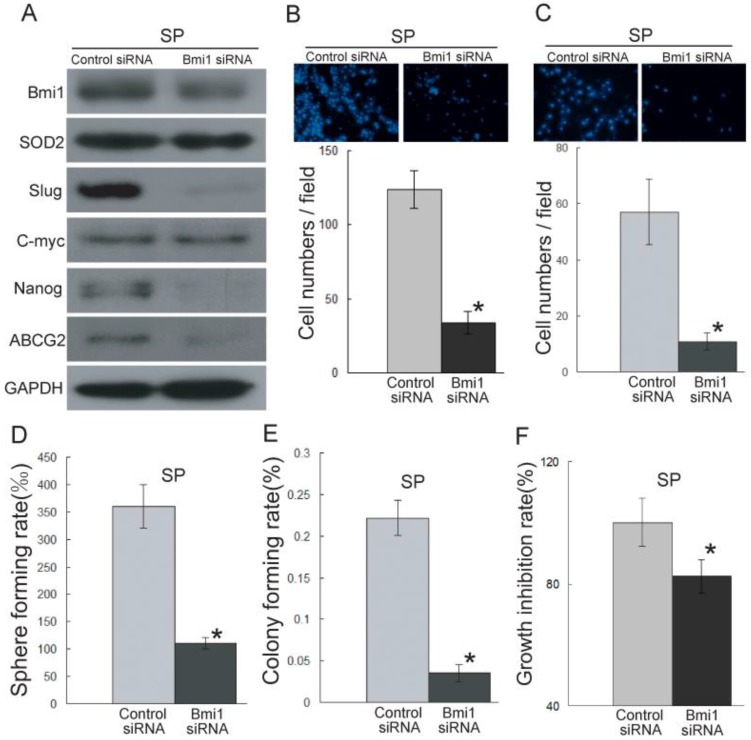
To further characterize the role of Bmi1 in the migration and invasion ability of cancer stem-like cells in TSCC, we knocked down the expression of Bmi1 (Figure 4A) in SP cells. As shown in Figure 4B-C, the SP cells transfected with Bmi1 siRNA displayed significantly decreased migration (Figure 4B) and invasion (Figure 4C) abilities compared to the control siRNA-transfected SP cells. Bmi1 knockdown resulted in significantly reduced sphere formation (Figure 4D), colony formation (Figure 4E), and proliferation (Figure 4F) in SP cells. We also found that the SP cells transfected with Bmi1 siRNA displayed significantly reduced expression levels of SOD2, Slug, and stem cell markers (ABCG2 and Nanog) (Figure 4A and S2C). Conversely, Bmi1 knockdown in UM1 and SP cells did not affect the expression of C-myc (Figure 3G, 4A, S2B and S2C). All of these data indicated that Bmi1-mediated TSCC migration and invasion is related to cancer stem-like cells. Furthermore, Bmi1 knockdown led to the down-regulation of SOD2 and Slug.
Figure 4.
Bmi1 knockdown inhibits the migration and invasion ability of cancer stem-like cells (SP cells) in TSCC. (A) The expression of Bmi1, SOD2, Slug, and stem cell markers (ABCG2, Nanog) was inhibited in SP cells transfected with Bmi1 siRNA. (B, C) The migration (B) and invasion (C) abilities were significantly inhibited in Bmi1 siRNA-transfected SP cells compared to control siRNA-transfected SP cells. (D-F) The SP cells transfected with Bmi1 siRNA had significantly decreased sphere formation (D, P=0.0005), colony formation (E, P=0.0002), and proliferation (F, P=0.001) compared to control siRNA-transfected SP cells. *P < 0.05.
C-myc directly targets the Bmi1-mediated migration and invasion of cancer stem-like cells in TSCC
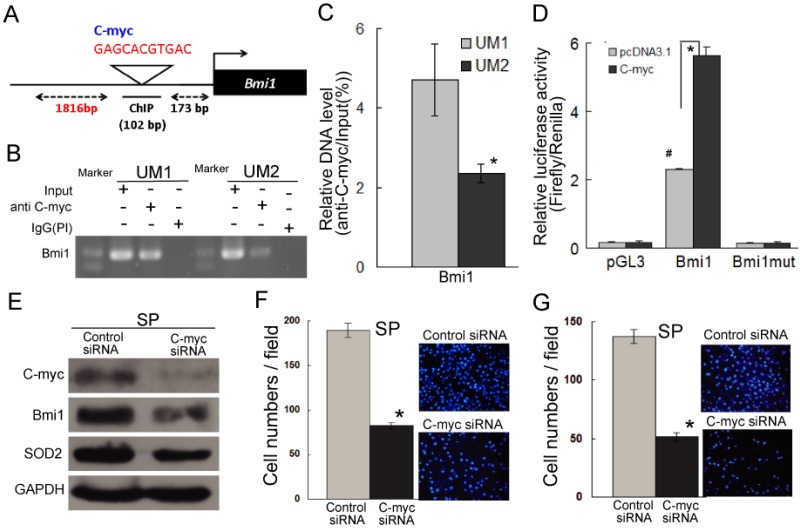
As shown in Figure 5A, a C-myc recognition site is present in the promoter of Bmi1 genes (http://genome.ucsc.edu/), suggesting that the expression of Bmi1 may be regulated by C-myc. To address whether C-myc directly binds to the promoter of Bmi1 in TSCC cells, ChIP assays were performed. As shown in Figure 5B and Figure 5C, a significantly higher level of C-myc binding to the promoter of Bmi1 was shown for the UM1 cells versus the UM2 cells. To further investigate whether Bmi1 is transcriptionally induced by C-myc, a dual luciferase assay was performed. UM2 cells expressing a C-myc expression vector, co-transfected with pGL-Bmi1, exhibited significantly increased Bmi1 promoter activity compared with UM2 cells expressing the control vector (Figure 5D). The luciferase activity in UM2 cells transfected with pGL-Bmi1mt did not respond to induction by C-myc (Figure 5D). Subsequently, we analyzed the regulation of Bmi1 by C-myc by knocking down C-myc expression in SP cells, which show a high expression of C-myc than non-SP cells (Figure S4). As expected, the expression of Bmi1 was inhibited in SP cells after transfection with C-myc siRNA (Figure 5E and S2D); the SP cells transfected with C-myc siRNA also displayed decreased migration and invasion abilities compared to the control siRNA-transfected cells (Figure 5F and 5G). Furthermore, SP cells transfected with C-myc siRNA displayed reduced SOD2 expression (Figure 5E and S2D). These results indicated C-myc directly regulates Bmi1 and mediates the migration and invasion of cancer stem-like cells in TSCC.
Figure 5.
C-myc directly targets Bmi1 and mediates the migration and invasion of SP cells in TSCC. (A) The predicted C-myc target sites in the promoter of Bmi1 (NM_005180) are presented. (B, C) Endogenous C-myc binding to the Bmi1 promoter was assessed by a CHIP assay. Rabbit IgG was used as the negative control (PI). Quantitative PCR (C) was carried out on immunoprecipitated chromatin (IP by C-myc) and input chromatin and expressed as 2Ct input - Ct IP × 100 %. (D) Bmi1 promoter activity induced by C-myc was detected by a luciferase assay. UM2 cells were transiently cotransfected with pGL-Bmi1 or pGL-Bmi1-mt with an expression construct for C-myc or an empty vector (#: compared with pcDNA3.1, P < 0.05; this may be related to the low level expression of C-myc in UM2 cells.). (E) The expression of C-myc, Bmi1, and SOD2 was inhibited in SP cells transfected with C-myc siRNA. (F, G) The migration (F) and invasion (G) abilities were significantly inhibited in C-myc siRNA-transfected SP cells compared to control siRNA-transfected SP cells. *P < 0.05.
Discussion
Bmi-1 is a stem cell marker associated with head and neck tumorigenesis. Studies have revealed that Bmi-1 expression is associated with the development of oral cancer in patients with oral leukoplakia (LP) 14, oral erythroplakia 11, and oral lichen planus 15. In clinical specimens of HNSCC, many researchers have found that the overexpression of Bmi-1 correlates with poor overall survival 13,16,31. Similarly, we also found that the expression of Bmi1 was higher in TSCC and correlated with a poor overall survival and was also associated with lymph node metastasis and clinical stage. Moreover, Bmi-1 overexpression was found in LP, with a significant difference between light LP and moderate and severe LP. Consistent results were reported in Kang`s research 32. These results indicate that Bmi-1 expression may occur at a very early stage in tongue carcinogenesis and that Bmi1 is an important factor in the development and progression of TSCC. Conversely, a statistically significant correlation was in reported by Hayry et al between lack of Bmi-1 immunoexpression and a poor prognosis in TSCC patients 17. The apparent difference may be explained by the fact that our TSCC patient cohort included all staged of TSCC, whereas that of Hayry et al only included pT1 and pT2 17.
Robust evidence suggests that Bmi1 is critical to invasive potential and contributes to the maintenance and self-renewal of CSCs in several tumor types, including HNSCC 7, 13, 33, 34. For example, Chou`s study found that Bmi1 overexpression drives stem-like properties associated with invasion, metastasis, and poor prognosis in HNSCC 13. As HNSCC includes a group of diverse cancers that develop from many different anatomic sites and is associated with different risk factors and genetic characteristics 35, it is necessary to clarify signature gene sets for tumor metastases of HNSCC originating from different anatomic sites. To date, few studies have focused on the relationship between Bmi1 and the metastasis of TSCC. In our study, we found that Bmi1 was overexpressed in UM1 cells and SP cells, both with higher migration and invasion abilities. The knockdown of Bmi1 in UM1 cells or SP cells inhibited cell proliferation and in vitro migration and invasion and decreased sphere formation, clone formation, and the expression of stem cell markers (Nanog and ABCG2, which had been found to be cancer stem cell markers of TSCC 36,37). SP cells from TSCC were found to possess the characteristics of cancer stem-like cells, possessing a higher proportion of sphere and colony formation and a higher expression of stem cell markers. These results indicate that the Bmi1-mediated migration and invasion of TSCC is related to cancer stem-like cells.
Our previous studies had found that the SOD2-dependent production of H2O2 contributes to the migration and invasion abilities of TSCC via the Snail signaling pathway 22-24. Many studies have reported that Snail promotes invasion in many types of cancers 16,28,38, including salivary adenoid cystic carcinoma 28 and TSCC 38. Yu`s study suggested that Bmi-1 plays a key role in regulating Snail expression and the cancer stemness properties of HNSCC-ALDH1(+) cells 16. However, the relationship between Bmi1 and the SOD2-Slug pathway in TSCC has not been reported. In this study, we found that the expression of Bmi1 was correlated with SOD2 in TSCC tissues. Bmi1 knockdown inhibited the expression of SOD2 and Slug in UM1 and SP cells. C-myc is a transcription factor that recognizes the E-box sequence and several related noncanonical sequences in target genes 19,39,40, and recent studies showed that Bmi1 is a direct transcriptional target of C-myc 8,21. In the present study, we also verified that C-myc binds to the E-box sequence in the promoter of Bmi1. As the knockdown of C-myc inhibited the expression of Bmi1 and SOD2 and the migration and invasion of SP cells, our results indicate that C-myc directly targets Bmi1 and that Bmi1 regulates the SOD2-Slug pathway in TSCC.
Altogether, the present findings reveal that Bmi1 plays an important role in the development of TSCC. The Bmi1-mediated migration and invasion of TSCC is related to cancer stem-like cells. C-myc directly targets and regulates the expression of Bmi1. Moreover, Bmi1 regulates the SOD2-Slug pathway and mediates the migration and invasion of TSCC.
Supplementary Material
Tables S1-S3, Figures S1 - S4.
Acknowledgments
This work was supported, in part, by grants from the National Nature Science Foundation of China (NSFC81472523, NSFC81272953), the Guangdong Natural Science Foundation (S2011020002325), the research fund for the doctoral program of Ministry of Education (20120171110050), the fundamental research funds for the Central Universities (11ykzd09), and the program for New Century Excellent Talents in University (NCET-10-0857).
Abbreviations
- Bmi1
B-cell-specific Moloney murine leukemia virus insertion site 1
- SOD2
manganese superoxide dismutase
- TSCC
tongue squamous cell carcinoma
- LP
leukoplakia
- OSCC
oral squamous cell carcinoma
- HNSCC
head and neck cancer
- RNA
Ribonucleic Acid
- CSCs
cancer stem cells
- SP
side population
- IHC
immunohistochemistry
- ChIP
Chromatin immunoprecipitation
- PCR
polymerase chain reaction.
References
- 1.Haddad RI, Shin DM. Recent advances in head and neck cancer. N Engl J Med. 2008;359:1143–54. doi: 10.1056/NEJMra0707975. [DOI] [PubMed] [Google Scholar]
- 2.Zou B, Sun S, Qi X. et al. Aldehyde dehydrogenase activity is a cancer stem cell marker of tongue squamous cell carcinoma. Mol Med Rep. 2012;5:1116–20. doi: 10.3892/mmr.2012.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rastogi P. Emergence of cancer stem cells in head and neck squamous cell carcinoma: A therapeutic insight with literature review. Dent Res J (Isfahan) 2012;9:239–44. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Ailles L, Prince M. Cancer stem cells in head and neck squamous cell carcinoma. Methods Mol Biol. 2009;568:175–93. doi: 10.1007/978-1-59745-280-9_11. [DOI] [PubMed] [Google Scholar]
- 5.Prince ME, Sivanandan R, Kaczorowski A. et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA. 2007;104:973–8. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chinn SB, Darr OA, Peters RD. et al. The role of head and neck squamous cell carcinoma cancer stem cells in tumorigenesis, metastasis, and treatment failure. Front Endocrinol (Lausanne) 2012;3:90. doi: 10.3389/fendo.2012.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Proctor E, Waghray M, Lee CJ. et al. Bmi1 enhances tumorigenicity and cancer stem cell function in pancreatic adenocarcinoma. PLoS One. 2013;8:e55820. doi: 10.1371/journal.pone.0055820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang R, Cheung NK, Vider J. et al. MYCN and MYC regulate tumor proliferation and tumorigenesis directly through BMI1 in human neuroblastomas. Faseb J. 2011;25:4138–49. doi: 10.1096/fj.11-185033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai LL, Yu CC, Chang YC. et al. Markedly increased Oct4 and Nanog expression correlates with cisplatin resistance in oral squamous cell carcinoma. J Oral Pathol Med. 2011;40:621–8. doi: 10.1111/j.1600-0714.2011.01015.x. [DOI] [PubMed] [Google Scholar]
- 10.Seo E, Basu-Roy U, Zavadil J. et al. Distinct functions of Sox2 control self-renewal and differentiation in the osteoblast lineage. Mol Cell Biol. 2011;31:4593–608. doi: 10.1128/MCB.05798-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng JQ, Xu ZY, Shi LJ. et al. Expression of cancer stem cell markers ALDH1 and Bmi1 in oral erythroplakia and the risk of oral cancer. J Oral Pathol Med. 2013;42:148–53. doi: 10.1111/j.1600-0714.2012.01191.x. [DOI] [PubMed] [Google Scholar]
- 12.Li Z, Wang Y, Yuan C. et al. Oncogenic roles of Bmi1 and its therapeutic inhibition by histone deacetylase inhibitor in tongue cancer. Lab Invest. 2014 doi: 10.1038/labinvest.2014.123. doi: 10.1038/labinvest. [DOI] [PubMed] [Google Scholar]
- 13.Chou CH, Yang NK, Liu TY. et al. Chromosome instability modulated by BMI1-AURKA signaling drives progression in head and neck cancer. Cancer Res. 2013;73:953–66. doi: 10.1158/0008-5472.CAN-12-2397. [DOI] [PubMed] [Google Scholar]
- 14.Liu W, Feng JQ, Shen XM. et al. Two stem cell markers, ATP-binding cassette, G2 subfamily (ABCG2) and BMI-1, predict the transformation of oral leukoplakia to cancer: a long-term follow-up study. Cancer. 2012;118:1693–700. doi: 10.1002/cncr.26483. [DOI] [PubMed] [Google Scholar]
- 15.Ma L, Wang H, Yao H. et al. Bmi1 expression in oral lichen planus and the risk of progression to oral squamous cell carcinoma. Ann Diagn Pathol. 2013;17:327–30. doi: 10.1016/j.anndiagpath.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Yu CC, Lo WL, Chen YW. et al. Bmi-1 Regulates Snail Expression and Promotes Metastasis Ability in Head and Neck Squamous Cancer-Derived ALDH1 Positive Cells. J Oncol. 2011 doi: 10.1155/2011/609259. doi: 10.1155/2011/609259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayry V, Makinen LK, Atula T. et al. Bmi-1 expression predicts prognosis in squamous cell carcinoma of the tongue. Br J Cancer. 2010;102:892–7. doi: 10.1038/sj.bjc.6605544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.den Hollander J, Rimpi S, Doherty JR. et al. Aurora kinases A and B are up-regulated by Myc and are essential for maintenance of the malignant state. Blood. 2010;116:1498–505. doi: 10.1182/blood-2009-11-251074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozawa N, Shichiri M, Fukai N. et al. Regulation of adrenomedullin gene transcription and degradation by the c-myc gene. Endocrinology. 2004;145:4244–50. doi: 10.1210/en.2003-0402. [DOI] [PubMed] [Google Scholar]
- 20.Bretones G, Acosta JC, Caraballo JM. et al. SKP2 oncogene is a direct MYC target gene and MYC down-regulates p27(KIP1) through SKP2 in human leukemia cells. J Biol Chem. 2011;286:9815–25. doi: 10.1074/jbc.M110.165977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang HB, Liu GH, Zhang H. et al. Sp1 and c-Myc regulate transcription of BMI1 in nasopharyngeal carcinoma. Febs J. 2013;280:2929–44. doi: 10.1111/febs.12299. [DOI] [PubMed] [Google Scholar]
- 22.Ye H, Wang A, Lee BS. et al. Proteomic based identification of manganese superoxide dismutase 2 (SOD2) as a metastasis marker for oral squamous cell carcinoma. Cancer Genomics Proteomics. 2008;5:85–94. [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Wang A, Lo Muzio L. et al. Deregulation of manganese superoxide dismutase (SOD2) expression and lymph node metastasis in tongue squamous cell carcinoma. BMC Cancer. 2010;10:365. doi: 10.1186/1471-2407-10-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z, Li S, Cai Y. et al. Manganese superoxide dismutase induces migration and invasion of tongue squamous cell carcinoma via H2O2-dependent Snail signaling. Free Radic Biol Med. 2012;53:44–50. doi: 10.1016/j.freeradbiomed.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding X, Zhang N, Cai Y. et al. Down-regulation of tumor suppressor MTUS1/ATIP is associated with enhanced proliferation, poor differentiation and poor prognosis in oral tongue squamous cell carcinoma. Mol Oncol. 2012;6:73–80. doi: 10.1016/j.molonc.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang A, Liu X, Sheng S. et al. Dysregulation of heat shock protein 27 expression in oral tongue squamous cell carcinoma. BMC Cancer. 2009;9:167. doi: 10.1186/1471-2407-9-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Gong LY, Song LB. et al. Oncoprotein Bmi-1 renders apoptotic resistance to glioma cells through activation of the IKK-nuclear factor-kappaB Pathway. Am J Pathol. 2010;176:699–709. doi: 10.2353/ajpath.2010.090502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Q, Zhou X, Li S. et al. MicroRNA-181a suppresses salivary adenoid cystic carcinoma metastasis by targeting MAPK-Snai2 pathway. Biochim Biophys Acta. 2013;1830:5258–66. doi: 10.1016/j.bbagen.2013.07.028. [DOI] [PubMed] [Google Scholar]
- 29.Yao CJ, Lai GM, Yeh CT. et al. Honokiol Eliminates Human Oral Cancer Stem-Like Cells Accompanied with Suppression of Wnt/ beta -Catenin Signaling and Apoptosis Induction. Evid Based Complement Alternat Med. 2013;2013:146136. doi: 10.1155/2013/146136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang A, Alimova IN, Luo P. et al. Loss of CAK phosphorylation of RAR{alpha} mediates transcriptional control of retinoid-induced cancer cell differentiation. Faseb J. 2010;24:833–43. doi: 10.1096/fj.09-142976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen H, Zhou L, Wan G. et al. BMI1 promotes the progression of laryngeal squamous cell carcinoma. Oral Oncol. 2011;47:472–81. doi: 10.1016/j.oraloncology.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 32.Kang MK, Kim RH, Kim SJ. et al. Elevated Bmi-1 expression is associated with dysplastic cell transformation during oral carcinogenesis and is required for cancer cell replication and survival. Br J Cancer. 2007;96:126–33. doi: 10.1038/sj.bjc.6603529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Major AG, Pitty LP, Farah CS. Cancer stem cell markers in head and neck squamous cell carcinoma. Stem Cells Int. 2013;2013:319489. doi: 10.1155/2013/319489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen YC, Chang CJ, Hsu HS. et al. Inhibition of tumorigenicity and enhancement of radiochemosensitivity in head and neck squamous cell cancer-derived ALDH1-positive cells by knockdown of Bmi-1. Oral Oncol. 2010;46:158–65. doi: 10.1016/j.oraloncology.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 35.Zhou X, Temam S, Oh M. et al. Global expression-based classification of lymph node metastasis and extracapsular spread of oral tongue squamous cell carcinoma. Neoplasia. 2006;8:925–32. doi: 10.1593/neo.06430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanamoto S, Kawasaki G, Yamada S. et al. Isolation and characterization of cancer stem-like side population cells in human oral cancer cells. Oral Oncol. 2011;47:855–60. doi: 10.1016/j.oraloncology.2011.06.501. [DOI] [PubMed] [Google Scholar]
- 37.Lim W, Choi H, Kim J, Expression of cancer stem cell marker during 4-nitroquinoline 1-oxide-induced rat tongue carcinogenesis. J Mol Histol. 2014. doi 10.1007/s10735-014-95 84-1. [DOI] [PubMed]
- 38.Wang C, Liu X, Huang H. et al. Deregulation of Snai2 is associated with metastasis and poor prognosis in tongue squamous cell carcinoma. Int J Cancer. 2012;130:2249–58. doi: 10.1002/ijc.26226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujioka Y, Taira T, Maeda Y. et al. MM-1, a c-Myc-binding protein, is a candidate for a tumor suppressor in leukemia/lymphoma and tongue cancer. J Biol Chem. 2001;276:45137–44. doi: 10.1074/jbc.M106127200. [DOI] [PubMed] [Google Scholar]
- 40.Guney I, Wu S, Sedivy JM. Reduced c-Myc signaling triggers telomere-independent senescence by regulating Bmi-1 and p16(INK4a) Proc Natl Acad Sci USA. 2006;103:3645–50. doi: 10.1073/pnas.0600069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1-S3, Figures S1 - S4.