Abstract
Low-dose metronomic (LDM) paclitaxel therapy displayed a stronger anti-angiogenic activity on breast tumors with fewer side effects. Upregulation of anti-angiogenic factor Thrombospondin-1 (TSP-1) accords for therapeutic potency of LDM paclitaxel, but its molecular mechanism has not been elucidated yet. microRNAs (miRNAs) have emerged as new important regulators of tumor growth and metastasis. Here, we hypothesize that miRNAs are involved in TSP-1 overexpression in paclitaxel LDM therapy of breast tumors. The miRNA profile of tumor tissues from control, LDM and MTD groups in 4T1 mouse breast cancer model was detected by microarray, and then verified by quantitative real-time PCR (qRT-PCR). Luciferase assay and western blot were employed to explore the mechanisms of miRNAs involved in this process. We found that let-7f, let-7a, miR-19b and miR-340-5p were reduced by >2 fold, and miR-543* and miR-684 were upregulated by at least 50% in paclitaxel LDM therapy. qRT-PCR verification revealed that let-7f level was reduced most significantly in LDM therapy. Computational prediction using TargetScan and miRanda suggested THBS1 which encodes TSP-1 as a potential target for let-7f. Luciferase activity assay further confirmed that let-7f may bind to 3'UTR of THBS1 gene and inhibit its activity. Moreover, forced expression of let-7f led to a decrease of TSP-1 at both mRNA and protein levels in MCF-7 cells. Contrastly, let-7f inhibition induced an increased expression of THBS1 mRNA and TSP-1 protein, but did not affect the proliferation and apoptosis of MCF-7 cells. Paclitaxel LDM therapy led to a decrease of let-7f and the elevation of TSP-1 protein expression in MCF-7 cells, while overexpression of let-7f may abolish LDM-induced the upregulation of TSP-1 in MCF-7 cells. In summary, let-7f inhibition contributed to the upregulation of TSP-1 in paclitaxel LDM therapy, independently of proliferation, cell cycle arrest and apoptosis of breast cancer. This study indicates let-7f as a potential therapeutic target for breast tumor.
Keywords: let-7f, Thrombospondin-1, Low-dose metronomic paclitaxel therapy.
Introduction
Paclitaxel is a member of the taxane class of chemotherapeutics and has been indicated as a first-line therapy for many malignancies including breast 1, lung 2, ovarian 3, as well as head and neck cancers 4. Even though paclitaxel is commonly and broadly used to prevent and treat cancers, some adverse effects such as severe anaphylactic hypersensitivity reactions 5 and peripheral neuropathy 6 have limited clinical application of paclitaxel in patients. Compared with maximum tolerable dose (MTD) chemotherapy, low-dose metronomic (LDM) chemotherapy represents a new therapeutic strategy for solid tumors 7, especially in combination with other anti-angiogenic agents. Conventional MTD therapy produced antitumor activities through the inhibition of cell proliferation and the induction of cell apoptosis, with unavoidable damages on normal tissues 7. Whereas, LDM therapy inhibited the progress and metastasis of cancers through its stronger anti-angiogenic and anti-lymphangiogenic activities 8 rather than inducing tumor cell apoptosis 9. Consistently, our previous studies revealed that paclitaxel LDM therapy displayed a higher therapeutic efficacy in suppressing growth and metastasis of breast tumors than paclitaxel MTD therapy by inhibiting tumor angiogenesis and lymphangiogenesis with less toxic effects 8, 10. It was explained by that paclitaxel LDM therapy caused the upregulation of TSP-1 protein and the downregulation of vascular endothelial growth factor receptor-2 (VEGFR2) 11. However, the underlying mechanisms of TSP-1 overexpressed in paclitaxel LDM therapy remain obscure.
microRNAs (miRNAs), as a category of endogenous non-coding small RNAs, have emerged as one of protagonist in gene expression regulation, and participate in many essential biological processes 12. miRNAs have displayed the activity in regulating the proliferation, apoptosis and migration of tumor cells 13. Some miRNAs were also shown to mediate angiogenesis or anti-angiogenesis of breast tumor in previous studies 14-15. Abnormal expression of miRNAs has been recognized as novel potential biomarkers or targets in the early diagnosis and treatment of tumors 16. However, little is known about whether abnormal expression of miRNAs is involved in paclitaxel LDM therapy.
The purpose of this study was to examine the changes of microRNA let-7f expression in paclitaxel LDM therapy and if this change contributes to TSP-1 overexpression and anti-angiogeneic activity.
Material and Methods
Reagents and antibodies
Paclitaxel was purchased from Hoffmann-La Roche Ltd., Basel, Switzerland. Polyoxyethylene series of castor oil (CrEL) was purchased from BASF Co., Ltd., Shanghai, China. The monoclonal antibodies against TSP-1 (sc-73158), and GAPDH (sc-32233) were purchased from Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA. CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS) was purchased from Promega, USA, and sterile dimethyl sulfoxide (DMSO) was purchased from ATCC, Rockville, USA.
Animal
6-8 weeks old BALB/c female mice were obtained from the Experimental Animal Center of the First Affiliated Hospital of Harbin Medical University, China. Animals were maintained in an air-conditioned room and on 12 hours dark-light artificial cycle, with food and water ad libitum. Animal model and treatment were prepared as described by Jiang 11. Briefly, Tumors were established by subcutaneous injection of 4 × 104 4T1 cells suspended in 50 μL of phosphate-buffered saline (PBS) into the left inguinal mammary fat pads of mice. Tumor volumes were estimated according to the following formula: π6 ×a2 × b (a is the short axis, and b is the long axis). When tumors reached around 100 mm3, the mice were randomly assigned to three groups (10 mice in each group): control (PBS, 200 μL/day, i.p.), paclitaxel LDM chemotherapy (1.3 mg/kg/day, i.p.) 17, and paclitaxel MTD chemotherapy (20 mg/kg/week, i.p.) 18. Forty-two days after tumor implantation, the mice were euthanized.
Cell culture
Human breast adenocarcinoma cell line Michigan Cancer Foundation-7 (MCF-7) was cultivated in Dulbecco's Modified Eagle's Medium-High Glucose (Hyclone, Cat. SH30022.01B) with 4 mM L-glutamine, 10% FBS (Hyclone, Cat. SH30087.01), and 100 U/mL penicillin/streptomycin (Hyclone, Cat. SH30010). Cells were grown in a humidified atmosphere of 5% CO2 in air at 37oC and passaged with 0.05% trysin-EDTA every 3-4 days. MCF-7 cells were transfected with 50 nM let-7f, AMO-let-7f or NC miRNAs (Guangzhou RiboBio Co., Ltd. Guangzhou, China) with Lipofectamine™ RNAiMAX (Life Technologies, Grand Island, NY, USA). 48 h after transfection, cells were collected for further total RNA or protein extraction.
Determination of apoptosis/necrosis by flow cytometry
MCF-7 cells were treated with NC, let-7f and AMO-let-7f for 24 h. After treatment, cells were harvested, washed with PBS, and resuspended in 500 μL of binding buffer containing 1.25 μL Annexin V-FITC and incubated in dark at room temperature for 15 mins. Centrifuged at 1000g for 5 mins and added 10 μL propidium lodide according to the instructions of the manufacture (Nanjing KeyGen Biotech. Co., LTD. China). After incubation, samples were immediately analyzed by flow cytometry (FACS Calibur, BD Biosciences, San Joes, CA, USA) using a 488 nm laser.
Cell proliferation assay
MCF-7 cells (1×105 cells/100 μL/well) were seeded in a sterile flat bottom 96-well plate and stabilized by incubation for 24 h at 37oC in an incubator containing 5% CO2. 100 μL PBS or 50 nM of each treatment (control, NC, let-7f, AMO-let-7f) was added to each well. Blank wells contained the media with no cells. After 24, 48 and 72 h of test compound incubation, 10 μL of the MTS reagent in combination with the electron coupling agent phenazine methosulfate was added to the wells and cells were incubated in CO2 incubator for 4 h. Absorbance at 490 nm (OD490) was monitored with a plate reader (multiscan MK3, Thermo Fisher Scientific, USA) to obtain the number of viable cells relative to the control population. Percentage of viability in the test compound treated cells is expressed as percentage compared to control. Data was obtained from three different experiments (n=6 per plate per time point).
Cell cycle analysis
MCF-7 cells (1 × 106 cells) in a six well plates were incubated (37oC, 5% CO2) with NC, let-7f, AMO-let-7f media for 48 h. Following trypsinization, cells were washed and centrifuged at 2000 ×g for 10 min and the pellet was resuspended in 1 mL PBS. Fixation was completed by adding 70% cold ethanol at 4oC overnight. The fixed cells were washed with PBS and resuspended in 0.5 mL PBS with propidium iodide (0.05 mg/mL), DNAase free RNAse A (100 μg/mL) and 0.2% Triton X-100. Cells were incubated at 4oC in dark for 30 mins. The cells were analyzed for cell cycle using flow cytometry (FACS Calibur, BD Biosciences, San Joes, CA, USA) with an excitation wave length of 488 nm and emission at 670 nm. DNA content was determined by ModFit software (Verity Software House, Topsham, ME), which provided histograms to evaluate cell cycle distribution.
RNA extraction
Total RNA was isolated using TRIzol (Life Technologies, Carlsbad, CA, USA) and miRNeasy mini kit (QIAGEN, Valencia, CA, USA) according to manufacturer's instructions, which efficiently recovered all RNA species, including miRNAs. RNA quality and quantity were measured using NanoDrop spectrophotometer (ND-1000, NanoDrop Technologies) and RNA integrity was determined by gel electrophoresis.
miRNA microarray
Total RNA of four tumor tissue samples from mouse model in each group was randomly selected, and microarray analysis was completed with the 6th generation of miRCURYTM LNA Array (v.16.0) (Exiqon, Inc. Woburn, MA, USA), which contains more than 1891 capture probes, covering all human, mouse and rat microRNAs annotated in miRBase 16.0.
RNA labeling, array hybridization and data analysis
After RNA isolation from the samples, the miRCURY™ Hy3™/Hy5™ Power labeling kit (Exiqon, Vedbaek, Denmark) was used according to the manufacturer's guideline for miRNA labeling. Each sample was labeled with Hy3™ fluorescent at 3'-end. Then, the Hy3™-labeled samples were hybridized on the miRCURY™ LNA Array (v.16.0) according to array manual. Following hybridization, the slides were achieved, washed several times using miRCURY LNA™ Array, washing buffer kit (Exiqon, Inc. Woburn, MA, USA), and finally dried by centrifugation for 5 min at 400 rpm. Then the slides were scanned using the Axon GenePix 4000B microarray scanner (Axon Instruments, Foster City, CA). Scanned images were imported into GenePix Pro 6.0 software (Axon) for grid alignment and data extraction. Replicated miRNAs were averaged and miRNAs that intensities >50 in all samples were chosen for calculating normalization factor. Expressed data were normalized using the median normalization. After normalization, differentially expressed miRNAs were identified through Fold Change filtering. Hierarchical clustering was performed using MEV software (version 4.6, TIGR).
ELISA assay
The level of TSP-1 protein in the supernatant of MCF-7 cells was measured by ELISA kit (IBL-America) according to the supplier's instructions.
Tube formation assay
Matrigel (BD Biosciences) were thawed at 4°C for overnight, and each well of prechilled 48-well plates was coated with 150 μL Matrigel and incubated at 37°C for 2 h. The cultured HUVECs (2× 104 cells) were resuspended with condition medium from each group, and then added in the plates. After incubation at 37°C for 6 h, tube formation of HUVECs was assessed with OLYMPUS inverted microscope. Tube structure was quantified by the counting of low power fields, and inhibition percentage was presented using untreated wells as 100%.
Western blot
Tumor tissues were homogenized in lysis buffer (20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton and 1× complete protease inhibitor cocktail), and spined extract 10 mins at 14,000g in 4oC centrifuge. Cells were lysed in lysis buffer, and sonicated briefly, centrifuged extract 10 mins at 14,000g in 4oC centrifuge. Total protein samples (30 μg-100 μg) were analyzed by 8% SDS-PAGE gel and transferred to polyvinylidene difluoride (PVDF) membranes by a wet blotting procedure (100 V, 120 mins, 4oC). After blocked with 5% blocking buffer, the membranes incubated with primary antibodies at 4oC overnight using the following concentration: TSP-1 (1:1200), GAPDH (1:3000), followed by alkaline phosphatase conjugated secondary antibody. Visualization of bound antibody was achieved with film (Kodak, Rochester, NY USA) by using 5-bromo-4-chloro-3-indolyl phosphate (BCIP)/nitro blue tetrazolium (NBT) (Beyotime Institute of Biotechnology, Jiangsu, China) at several time points (2 mins, 10 mins and 2 h). We chose the optimal conditions of TSP-1/GAPDH signal image respectively and presented here. GAPDH acted as the internal control for normalization of gene expression data.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from tumor tissue and MCF-7 cells using TRIzol (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions. Each sample for real-time PCR consisted of one microgram of total RNA. cDNA was assessed with SYBR Green PCR Master Mix (TOYOBO) by ABI PRISM® 7500 Sequence Detection System (Applied Biosystems). The PCR amplification program was 95oC 5 min, (95oC 15 sec, 65oC 15 sec, 72oC 32 sec, 40 cycles) and followed by dissociation curve protocol (95oC 15 sec, 60oC 1 min, 95oC 15 sec, and 60oC 15 sec). The U6 and 18S were used as internal standards for miRNA and mRNA respectively. The primer pairs are listed in table 1. We determined the appropriate cycle threshold (Ct) using the automatic baseline determination feature and analyzed data by relative quantitative analysis method. The mean Ct values (± SEM) of three independent experiments are presented.
Table 1.
Primers for qRT-PCR.
| Primer | Primer Sequence (5'-3') | |
|---|---|---|
| mmu-let-7a | RT | CTCAACTGGTGTCGTGGAGTCGGCAAGAGTCGGCAATTCAGTTGAGAACTATACAAC |
| Forward | ACACTCCAGCTGGGTGAGGTAGTAGGTTGTATA | |
| mmu-let-7f | RT | CTCAACTGGTGTCGTGGAGTCGGCAAGAGTCGGCAATTCAGTTGAGAACTATACAAT |
| Forward | ACACTCCAGCTGGGTGAGGTAGTAGATTG | |
| mmu-miR-19b | RT | CTCAACTGGTGTCGTGGAGTCGGCAAGAGTCGGCAATTCAGTTGAGTCAGTTTTGCATG |
| Forward | ACACTCCAGCTGGGTGTGCAAATCCATGCAAAAC | |
| Mmu-miR-340-5p | RT | CTCAACTGGTGTCGTGGAGTCGGCAAGAGTCGGCAATTCAGTTGAGAATCAGTCTCAT |
| Forward | ACACTCCAGCTGGGTTATAAAGCAATGAGACT | |
| mmu-miR-543* | RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCGAAAAAC |
| Forward | ACACTCCAGCTGGGAAGTTGCCCGCGTGTTTTT | |
| mmu-miR-684 | RT | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTTGACTT |
| Forward | ACACTCCAGCTGGGAGTTTTCCCTTCAAGT | |
| miRNA | Reverse | CTCAACTGGTGTCGTGGA |
| U6 | RT | AACGCTTCACGAATTTGCGT |
| Forward | CTCGCTTCGGCAGCACA | |
| Reverse | AACGCTTCACGAATTTGCGT | |
| Thbs1 | Forward | CAAGGGCTCAGGGATACTCA |
| Reverse | TGAGACGCCATCTGTATGCA | |
| 18S | Forward | CCTGGATACCGCAGCTAGGA |
| Reverse | GCGGCGCAATACGAATGCCCC |
Note: miRNA Reverse as a common reverse primer used for all miRNAs detected by qRT-PCR in our study, which is matched with all miRNAs qRT-PCR forward primers.
Luciferase assays
We synthesized fragments of THBS1 3'UTR containing the exact target site for let-7f or the mutated target site using the Taq PCR amplification. The primer pairs are listed in table 2. We insert the THBS1 3'UTR fragments into the multiple cloning sites (SgfI and XhoI) of the psiCHECKTM-2 luciferase miRNA expression reporter vector (Promega, Madison, WI, USA). The HEK293T cells (2×104 pre well) were transfected with let-7f (50 nM) or AMO-let-7f (50 nM) and 0.5 μg plasmid by Lipofectamine™ RNAiMAX, according to the manufacturer's instructions. Luciferase activities were measured 48 h after transfection with a Dual-Luciferase Reporter Assay System (E1910, Promega) on a luminometer (GloMax, Promega).
Table 2.
Primers for THBS1 3'UTR-luciferase reporter clone.
| Primer Sequence (5'-3') | ||
|---|---|---|
| THBS1 | Forward | (SgfI) gaatgcgatcgcTCATCAGCTGCCAATCATAAC |
| Reverse | ( XhoI) ccgctcgagTATAAATATAAACTTCCATATGAT | |
| mutTHBS1 | Forward | GTGGAACTTCTATACAAATATATGGAGTTTTGTTGTGTGACTGAGTAA |
| Reverse | TTACTCAGTCACACAACAAAACTCCATATATTTGTATAGAAGTTCCAC |
Note: SgfI and XhoI are name of restriction enzyme.
Statistical analysis
Student's t-test was used to compare the difference between two groups. Statistical comparisons among multiple groups were performed by Tukey's post hoc test and One-Way ANOVA. All data are presented as Mean ± SEM. All of the figures were plotted using GraphPad Prism (GraphPad Software Inc., San Diego, USA). P<0.05 was considered as significantly different.
Results
let-7f was reduced by paclitaxel LDM therapy in 4T1 breast cancer
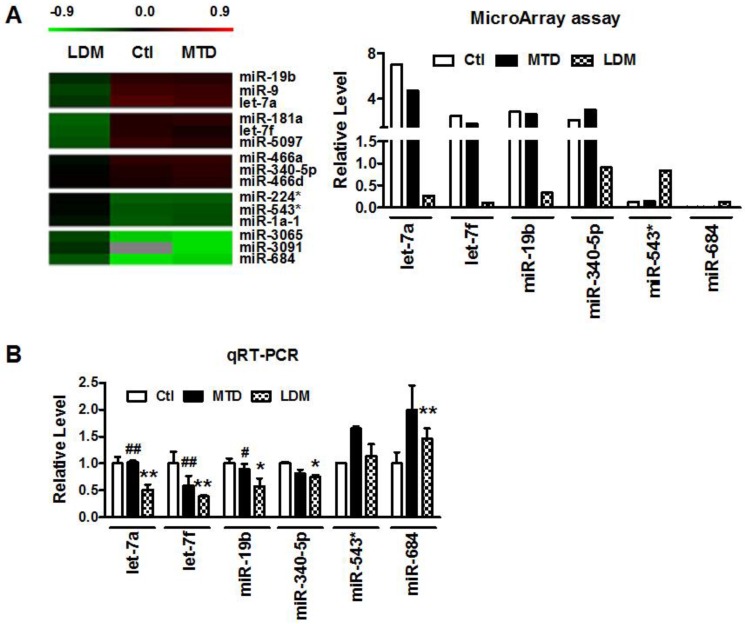
Our previous studies demonstrated that low-dose metronomic (LDM) paclitaxel therapy displayed a stronger anti-angiogenic activity in breast tumor via upregulating TSP-1 expression. Here, we further studied whether miRNAs were involved in the process of LDM paclitaxel therapy of breast tumor. Firstly, we established a xenograft breast cancer model by subcutaneous injection of 4T1 tumor cells into BALB/c female mice. When tumor size reached around 100 mm3, the mice were treated daily with PBS (control group), low-dose metronomic paclitaxel (LDM group) and maximum tolerable dose paclitaxel (MTD group), respectively. Then microarray assay was used to screen out differential expressed microRNAs among three groups. As illustrated in Figure 1A, let-7f, let-7a, miR-19b and miR-340-5p were found to be downregulated by >2 fold, whereas miR-543* and miR-684 were upregulated by at least 50% in LDM group compare with control. Furthermore, qRT-PCR verification showed that let-7f, let-7a, miR-19b and miR-340-5p were decreased, and miR-684 was increased in LDM group (Figure 1B). Interestingly, paclitaxel LDM therapy induced a more significant reduction of let-7f and let-7a than control. Particularly, let-7f demonstrated the most pronounced change in paclitaxel LDM therapy. It suggests that let-7f is involved in pacliaxel LDM therapy of breast cancer.
Figure 1.
Paclitaxel LDM therapy reduced let-7f expression in 4T1 breast tumors. A, The heat map of hierarchical clustering showed differential expression profiling of miRNAs from control, paclitaxel LDM therapy and MTD therapy groups. Red represents upregulated miRNAs, and green represents downregulated miRNAs (left). Altered expression profile of miRNAs in 4T1 breast tumors in three groups was analyzed by miRNA microarray (right). B, Quantitative real-time RT-PCR (qRT-PCR) verification of the miRNA expression profile in mouse models. ** P<0.01 vs control, ## P<0.01 vs MTD group. n=6 independent samples for each group.
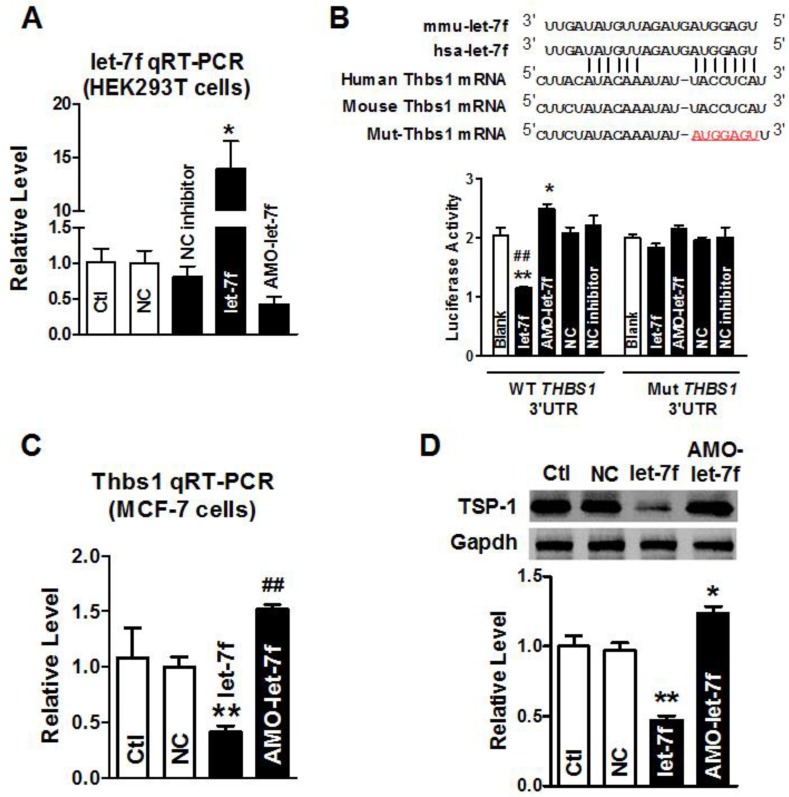
let-7f targeted at THBS1 gene via binding to its 3'UTR
Our previous study has shown that paclitaxel LDM therapy upregulated TSP-1 expression in breast cancer, which conferred antiangiogenesis activity of LDM. Computational prediction using TargetScan and miRnada predicted THBS1 gene which encodes thrombospondin-1 (TSP-1) in tumor cells, as a potential target for let-7f. Then, we employed the dual-luciferase activity assay to determine whether let-7f regulates TSP-1 expression via binding to THBS1 3'UTR. We engineered two versions of the full-length THBS1 3'UTR sensor: one containing the WT let-7f seed homology sequence, and the other carrying a 7-nt substitution of let-7f seed site (Figure 2B). It was verified that let-7f was successfully transfected into HEK293T cells by qRT-PCR (Figure 2A) before luciferase reporter activity assay was carried out. As illustrated in Figure 2B, transfection of let-7f (50 nM) caused a substantial reduction of luciferase activity of constructs carrying fragment of WT THBS1 3'UTR to 55% (P<0.01) compared with NC group (Figure 2B). In contrast, AMO-let-7f (50 nM) enhanced the luciferase activity in constructs of WT THBS1 3'UTR. But, transfection of let-7f or AMO-let-7f did not affect the activity of luciferase reporter with seed sequence mutation.
Figure 2.
THBS1 as a target gene of let-7f. A, qRT-PCR confirmed transfection efficiency of let-7f and AMO-let-7f in HEK293T cells. After transfected for 48 h, let-7f expression was increased, and AMO-let-7f inhibited let-7f expression to 42% of NC group. * P<0.05 vs NC, n=6 independent batches cells. B, Luciferase assay in HEK293T cells for post-transcriptional inhibition of THBS1 (gene encoding TSP-1 protein). WT THBS1 3'UTR luciferase reporter vector was in structure with full length of THBS1 3'UTR including let-7f binding stie; Mut THBS1 3'UTR luciferase reporter vector was insert with fragments of mutant 7 nucleotide of let-7f binding site in full length THBS1 mRNA 3'UTR (upper). HEK293T cells were transfected with miR-let-7f, AMO-let-7f or a scrambled miRNA (NC) and NC inhibitor as negative control by Lipofectamine™ RNAiMAX. Blank group was treated with Lipofectamine™ RNAiMAX alone. * P<0.05 vs NC, ** P<0.01 vs NC, ## P<0.01 vs let-7f alone; n=6 independent batches of cells for each group. C, Let-7f and AMO-let-7f regulated THBS1 mRNA expression in MCF-7 cells. After transfected with let-7f or AMO-let-7f in MCF-7 cells for 48 hours. THBS1 mRNA expression was significantly decreased to 41% of NC group. Consistently, AMO-let-7f increased THBS1 mRNA expression as well. ** P<0.01 vs NC group, ## P<0.01 vs let-7f alone, n=6 independent batches cells in each group. D, The effect of let-7f or AMO-let-7f on TSP-1 protein expression in MCF-7 cells. Downregulation of TSP-1 protein was observed in let-7f group, and AMO-let-7f increased the expression level of TSP-1 in MCF-7 cells. ** P<0.01 vs NC, ## P<0.01 vs let-7f alone, n=6 independent batches of cells for each group.
Western blot and qRT-PCR were further used to examine the role of let-7f in regulating the expression of THBS1 gene, and the result showed that the level of THBS1 mRNA was remarkably decreased in MCF-7 cells transfected with let-7f (Figure 2C). In contrast, AMO-let-7f induced an increase of THBS1 mRNA expression in MCF-7 cells. Figure 2D showed that AMO-let-7f had the ability to increase the expression of TSP-1 protein in MCF-7 cells. The above data suggests that let-7f targets at THBS1 gene, and let-7f reduction leads to the upregulation of TSP-1 protein in breast cancer.
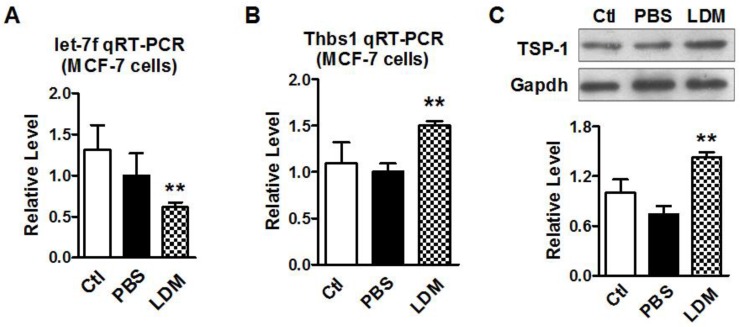
Paclitaxel LDM therapy reduced let-7f and upregulated TSP-1 expression
We then verified if paclitaxel LDM therapy led to a reduction of let-7f and increase of TSP-1 expression in MCF-7 cells. MCF-7 cells were divided into three groups: control group (blank MCF-7 cells), PBS group (treat with equal volume of PBS), and LDM group (paclitaxel 3.6 μg/ml). We firstly tested if let-7f was markedly reduced by paclitaxel LDM therapy in MCF-7 cells. The treatment with pacliaxel LDM therapy for 48 h led to about 40% decrease of let-7f expression in MCF-7 cells (Figure 3A). Figure 3B showed that THBS1 mRNA level was increased by 1.5 folds in LDM group (P<0.05) compared with PBS group. TSP-1 protein level was increased by about 2 folds as well after LDM treatment (Figure 3C, P<0.05), whereas MCF-7 Cells treated with PBS showed no significant difference from cells in control group. These results support that paclitaxel LDM therapy upregulates TSP-1 expression via reducing let-7f level in breast cancer.
Figure 3.
Paclitaxel LDM treatment inhibited let-7 expression and upregulated TSP-1 expression in MCF-7 cells. A, qRT-PCR verification of let-7f expression in MCF-7 cells treated with pacliaxel LDM therapy. ** P<0.01 vs PBS group. n=6 independent batches. B, qRT-PCR analysis of THBS1 mRNA expression in MCF-7 cells of control, PBS and paclitaxel LDM treated groups. LDM paclitaxel treatment increased mRNA level of THBS1. ** P<0.01 vs PBS groups, n=6 samples for each group. C, Western blot detected TSP-1 protein expression in MCF-7 cells of control, PBS and paclitaxel LDM groups. TSP-1 protein was significantly increased in paclitaxel LDM therapy compared with PBS treated group. ** P<0.01 vs PBS, n=6 samples for each group.
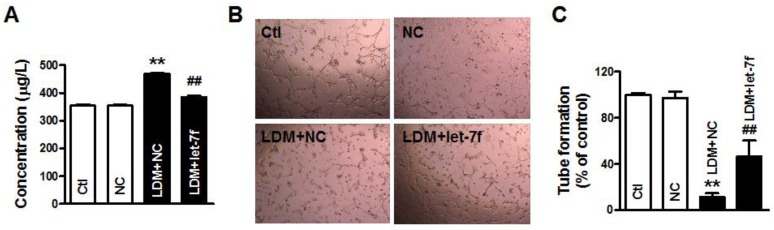
Overexpression of let-7f antagonized LDM-induced the upregulation of TSP-1
To further confirm that let-7f plays an important role in LDM therapy, we studied the effects of let-7f overexpression on LDM-induced the upregulation of TSP-1. As shown in Fig. 4A, we found that the level of TSP-1 protein was significant increased in the supernatant of MCF-7 cells after LDM treatment, but let-7f overexpression in MCF-7 cells can inhibit the augment of TSP-1 level in the supernatant induced by LDM treatment. It indicates that let-7f is directly involved in TSP-1 expression increase by LDM. Furthermore, we collected the conditioned medium of each group and studied the influence of conditioned medium on tube formation. We found that overexpression of let-7f attenuated the inhibition of angiogenesis by LDM (Fig. 4B-4C). It means that let-7f plays an essential role in LDM-induced inhibition of angiogenesis.
Figure 4.
let-7f overexpression attenuated LDM-induced inhibition of angiogenesis. A, The TSP-1 level in the culture medium was measured by ELISA assay. Let-7f overexpression inhibited the increase of TSP-1 in the medium after treated by LDM. ** P<0.05 vs NC groups, n=6 batches cells for each group. B and C, LDM treatment suppressed the tube formation and resulted in the inhibition of angiogenesis, but let-7f overexpression abolished LDM-induced the inhibition of angiogenesis. ** P<0.05 vs NC groups, n=6 batches cells for each group.
Decrease of let-7f did not affect the proliferation of MCF-7 cells
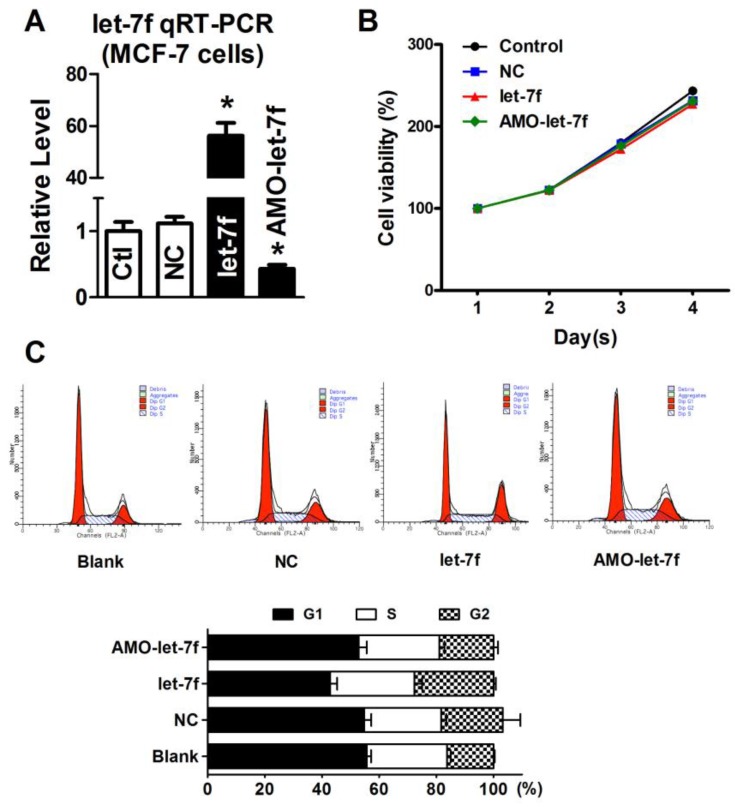
Our previous study has reported that paclitaxel LDM therapy inhibited the angiogenesis by upregulating TSP-1, but had no effects on cellular proliferation and apoptosis. Therefore, it is reasonable to speculate that the decrease of let-7f did not affect the proliferation and apoptosis of breast cancer cell. So, we transfected let-7f and AMO-let-7f into MCF-7 cells to determine if let-7f affects the proliferation of breast cancer cells. qRT-PCR was used to detect the transfection efficiency of let-7f and its inhibitor. As shown in Figure 5A, the expression of let-7f was increased by 56 folds than NC miRNA transfected cells, and AMO-let-7f inhibited 57% endogenous let-7f in MCF-7 cells. Then, MTT assay was performed to assess the proliferation of MCF-7 cells. As shown in Figure 5B, MCF-7 cells showed the similar increase speed in the viability after treated with NC miRNA, let-7f or AMO-let-7f for 1day, 2 days, 3 days, and 4 days, indicating that let-7f inhibition did not affect the proliferation of MCF-7 tumor cells in vitro study. Furthermore, the effect of let-7f on cell cycle distribution was examined by flow cytometry. At 48 h of treatment with let-7f or AMO-let-7f (50 nM), there were no significant changes in S phase, G0/G1 phase and G2/M phase (p>0.05) (Figure 5C). The data indicates that let-7f inhibition does not affect the proliferation and cell cycle of tumor cells directly.
Figure 5.
let-7f reduction did not affect the proliferation of MCF-7 cells. A, qRT-PCR analysis of let-7f expression in MCF-7 cells treated with let-7f, AMO-let-7f or with NC miRNA. After transfected let-7f into MCF-7 cells for 48 hours, let-7f expression was increased by over 56 folds compared with NC group. AMO-let-7f transfection inhibited let-7f expression by 62% compared with NC group. * P<0.05, n=6 batches cells for each group. B, MCF-7 cells were treated with let-7f, AMO-let-7f or NC miRNA for 0, 24, 48 and 72 hours, and MTT assay was then performed to assess the proliferation of MCF-7 cells. MCF-7 cells showed the similar increase speed in the viability after treated with each group for 1day, 2 days, 3 days, and 4 days. C, Cell cycle analysis of MCF-7 cells after let-7f, AMO-let-7f or NC miRNA treatment by flow cytometry. After exposure of MCF-7 cells to let-7f, AMO-let-7f or NC miRNA for 48 hours, cells were analyzed by flow cytometry. There were no significantly changes in S phase, G0/G1 phase and G2/M phase (P>0.05). Data are expressed as mean ± SEM, Flow cytometric histograms are representative of three separate experiments.
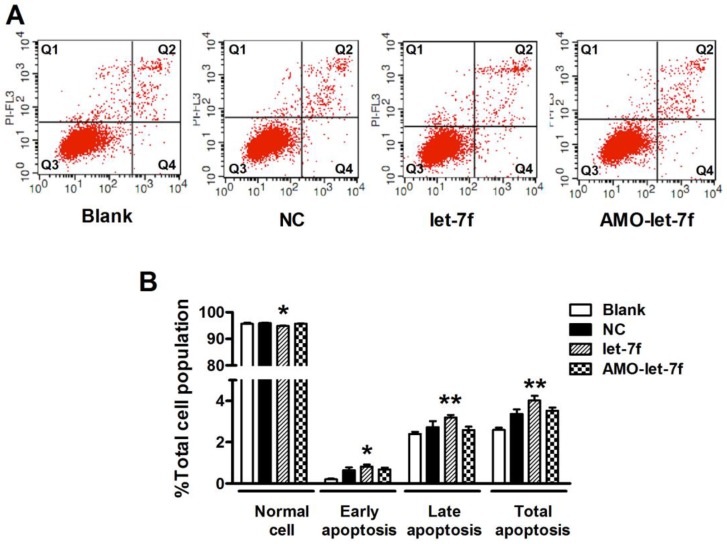
let-7f inhibition did not affect the apoptosis of MCF-7 cells
Then the effects of let-7f and AMO-let-7f on apoptosis were evaluated by using Annexin V/PI staining. As shown in Figure 6A, there was no significant change in the number of living cells after treated with AMO-let-7f for 48 h, but the percentages of apoptotic cells were significantly increased by let-7f in both early apoptosis stage (up to 0.82±0.1%, P<0.05) and late apoptosis/necrotic cells (up to 3.2±0.12%, P<0.01). Figure 6B showed that let-7f induced apoptosis of MCF-7 cells, while AMO-let-7f did not produce any effects on apoptosis of MCF-7 cells compared with NC miRNA treatment. This result is in consistent with the paclitaxel LDM therapy has no effects on tumor cells in our in vivo study 8, 10.
Figure 6.
let-7f did not induce apoptosis of MCF-7 cells. A, The effects of let-7f and AMO-let-7f on apoptosis were evaluated by Annexin V/PI staining using flow cytometry. MCF-7 tumor cells were treated with let-7f, AMO-let-7f or NC siRNA for 24 hours. Lower right quadrant (Q4) Annexin V positive/PI negative cells denote early apoptosis and upper right quadrant (Q2) Annexin V positive/PI positive cells denote late apoptosis or necrosis cells. Images are the representative of three independent experiments. B, Statistic analysis of the results of apoptosis assay. let-7f induced apoptosis of MCF-7 cells including early apoptosis (P<0.05) and late apoptosis/necrosis cells (P<0.01), while AMO-let-7f did not produce any effects on apoptosis of MCF-7 cells (P>0.05). * P<0.05 vs NC, ** P<0.01 vs NC.
Discussion
In the present study, we uncovered that paclitaxel LDM therapy led to the reduction of let-7f which in turn caused the increase of TSP-1 expression in breast cancer via acting on 3'UTR of THBS1. It suggests that let-7f mediates the increase of TSP-1 in paclitaxel LDM therapy. This study will help us better understand the molecular mechanism of anti-angiogenic activity of paclitaxel LDM therapy.
Breast cancer is the first leading cancers in women all over the world, and more than 200,000 females were diagnosed as breast cancer per year in the United States 19. In additional to surgery, chemotherapy is an important strategy for the treatment of breast cancer. Paclitaxel is one of the most effective chemotherapy drugs to prevent the metastasis of breast cancer 20. However, severe side effects such as anaphylactic hypersensitivity reactions and peripheral neuropathy during paclitaxel therapy were often observed in patients 5, 21. So, it is necessary to develop a better therapeutic strategy to improve the efficacy of paclitaxel and reducing its toxicity.
Low-dose metronomic (LDM) chemotherapy is defined as the frequent and continuous low-dose chemotherapeutics, and has attracted more and more attention as an alternative scheme for the treatment of many cancers 22. Compared with conventional MTD therapy, LDM was well characterized by high efficiency, less toxicity and reduced drug resistance in clinics 23, which was mainly attributed to its anti-angiogenic activity. LDM chemotherapy of paclitaxel has been suggested as a novel potential approach to avoiding or reducing adverse effects of paclitaxel. For example, a clinical trial showed the evidence that paclitaxel LDM therapy in combination with other anti-cancer drugs was beneficial and safe in patients with metastatic melanoma 24. Our previous study reported that LDM therapy displayed a stronger anti-tumor activity in suppressing primary and metastatic breast tumors with fewer side effects 8, 10. Paclitaxel LDM therapy has stronger anti-angiogenic and anti-lymphangiogenic activities than MTD therapy, whereas paclitaxel MTD therapy showed stronger pro-apoptotic and anti-proliferative activities. Upregulation of TSP-1, an important angiogenesis inhibitor was defined as the underlying mechanism of the anti-tumor activity of paclitaxel LDM therapy 10. However, how LDM chemotherapy paclitaxel caused the upregulation of TSP-1 has not been elucidated yet.
miRNAs are a category of endogenous noncoding small RNAs, and negatively regulate target gene expression in a post-transcriptional manner 12. Accumulating evidence uncover that miRNAs play an important role in cellular apoptosis, differentiation, proliferation, aging, etc 12, 16. Especially, some miRNAs called "anti-oncomirs" serve as natural inhibitors of oncogene and may suppress the growth and metastasis of tumors 13. In contrast, some miRNAs acts as “oncomirs” and promote the proliferation and migration of tumor cells 25-26. Abnormal expression of miRNAs might be clinically valuable as a therapeutic target in the diagnosis, treatment, and prognosis of tumors 27. The changes of miRNAs expression were also shown to be related to angiogenesis of breast tumor in previous studies 28. However, little is known concerning the role of miRNAs in paclitaxel LDM therapy of breast cancer.
We found that a variety of miRNAs such as let-7f, let-7a, miR-19b, miR-543*, miR-224, miR-5097, miR-340-5p and miR-684 were changed after paclitaxel LDM therapy by microarray screening. qRT-PCR verification demonstrated that let-7f, let-7a, miR-19b and miR-340-5p were reduced, and miR-684 was upregulated in tumor tissues after LDM treatment. In particular, let-7f has a most pronounced reduction in both LDM group and MTD group as compared to other miRNAs. In vitro, MCF-7 cells treatment with paclitaxel resulted in a decrease of let-7f as well. So, we firstly studied the role of let-7f in paclitaxel LDM therapy in this study. It has been reported that TSP-1 protein was a critical target of anti-angiogenesis of paclitaxel LDM therapy in breast cancers. Interestingly, computation analysis predicted that TSP-1 encoding gene THBS1 is the target of let-7f, which was then confirmed by luciferase assay. The mutation of binding sites can abolish the inhibition of luciferase activity by let-7f. As a highly conserved noncoding RNA, let-7f has the same RNA sequences cross many species such as human, mouse and rat. We then transfected the synthesized let-7f into MCF-7 cells and studied its effects on TSP-1 expression. The results showed that let-7f overexpression inhibited TSP-1 expression, but let-7f knockdown upregulated TSP-1 level in MCF-7 cells. Consistently, previous study has shown that endogenous angiogenesis inhibitor TSP-1 was the target for let-7f in endothelial cells 29.
Our previous study showed that LDM can inhibit the lymphangiogenic and angiogeneic activity via upregulating TSP-1 expression, but did not affect the proliferation and apoptosis of breast cancer. So, we further investigated whether the inhibition of let-7f by LDM therapy did not affect the proliferation and apoptosis of MCF-7 cells. It was shown that neither let-7f nor AMO-let-7f affected the proliferation of MCF-7 cells. The cell cycle of MCF-7 was not altered after let-7f or AMO-let-7f treatment. Consistent with this data, recent studies reported that let-7 inhibition did not enhance cellular proliferation 30-31. Moreover, the knockdown of let-7f did not raise the number of apoptotic tumor cells, but let-7f overexpression induced an increase of apoptosis in MCF-7 cells. It means that LDM paclitaxel-induced the knockdown of let-7f did not increase the apoptosis of MCF-7 cells. MTD therapy induced the apoptosis of breast cancer independently of let-7f.
Other microRNAs, let-7a and miR-19b were also decreased after LDM treatment. In order to evaluate the contribution of let-7a and miR-19b to TSP-1 upregulation, we also transfected let-7a and miR-19b level into MCF-7 cells, and observed their effects on luciferase activity of THBS 3'UTR vector. We found that let-7a and miR-19b also showed an inhibitory effect on TSP-1 expression, but the inhibitory effect of let-7a or miR-19b on TSP-1 was less than let-7f (supplementary Fig. 1).
Limitation of this study: Other factors including transcription factors, translation efficiency, methylations, etc, also possibly contribute to the increase of TSP-1 after LDM treatment. In this study, we just confirmed that let-7f reduction may lead to the augment of TSP-1 expression in breast cancer cells. Whether other factors were involved in this process need further studies.
In summary, we found that LDM therapy caused the downregulation of let-7f in breast cancer, which contributed to the upregulation of TSP-1 in breast cancer. This study confer us a better understanding of molecular mechanisms of paclitaxel LDM therapy.
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China (81101997/H1622), Foundation of Educational Commission of Heilongjiang Province (12521280), Heilongjiang Postdoctoral Fund (LBH-Z11085), China Postdoctoral Science Foundatiion (2011M500694), Specialized Research Fund for the Doctoral Program of Higher Education of China for new teacher (20112307120019) and the funding of The Affiliated Tumor Hospital of Harbin Medical University (JJZ2010-14).
References
- 1.Ueda S, Kuji I, Shigekawa T, Takeuchi H, Sano H, Hirokawa E, Optical Imaging for Monitoring Tumor Oxygenation Response after Initiation of Single-Agent Bevacizumab followed by Cytotoxic Chemotherapy in Breast Cancer Patients. PLoS One. 9: e98715. doi:10.1371/journal.pone.0098715 PONE-D-13-48502 [pii] [DOI] [PMC free article] [PubMed]
- 2.Mita M, Gordon M, Rosen L, Kapoor N, Choy G, Redkar S, Phase 1B study of amuvatinib in combination with five standard cancer therapies in adults with advanced solid tumors. Cancer Chemother Pharmacol. doi:10.1007/s00280-014-2481-1. [DOI] [PubMed]
- 3.Gonzalez-Martin A, Sanchez-Lorenzo L, Bratos R, Marquez R, Chiva L. First-line and maintenance therapy for ovarian cancer: current status and future directions. Drugs. 2014;74:879–89. doi: 10.1007/s40265-014-0221-9. doi:10.1007/s40265-014-0221-9. [DOI] [PubMed] [Google Scholar]
- 4.Misiukiewicz K, Gupta V, Bakst R, Posner M. Taxanes in cancer of the head and neck. Anticancer Drugs. 2014;25:561–70. doi: 10.1097/CAD.0000000000000086. doi:10.1097/CAD.0000000000000086. [DOI] [PubMed] [Google Scholar]
- 5.A fatal anaphylactic reaction to paclitaxel is described, which was preceded by a possible delayed reaction to the initial infusion. Allergy Asthma Proc. 2011;32:79.. doi: 10.2500/aap.2011.32.3409. doi:10.2500/aap.2011.32.3409. [DOI] [PubMed] [Google Scholar]
- 6.Flatters SJ, Xiao WH, Bennett GJ. Acetyl-L-carnitine prevents and reduces paclitaxel-induced painful peripheral neuropathy. Neurosci Lett. 2006;397:219–23. doi: 10.1016/j.neulet.2005.12.013. doi:S0304-3940(05)01407-2 [pii] 10.1016/j.neulet.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong LH, Chen XX, Wang H, Jiang QW, Pan SS, Qiu JG. et al. Piperlongumine induces apoptosis and synergizes with cisplatin or paclitaxel in human ovarian cancer cells. Oxid Med Cell Longev. 2014:906804.. doi: 10.1155/2014/906804. doi:10.1155/2014/906804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang H, Tao W, Zhang M, Pan S, Kanwar JR, Sun X. Low-dose metronomic paclitaxel chemotherapy suppresses breast tumors and metastases in mice. Cancer Invest. 2010;28:74–84. doi: 10.3109/07357900902744510. doi:10.3109/07357900902744510. [DOI] [PubMed] [Google Scholar]
- 9.Bocci G, Di Paolo A, Danesi R. The pharmacological bases of the antiangiogenic activity of paclitaxel. Angiogenesis. 2013;16:481–92. doi: 10.1007/s10456-013-9334-0. doi:10.1007/s10456-013-9334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M, Tao W, Pan S, Sun X, Jiang H. Low-dose metronomic chemotherapy of paclitaxel synergizes with cetuximab to suppress human colon cancer xenografts. Anticancer Drugs. 2009;20:355–63. doi: 10.1097/CAD.0b013e3283299f36. doi:10.1097/CAD.0b013e3283299f36. [DOI] [PubMed] [Google Scholar]
- 11.Jiang H, Tao W, Zhang M, Pan S, Kanwar JR, Sun X. Low-dose metronomic paclitaxel chemotherapy suppresses breast tumors and metastases in mice. Cancer Invest. 2010;28:74–84. doi: 10.3109/07357900902744510. doi:10.3109/07357900902744510. [DOI] [PubMed] [Google Scholar]
- 12.Maqbool R, Hussain MU. MicroRNAs and human diseases: diagnostic and therapeutic potential. Cell Tissue Res. 2014. doi:10.1007/s00441-013-1787-3. [DOI] [PubMed]
- 13.Le XF, Merchant O, Bast RC, Calin GA. The Roles of MicroRNAs in the Cancer Invasion-Metastasis Cascade. Cancer Microenviron. 2010;3:137–47. doi: 10.1007/s12307-010-0037-4. doi:10.1007/s12307-010-0037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Yu H, Lou JR, Zheng J, Zhu H, Popescu NI. et al. MicroRNA-19 (miR-19) regulates tissue factor expression in breast cancer cells. J Biol Chem. 2011;286:1429–35. doi: 10.1074/jbc.M110.146530. doi:M110.146530 [pii] 10.1074/jbc.M110.146530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattiske S, Suetani RJ, Neilsen PM, Callen DF. The oncogenic role of miR-155 in breast cancer. Cancer Epidemiol Biomarkers Prev. 2012;21:1236–43. doi: 10.1158/1055-9965.EPI-12-0173. doi:1055-9965.EPI-12-0173 [pii] 10.1158/1055-9965.EPI-12-0173. [DOI] [PubMed] [Google Scholar]
- 16.Sandhu S, Garzon R. Potential applications of microRNAs in cancer diagnosis, prognosis, and treatment. Semin Oncol. 2011;38:781–7. doi: 10.1053/j.seminoncol.2011.08.007. doi:S0093-7754(11)00215-6 [pii] 10.1053/j.seminoncol.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 17.Ng SS, Sparreboom A, Shaked Y, Lee C, Man S, Desai N. et al. Influence of formulation vehicle on metronomic taxane chemotherapy: albumin-bound versus cremophor EL-based paclitaxel. Clin Cancer Res. 2006;12:4331–8. doi: 10.1158/1078-0432.CCR-05-2762. doi:12/14/4331 [pii] 10.1158/1078-0432.CCR-05-2762. [DOI] [PubMed] [Google Scholar]
- 18.Kanwar JR, Palmano KP, Sun X, Kanwar RK, Gupta R, Haggarty N. et al. 'Iron-saturated' lactoferrin is a potent natural adjuvant for augmenting cancer chemotherapy. Immunol Cell Biol. 2008;86:277–88. doi: 10.1038/sj.icb.7100163. doi:7100163 [pii] 10.1038/sj.icb.7100163. [DOI] [PubMed] [Google Scholar]
- 19.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. doi:10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 20.Montero AJ, Adams B, Diaz-Montero CM, Gluck S. Nab-paclitaxel in the treatment of metastatic breast cancer: a comprehensive review. Expert Rev Clin Pharmacol. 2011;4:329–34. doi: 10.1586/ecp.11.7. doi:10.1586/ecp.11.7. [DOI] [PubMed] [Google Scholar]
- 21.Emmenegger U, Man S, Shaked Y, Francia G, Wong JW, Hicklin DJ. et al. A comparative analysis of low-dose metronomic cyclophosphamide reveals absent or low-grade toxicity on tissues highly sensitive to the toxic effects of maximum tolerated dose regimens. Cancer Res. 2004;64:3994–4000. doi: 10.1158/0008-5472.CAN-04-0580. doi:10.1158/0008-5472.CAN-04-0580 64/11/3994 [pii] [DOI] [PubMed] [Google Scholar]
- 22.Lien K, Georgsdottir S, Sivanathan L, Chan K, Emmenegger U. Low-dose metronomic chemotherapy: a systematic literature analysis. Eur J Cancer. 2013;49:3387–95. doi: 10.1016/j.ejca.2013.06.038. doi:S0959-8049(13)00541-8 [pii] 10.1016/j.ejca.2013.06.038. [DOI] [PubMed] [Google Scholar]
- 23.Loven D, Hasnis E, Bertolini F, Shaked Y. Low-dose metronomic chemotherapy: from past experience to new paradigms in the treatment of cancer. Drug Discov Today. 2013;18:193–201. doi: 10.1016/j.drudis.2012.07.015. doi:S1359-6446(12)00274-7 [pii] 10.1016/j.drudis.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 24.Bhatt RS, Merchan J, Parker R, Wu HK, Zhang L, Seery V. et al. A phase 2 pilot trial of low-dose, continuous infusion, or "metronomic" paclitaxel and oral celecoxib in patients with metastatic melanoma. Cancer. 2010;116:1751–6. doi: 10.1002/cncr.24902. doi:10.1002/cncr.24902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–78. doi: 10.1007/s10555-009-9188-5. doi:10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 26.Goga A, Benz C. Anti-oncomir suppression of tumor phenotypes. Mol Interv. 2007;7:199–202. doi: 10.1124/mi.7.4.6. 180. doi:10.1124/mi.7.4.6. [DOI] [PubMed] [Google Scholar]
- 27.Fiorucci G, Chiantore MV, Mangino G, Percario ZA, Affabris E, Romeo G. Cancer regulator microRNA: potential relevance in diagnosis, prognosis and treatment of cancer. Curr Med Chem. 2012;19:461–74. doi: 10.2174/092986712798918798. doi:BSP/CMC/E-Pub/2012/043 [pii] [DOI] [PubMed] [Google Scholar]
- 28.Plummer PN, Freeman R, Taft RJ, Vider J, Sax M, Umer BA. et al. MicroRNAs regulate tumor angiogenesis modulated by endothelial progenitor cells. Cancer Res. 2013;73:341–52. doi: 10.1158/0008-5472.CAN-12-0271. doi:0008-5472.CAN-12-0271 [pii] 10.1158/0008-5472.CAN-12-0271. [DOI] [PubMed] [Google Scholar]
- 29.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. doi:CIRCRESAHA.107.153916 [pii] 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 30.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–6. doi: 10.1038/nature08725. doi:nature08725 [pii] 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Worringer KA, Rand TA, Hayashi Y, Sami S, Takahashi K, Tanabe K. et al. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell. 2014;14:40–52. doi: 10.1016/j.stem.2013.11.001. doi:S1934-5909(13)00492-X [pii] 10.1016/j.stem.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.