Abstract
A method for genotyping hepatitis B virus by partial HBsAg gene sequencing with primers common to all known genotypes was developed. Mutations related to anti-HBs resistance are also detected with this method. Samples from 103 Brazilian patients were analyzed. Precore and core region of these viruses were also sequenced in 101 patients. Genotypes A, B, C, D, and F were found with frequencies of 49.5, 2.9, 13.6, 24.3, and 9.7%, respectively. Genotypes B and C were found only in Asian patients, whereas genotypes A, D, and F were more common in patients without an Asian background. Precore mutants were found in 32 (31.7%) of 101 patients, with a higher frequency in those infected with genotype D (22 of 25 [88.0%]). Analysis of nucleotide 1858 showed presence of thymine in all patients with genotypes B, C, and D and in a few patients with genotypes A (10.0%) and F (30.0%), who showed more frequently the presence of cytosine. This nucleotide was closely related to the presence of precore mutants. Mutations in the basal core promoter were found in 64 of 101 (63.4%) samples. These mutations were more frequent in patients infected with genotype F (90.0%) and less frequent in patients infected with genotype B (33.3%). Deletions in this region were found in two genotype C-infected patients.
Hepatitis B virus (HBV) infection is very common worldwide, and more than 350 million people are chronic carriers (22). HBV infection is associated with different clinical pictures and leads to chronic carrier state in 5 to 10% patients infected in adult life and 85 to 90% of those infected in infancy (13).
HBV was formerly classified into four different subtypes that were afterward subdivided according to the antigenic determinants of HBsAg in adw (adw2 and adw4), ayw (ayw1, ayw2, ayw3, and ayw4), adr (adrq+ adrq−), and ayr (33). Subtype a is common to the majority of viruses and is related to a neutralizing epitope. Divergence of the complete genome in a same subtype is ca. 8%, similar to the one found between different subtypes (38).
Another classification reflecting the phylogenetic origin of the virus isolates was later proposed dividing HBV into six genotypes (36-38), designated A to F. These genotypes were differentiated by a sequence divergence in the entire genome exceeding 8%. Norder et al. (35) demonstrated that HBV genotyping could also be performed by sequencing the S gene. Recently, seventh and eighth genotypes were reported: genotype G, which has an insertion of 36 nucleotides (nt) in the core gene and was discovered in France and United States (49), and genotype H, which was found in Nicaragua, Mexico, and California and has probably split off from genotype F within the New World (2). Previous studies have shown the geographical distribution of HBV genotypes worldwide but the results are still incomplete. Few data are reported from Brazil, where genotypes A, D, and F have been found in Rio de Janeiro, Goiás, and Santa Catarina states (8, 32, 52).
HBV vaccination, anti-HBV hyperimmunoglobulin (HBIg), and anti-HBs monoclonal antibody may induce resistant mutations, as previously shown by other researchers (5-7, 11, 40-42, 46, 53). These mutations are especially common in regions where HBV is endemic and where widespread anti-HBV vaccination have already been established and in patients undergoing liver transplantation due to hepatitis B who are using specific prophylaxis with HBIg or anti-HBs monoclonal antibodies.
Precore mutants were first described in Italy (4) related to cases showing negative HBeAg but with detectable HBV-DNA, HBsAg, anti-HBe, and anti-HBc. These strains have mutations in the nt 1896 (G→A) and 1899 (C→A) in the precore region, leading to a stop codon that prevents HBeAg synthesis. Precore region also contains the ɛ encapsidation signal, a secondary structure maintained by hydrogen bonds among bases in the same strain. Precore mutations are especially rare in genotypes that usually contain C at position 1858, since this nucleotide pairs with the nt 1896 in the secondary structure of ɛ encapsidation sign (23). Precore mutants have been associated with more severe hepatitis B evolution. This effect was related to the absence of the immune response modulator role from HBeAg and to alterations in the binding of the ɛ signal, leading to more efficient viral replication (50), but the overall pathogenic meaning of these mutations remains unclear (15, 28, 39). Since HBV genotypes are not uniformly distributed around the world, precore mutants also have a particular distribution, being more frequent in geographic regions such as Asia and the Mediterranean basin, where genotypes B, C, or D are predominant, and rare in North America and Europe, where genotype A is commonly found (24, 45). In South America, the frequency of this mutation is poorly studied, but Castro et al. (9) found a low frequency of precore mutations (7 of 29 [24%]) in anti-HBe-positive Brazilian carriers.
HBsAg, anti-HBe, and HBV-DNA positive serological profile can also be related to mutations affecting the basal core promoter (BCP), especially A1762T and G1764A. These mutations have been associated with active hepatitis, severe disease after liver transplantation, development of hepatocellular carcinoma, and fulminant hepatitis (3, 51).
Recently, several methods based on sequencing have been described that allow determination of viral features related to prognosis and therapeutic response, such as genotype, precore and BCP mutants, and drug resistance.
We describe here a new set of primers for determining HBV genotypes with the same reagents by nested PCR and automated DNA sequencing. We also verified the frequency of HBV genotypes and precore and BCP mutations in Brazilian patients.
MATERIALS AND METHODS
Patients.
We studied 103 samples from HBV chronic carriers (determined by two consecutive HBsAg-positive results from samples obtained at least 6 months apart) from different Brazilian regions that were sent to Laboratório Bioquímico Jardim Paulista, São Paulo, Brazil, from 1998 to 2001. Serum samples were stored at −20°C and thawed immediately before use. The Ethical Committee from the University of São Paulo School of Medicine approved this protocol (research protocol 145/00).
Detection of HBV-DNA by PCR.
Extraction and amplification of HBV-DNA were carried out by nested PCR as described by Kaneko et al. (16). All reagents utilized were obtained from Life Technologies do Brazil, São Paulo, Brazil.
Analysis of HBV sequences from different genotypes for primer design.
For HBV genotyping, we developed a system amplifying a 417-bp fragment covering a region of S gene with marked heterogeneity among genotypes, flanked by a conserved region to allow equal amplification of them. In this way, an alignment was made with S gene sequences from the different genotypes already described. We used the MegAlign program from Lasergene package (DNASTAR, Inc., Madison, Wis.) to align 49 sequences corresponding to the gene codifying HBsAg of different HBV isolates obtained from GenBank and corresponding to the known HBV genotypes (Table 1). Alignment was carried out by using the CLUSTAL method (14) and the weight residues table PAM250.
TABLE 1.
Partial S-gene sequences utilized in the alignment carried out to distinguish different HBV genotypes
| Genotype | Sequence (GenBank accession no.) |
|---|---|
| A | X75666, X75669, X65258, M54898, J02205, X51970, and M74498 |
| B | X75660, D23677, D23678, D00329, D00330, D00331, M54923, S74815, and S74867 |
| C | X75667, X75665, X75792, X75656, X01587, D23680, D23682, D23681, D16665, S62754, S75184, S81945, S81946, and U19777 |
| D | X75668, X75662, M32138, X77309, X65259, X59795, X68292, X77308, J02202, V01460, X02496, and X77310 |
| E | X75657, X75664, and L24071 |
| F | X75658, X75661, and X69798 |
| G | AF369533 and AF160501 |
| H | U91819, U91827, and AY090460 |
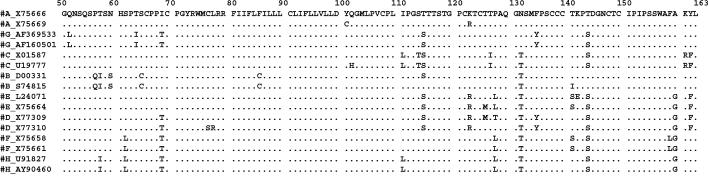
Selected primers corresponded to conserved regions among the different genotypes, surrounding heterogeneous regions between them, allowing the distinction of HBV genotypes, as shown in Fig. 1. The selected region for amplification also covers the amino acid loop corresponding to a, d/y, and w/r specificities and mutations related to HBIg, anti-HBs monoclonal antibody, and vaccine resistance. Selected primers were as follows: FHBS1, 5′-GAG TCT AGA CTC GTG GTG GAC TTC-3′; FHBS2, 5′-CGT GGT GGA CTT CTC TCA ATT TTC-3′; RHBS1, 5′-AAA TKG CAC TAG TAA ACT GAG CCA-3′; and RHBS2, 5′-GCC ARG AGA AAC GGR CTG AGG CCC-3′. The primer positions in the HBV genome (strain HBVADW; GenBank accession number V00866) were as follows: HBS1F (positions 244 to 267), HBS2F (positions 255 to 278), HBS2R (positions 648 to 671), and HBS1R (positions 668 to 691).
FIG. 1.
Schematic representation of the HBsAg coding region (amino acids 50 to 163) corresponding to the region amplified for genotyping. GenBank accession number sequence X75666 is shown in detail. For each known HBV genotype, two sequences are shown, and only amino acids differing from X75666 are indicated. Dots indicate that the same amino acid was observed in the sequence. Mutations specific for HBV subtypes: 122 (K = d, R = y), 160 (K = w, R = r), 127 (L = w4), and 140 (S = w4). Mutations related to resistance to HBIg, anti-HBs monoclonal antibody, or vaccination: 107 (Y), 110 (V), 111 (T), 114 (R), 115 (K or S), 116 (S), 117 (R), 118 (A, V, or S), 120 (E, T, S, or N insertion), 121 (T or R inser-tion), 122 (NT insertion or RA insertion), 123 (N or A or NSTGPCTT insertion), 124 (R or Y), 125 (A), 126 (S, N, or A), 127 (S or T), 129 (H), 130 (R or N), 131 (S or I), 132 (P), 133 (T, L, or I), 134 (L, I, S, or N), 135 (S), 137 (Y), 138 (Y or S), 140 (R, I, and P), 141 (E), 142 (S), 143 (W, R, M, or L), 144 (H, N, A, or E), 145 (R, K, or A), 146 (S), 147 (R), 148 (I), 149 (R or W), 154 (T or W), 155 (Y or P), 156 (L), and 157 (R).
HBsAg gene partial amplification.
Samples with detectable HBV-DNA were also subjected to S-gene partial amplification for HBV genotyping by sequencing. Serum samples were treated as described above and subjected to two rounds of amplification with sequentially outer (FHBS1 and RHBS1) and inner (FHBS2 and RHBS2) primers. The amplification conditions for the two rounds of the nested PCR were as follows: initial denaturation at 94°C for 20 s, followed by 30 cycles of 94°C for 20 s, 56°C for 20 s, and 72°C for 30 s, with a final extension step at 72°C for 1 min in a PTC-200 Thermocycler (MJ Research, Watertown, Mass.).
Detection of BCP and precore mutants.
For the detection of BCP and precore mutants, HBV-DNA-positive samples were amplified by using the primers described by Takahashi et al. (51).
Sequencing reaction.
PCR products were subjected to cycle sequencing reactions as described previously (48) by using the second-round primers and the ABI Prism BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, Calif.). After purification, samples were denatured and loaded in a 5% Long Ranger 6 M urea gel (Long Ranger gel solution; FMC) in an automated ABI Prism 377 DNA sequencer (Applied Biosystems).
Sequence analysis.
Genotyping, BCP, and precore mutant analysis were carried out by sequence comparison with other known sequences from different HBV genotypes described previously and were aligned as described above. For this purpose, EditSeq and MegAlign programs from the DNAstar package (LaserGene, Inc.) were used.
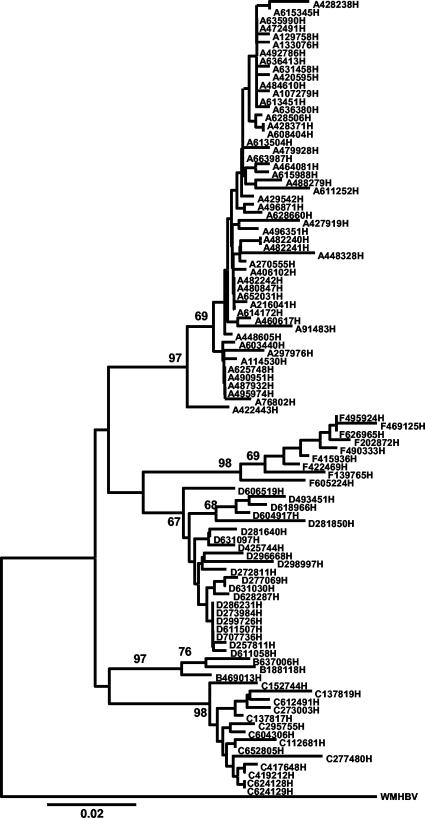
Phylogenetic and molecular evolutionary analyses were conducted by using MEGA version 2.1 (21). Sequences were aligned by using CLUSTAL X version 1.81 (54) and then analyzed by using the neighbor-joining method by a distance matrix calculated according to the Kimura two-parameter model (19). Woolly monkey hepatitis B virus (GenBank accession number AF046996) was utilized as an outgroup. A bootstrap test of phylogeny was performed with 1,000 replicates. and values ≥67 are indicated on the branches.
Statistical analysis.
For statistical analysis, we used the EpiInfo program (version 6.04; Centers for Disease Control and Prevention, Atlanta, Ga., and World Health Organization, Geneva, Switzerland).
GenBank accession numbers.
Sequences from the S and preC/C genes from Brazilian cases were deposited in the GenBank database under accession numbers AY323306 to AY323408 and AY329494 to AY329594, respectively.
RESULTS
HBsAg gene partial amplification analysis.
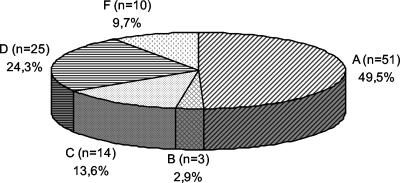
As shown in the phylogenetic analysis (Fig. 2), genotypes A, B, C, D, and F were found among our samples. The frequency of genotypes in the 103 patients is shown in Fig. 3. The following subtypes were found: adw, ayw, adr, and adw4. Genotypes A and B had only subtype adw, and genotype F had only adw4. Genotype C showed subtype adr more frequently, with one patient infected with subtype adw. Genotype D samples were all subtype ayw except for one (adw). In two Asian patients, mutations in the epitope “a” region were found. One genotype B sample had a T126S mutation, previously associated HBIg resistance, and the other one (genotype C) had a T118A mutation, which was associated with vaccine resistance.
FIG. 2.
Neighbor-joining tree of a 339-nt fragment of HBV-DNA (S gene) from isolates obtained in the studied population with woolly monkey hepatitis virus (GenBank accession number AF046996) used as an outgroup.
FIG. 3.
HBV genotype distribution in Brazilian patients (n = 103).
Genotypes, age, gender, and ethnicity.
The gender, age mean, and ethnicity ranges according to genotypes can be seen in Table 2. Most of the patients studied had a Western ethnicity (n = 83, 80.6%). Genotype A was found mainly in such patients (94.1%), and genotypes D and F were exclusive to them. The only three Asian patients infected with genotype A belonged to the same family (father, mother, and son). Among the 20 (19.4%) Asian patients, 17 (85.0%) had genotypes B or C. None of non-Asian patients had genotype B or C. Statistical analysis showed that infection with genotypes B and C is significantly more frequent among Asian patients in our population (P = 0.01 and P < 0.0001, respectively).
TABLE 2.
HBV genotypes and their relationships to patients and viral featuresa
| HBV genotype | Patient feature
|
BCP and precore region mutations (no. [%])
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| No. male (%) | Mean age (yr) (SD) | No. Asian (%) | BCP
|
nt 1858
|
Precore region
|
||||
| WT | Mutant | C | T | WT | Mutant | ||||
| A | 35 (68.6) | 40.7 (11.6) | 3 (5.9) | 19 (38.0) | 31 (62.0) | 45c (90.0) | 5 (10.0) | 46 (92.0) | 4 (8.0) |
| B | 3 (100) | 49.0 (9.87) | 3 (100)† | 2 (66.7) | 1 (33.3) | 0 | 3 (100) | 2 (66.7) | 1 (33.3) |
| C | 9 (64.3) | 33.5 (16.5) | 14 (100)* | 3 (23.1) | 10b (76.9) | 0 | 13 (100) | 10 (76.9) | 3 (23.1) |
| D | 19 (76.0) | 51.1 (12.9) | 0 | 12 (48.0) | 13 (52.0) | 0 | 25 (100) | 3 (12.0) | 22 (88.0) |
| F | 10 (100) | 33.0 (7.12) | 0 | 1 (10.0) | 9 (90.0) | 7 (70.0) | 3 (30.0) | 8 (80.0) | 2 (20.0) |
| Total | 76 (73.8) | 42.0 (13.9) | 20 (19.4) | 37 (36.6) | 64 (63.4) | 52 (51.5) | 49 (48.5) | 69 (68.3) | 32 (31.7) |
*, P < 0.0001; †, P = 0.01. WT, wild type.
Includes two cases with a deletion in the BCP region.
Includes two cases with an adenine at nt 1858.
Genotypes, precore and BCP mutants, and nt 1858 analysis.
In 101 patients, we also analyzed the BCP and precore region of HBV. In the other two patients, it was not possible to amplify this region. The frequency of mutants in these regions among the different genotypes, as well as the results of the nt 1858 analysis, are shown in Table 2. Precore mutants were more frequent in patients infected with genotype D virus (P < 0.0001).
Analysis of nt 1858 showed the presence of thymine in all patients with genotypes B, C, and D and in a few patients with genotypes A (5 of 50 [10.0%]) and F (3 of 10 [30.0%]). This nucleotide was closely related to the presence of precore mutants (P < 0.0001) since, of 49 isolates with nt 1851, 32 (65.3%) also had the stop codon mutation at nt 1896, whereas none of the isolates with a cytosine at nt 1858 showed this mutation. All patients with genotypes A and F that had a mutation in the precore region also had a thymine at position 1858.
BCP mutants were less frequent among genotype B-infected patients (33.3%) and more frequent among genotype F-infected patients (90.0%), but no statistical significant difference was found. Interestingly, in two patients infected with HBV genotype C, we found deletions in the BCP encompassing the studied mutation. These deletions extended from nt 1760 to 1770 in one patient and from nt 1760 to 1769 in the other.
DISCUSSION
HBV genotyping has been extensively analyzed recently. Better responses to treatment have been reported for genotypes A and B compared to genotypes D and C, respectively (44, 47, 56). On the other hand, evolution to chronic hepatitis or to more severe diseases such as hepatocellular carcinoma has been related to genotypes A and C, respectively (12, 30, 43, 44). Few data regarding genotype F have been reported, but death related to liver disease was more frequently associated with genotype F than with genotypes A or D in one study (47).
The genotyping method described here has the advantage of being very fast and straightforward and uses the same reagents to amplify all known HBV genotypes. It also allows analysis of subtype and other mutations, such as in the epitope a region of the same sequence. Other methods for HBV genotyping, such as restriction length fragment polymorphism analysis (24, 25, 29) or line probe assay (52), can distinguish the different genotypes, but they do not allow detection of subtypes or other point mutations specific to vaccine or gamma globulin resistance.
When we used this genotyping method in Brazilian patients, we detected genotypes A, B, C, D, and F. Genotype A was the most common and was found among 49.5% patients. Moraes et al. (32) found genotypes A, D, and F in Rio de Janeiro, Brazil. These same genotypes were also described in populations from GoiÂnia and Santa Catarina (8, 52). To our knowledge, the present study is the first time that genotypes B and C have been detected in Brazil, probably due to the low percentage of Asian subjects in the Brazilian states studied previously, although one of our patients infected with genotype C virus was from Rio de Janeiro.
A total of 85% of Asian patients had genotype B or C. Only three of these patients had genotype A, and all three individuals belonged to the same family. Several studies carried out in Eastern Asia have shown that genotypes B and C are the most common in this region. It was also reported that patients infected with these genotypes have a worse prognosis with a higher frequency of progression to cirrhosis and hepatocellular carcinoma (17). In China, Ding et al. (12) showed that genotype C infections were more severe than genotype B infections. Similar results have also been found in other studies in China and Japan, and it has been hypothesized that the higher frequency of BCP mutants in genotype C could be associated with the higher pathogenicity of this genotype (18, 44). Studies addressing the evolution of hepatitis B and the viral genotypes should also be carried out among Brazilian patients.
Genotypes E, G, and H were not found in our study population. Genotype E has only been found in some regions of Africa (31), and only one study outside Africa found a Haitian child infected with this genotype in Belgium (26). Genotype G has been found in ca. 10% of studied cases from France and United States (49). Genotype H seems to be a divergent clade from genotype F and was described after our primer selection and sample processing, but a later analysis with three sequences from GenBank (U91819, U91827, and AY090460) showed that this method is also able to distinguish this new genotype.
Although there are many African descendants in Brazil due to slaves being brought from Africa from the 16th to the 19th centuries, we did not find genotype E in our sample. There were few patients of known African descent in the studied population. Other studies in Brazilian states with a higher percentage of African descendants, such as Bahia, should be carried out to better evaluate this issue.
Different subtypes were also found in our samples. As expected, it would not be possible to classify the genotypes only based on that.
Mutations related to epitope a recognition were found in only two patients. Mutation T126S has already been described and associated with vaccination resistance (40, 55). Mutation T118A, on the other hand, is outside the main region of epitope a and was related to HBIg resistance (11). Unfortunately, we were not able to obtain more information about the progress of the disease in these two patients, but it is noteworthy that both had genotypes common to Asiatic regions (43). The finding of these mutations among our cases highlights the need for a continued surveillance to address the appearance of such cases and to better analyze their real meaning.
Precore mutants had a low frequency in our population (31.7%). Such mutants were found more frequently in patients infected with genotype D, with intermediate frequency in patients infected with genotypes B and C, and at a very low frequency in genotype A- and F-infected patients. The occurrence of this mutation is dependent upon the nucleotide (cytosine or thymine) at position 1858, which forms a base pair with nt 1896 in the pregenomic RNA loop at the ɛ encapsidation sign. A thymine at position 1858 is particularly common in genotype D viruses. The presence of a cytosine at position 1858 precludes the G-to-A mutation at nt 1896, since this would destabilize the stem-loop structure of the RNA encapsidation signal (27). Interactions of this encapsidation signal with the viral DNA polymerase is an essential step in the viral replication cycle, and it has been hypothesized that the increased strength of the guanine-to-cytosine base pairing found in precore mutants would implicate in a stronger ɛ encapsidation sign, allowing a more efficient replication of these strains. Genotype A usually shows a cytosine at this position (23), whereas genotype F may present thymine or cytosine (1). Our results from nt 1858 analysis corroborate with these data, since we also have found the presence of cytosine only in genotypes A and F, which showed a low frequency of precore mutants.
The frequency of this mutation varies widely around the world. Castro et al. (9) studied Brazilian patients and found the precore stop codon mutation in only 24% of them—a result similar to ours. These authors also demonstrated the higher frequency of the stop codon mutation at nt 1896 in the isolates that had T1858. On the other hand, the prevalence of this mutation in China was 86% (10). This discrepancy may reflect differences in genotype distributions in each studied population, since genotype A and D viruses are common in Brazilian populations, whereas in China genotypes B and C are more common. Furthermore, in our population, the frequencies of precore mutants were low even in genotype B- and C-infected patients (33.3 and 23.1%, respectively).
The BCP mutation was found in 63.4% of our patients and was present in all genotypes. BCP mutants frequency ranged from 33.3% in genotype B to 90.0% in genotype F. Changes at positions 1762 and 1764 have been described to be especially common in patients with C1858 (24), but we did not find any correlation between them. Among our patients, the frequency of BCP mutants was high in genotypes A and F patients from whom C1858 is especially common, but genotype C- and D-infected patients, in whom T1858 is common, have similar frequencies. On the other hand, other authors have reported that the presence of these mutants is not related to the presence of precore mutants (9). This point should be further analyzed in our population.
Two genotype C-infected patients in the present study had a deletion in this region. Other authors have reported a number of point mutations, as well as deletions and insertions of nucleotides in the precore region leading to initiation failure or premature termination. The positions of these mutations varied and were normally restricted from 8 to 21 nt, flanking AT-rich regions and overlapping transcription factor binding sites (10, 20, 34). An 8-bp deletion from nt 1768 to 1775 was also found in precore promoter region before exacerbation, but in a later sample this deletion was no longer found (34). The deletions reported here are different from the ones described previously, but they were always in the same region.
In conclusion, we have developed a method for HBV genotyping that can amplify isolates from all of the HBV genotypes found in Brazil. Furthermore, we have characterized by this approach the HBV genotypes in our population and their relationship to precore and BCP mutants. We have shown that genotypes E, G, and H were not found in Brazil until now. Genotype A was the most common in our study population. Genotypes A, D, and F are found in non-Asian subjects, whereas genotypes B and C are found among Asian patients. Precore mutants are more common among genotype D-positive patients, whereas BCP mutants were present in all of the genotypes, with a higher frequency among genotype F-infected patients.
Acknowledgments
We are deeply indebted to Simone Campiotto, Marcilio F. Lemos, and Rúbia A. Santana for technical assistance during the development of this study and to André C. Lyra for help in handling the MEGA2 package.
This study was supported by Laboratório Bioquímico Jardim Paulista, CAPES, and FAPESP (99/09551-0). Roberta Sitnik and Dennis Armando Bertolini were Ph.D. students from Faculdade de Medicina de Universidade de São Paulo and Escola Paulista de Medicina, respectively.
REFERENCES
- 1.Arauz-Ruiz, P., H. Norder, K. A. Visona, and L. O. Magnius. 1997. Genotype F prevails in HBV-infected patients of Hispanic origin in Central America and may carry the precore stop mutant. J. Med. Virol. 51:305-312. [PubMed] [Google Scholar]
- 2.Arauz-Ruiz, P., H. Norder, B. H. Robertson, and L. O. Magnius. 2002. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J. Gen. Virol. 83:2059-2073. [DOI] [PubMed] [Google Scholar]
- 3.Baumert, T. F., S. A. Rogers, K. Hasegawa, and T. J. Liang. 1996. Two core promotor mutations identified in a hepatitis B virus strain associated with fulminant hepatitis result in enhanced viral replication. J. Clin. Investig. 98:2268-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunetto, M. R., M. Stemler, F. Bonino, F. Schodel, F. Olivieri, M. Rizzetto, G. Verme, and H. Will. 1990. A new hepatitis B virus strain in patients with severe anti-HBe positive chronic hepatitis B. J. Hepatol. 10:258-261. [DOI] [PubMed] [Google Scholar]
- 5.Carman, W. F., J. Korula, L. Wallace, R. MacPhee, L. Mimms, and R. Decker. 1995. Fulminant reactivation of Hepatitis B due to envelope protein mutant that escaped detection by monoclonal HBsAg ELISA. Lancet 345:1406-1407. [DOI] [PubMed] [Google Scholar]
- 6.Carman, W. F., H. C. Thomas, A. J. Zuckerman, and T. J. Harrison. 1998. Molecular variants of hepatitis B virus, p. 141-177. In A. J. Zuckerman and H. C. Thomas (ed.), Viral hepatitis, 2nd ed. Churchill Livingstone, London, United Kingdom.
- 7.Carman, W. F., A. Owsianka, L. A. Wallace, B. C. Dow, and D. J. Mutimer. 1999. Antigenic characterisation of pre- and post-liver transplant hepatitis B surface antigen sequences from patients treated with hepatitis B immune globulin. J. Hepatol. 31:195-201. [DOI] [PubMed] [Google Scholar]
- 8.Carrilho, F. J., C. R. Moraes, J. R. R. Pinho, R. Moreira, D. Bertolini, I. M. Mello, S. Gatura, L. Bassit, R. A. Cardoso, L. C. Da Silva, and A. A. Laudanna. 2001. Hepatitis B virus infection in haemodialysis centers from Santa Catarina State, Southern Brazil: predictive factors for infection and molecular epidemiology. Hepatology 34(Pt. 4):612A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castro, L., C. Niel, and S. A. Gomes. 2001. Low frequency of mutations in the core promoter and precore regions of hepatitis B virus in anti-HBe positive Brazilian carriers. BMC Microbiol. 1:10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, H. L. Y., M. Hussain, and A. S. Lok. 1999. Different hepatitis B virus genotypes are associated with different mutations in the core promoter and precore regions during hepatitis Be antigen seroconversion. Hepatology 29:976-984. [DOI] [PubMed] [Google Scholar]
- 11.Chong-Jin, O., C. Wei Ning, K. Shiuan, and G. L. Keow. 1999. Identification of hepatitis B surface antigen variants with alterations outside the “a” determinant in immunized Singapore infants. J. Infect. Dis. 179:259-263. [DOI] [PubMed] [Google Scholar]
- 12.Ding, X., M. Mizokami, G. Yao, B. Xu, E. Orito, and M. Nakanishi. 2001. Hepatitis B virus genotype distribution among chronic hepatitis B virus carriers in Shangai, China. Intervirology 44:43-47. [DOI] [PubMed] [Google Scholar]
- 13.Gust, I., and S. Crowe. 1986. The global importance of viral hepatitis. Clin. Trop. Med. Commun. Dis. 1:281-301. [Google Scholar]
- 14.Higgins, D. G., and P. M. Sharp. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 15.Hsu, H. Y., M. H. Chang, C. Y. Lee, K. H. Hsieh, Y. H. Ni, P. J. Chen, and D. S. Chen. 1995. Precore mutant of hepatitis B virus in childhood fulminant hepatitis B: an infrequent association. J. Infect. Dis. 171:776-781. [DOI] [PubMed] [Google Scholar]
- 16.Kaneko, S., S. M. Feinstone, and R. H. Miller. 1989. Rapid and sensitive method for the detection of serum hepatitis B virus DNA using the polymerase chain reaction technique. J. Clin. Microb. 27:1930-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kao, J. H., P. J. Chen, M. Y. Lai, and D. S. Chen. 2000. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology 118:554-559. [DOI] [PubMed] [Google Scholar]
- 18.Kao, J. H., N. H. Wu, P. J. Chen, M. Y. Lai, and D. S. Chen. 2000. Hepatitis B genotypes and the response to interferon therapy. J. Hepatol. 33:998-1002. [DOI] [PubMed] [Google Scholar]
- 19.Kimura, M. 1980. A simple method for estimating evolutionary rates of bases substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 20.Kramvis, A., and M. C. Kew. 1999. The core promoter of hepatitis B virus. J. Viral Hepat. 6:415-427. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 22.Lee, W. M. 1997. Hepatitis B virus infection. N. Engl. J. Med. 337:1733-1745. [DOI] [PubMed] [Google Scholar]
- 23.Li, J. S., S. P. Tong, Y. M. Wen, L. Vitvitski, Q. Zhang, and C. Trepo. 1993. Hepatitis B virus genotype A rarely circulates as an HBe-minus mutant: possible contribution of a single nucleotide in the precore region. J. Virol. 76:5402-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindh, M., A. S. Andersson, and A. Gusdal. 1997. Genotypes, nt1858 variants, and geographic origin of hepatitis B virus: large-scale analysis using a new genotyping method. J. Infect. Dis. 175:1285-1293. [DOI] [PubMed] [Google Scholar]
- 25.Lindh, M., J. E. Gonzalez, G. Norkrans, and P. Horal. 1998. Genotyping of hepatitis B virus by restriction pattern analysis of a Pre-S amplicon. J. Virol. Methods 72:163-174. [DOI] [PubMed] [Google Scholar]
- 26.Liu, H. F., E. Sokal, and P. Goubau. 2001. Wide variety of genotypes and geographic origins of hepatitis B virus in Belgian children. J. Pediatr. Gastroenterol. Nutr. 32:244-247. [DOI] [PubMed] [Google Scholar]
- 27.Lok, A. S., U. Akarca, and S. Greene. 1994. Mutations in the pre-core region of hepatitis B virus serve to enhance the stability of the secondary structure of the pre-genome encapsidation signal. Proc. Natl. Acad. Sci. USA 91:4077-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lok, A. S., E. J. Heathcote, and J. H. Hoofnagle. 2001. Management of hepatitis B: summary of a workshop. Gastroenterology 120:1828-1853. [DOI] [PubMed] [Google Scholar]
- 29.Magnius, L. O., and H. Norder. 1995. Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S gene. Intervirology 38:24-34. [DOI] [PubMed] [Google Scholar]
- 30.Mayerat, C., A. Mantegani, and P. C. Frei. 1999. Does hepatitis B virus (HBV) genotype influence the clinical outcome of HBV infection? J. Viral Hepat. 6:299-304. [DOI] [PubMed] [Google Scholar]
- 31.Mizokami, M., T. Nakano, E. Orito, Y. Tanaka, H. Sakugawa, M. Mukaide, and B. H. Robertson. 1999. Hepatitis B virus genotype assignment using restriction fragment length polymorphism patterns. FEBS Lett. 450:66-71. [DOI] [PubMed] [Google Scholar]
- 32.Moraes, M. T., A. S. Gomes, and C. Niel. 1996. Sequence analysis of the pre-S/S gene of hepatitis B virus strains genotypes A, D, and F isolated in Brazil. Arch. Virol. 114:1767-1773. [DOI] [PubMed] [Google Scholar]
- 33.Neurath, A. R., and Y. Thanavala. 1990. Hepadnaviruses, p. 403-458. In M. H. V. Van Regenmortel and A. R. Neurath (ed.), Immunochemistry of viruses. II. The basis for serodiagnosis and vaccines. Elsevier, Amsterdam, The Netherlands.
- 34.Nishizono, A., K. Kohno, Y. Takita-Sonoda, M. Hiraga, H. Terao, T. Fujioka, M. Nasu, and K. Mifune. 1997. Sequential analyses of the mutations in the core upstream and precore regions of hepatitis B virus genome in anti-HBe positive-carriers developing acute exacerbation. J. Med. Virol. 53:266-272. [PubMed] [Google Scholar]
- 35.Norder, H., B. Hammas, S. Löfdal, A. M. Courouce, and L. O. Magnius. 1992. Comparison of the amino acid sequences of nine different serotypes of hepatitis B surface antigen and genomic classification of the corresponding hepatitis B virus strains. J. Virol. 73:1201-1208. [DOI] [PubMed] [Google Scholar]
- 36.Norder, H., A. M. Courouce, and L. O. Magnius. 1992. Molecular basis of hepatitis B virus serotype variations within the four major subtypes. J. Gen. Virol. 73:3141-3145. [DOI] [PubMed] [Google Scholar]
- 37.Norder, H., A. M. Courouce, and L. O. Magnius. 1994. Complete genomes, phylogenic relatedness and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology 198:489-503. [DOI] [PubMed] [Google Scholar]
- 38.Okamoto, H., F. Tsuda, H. Sakugawa, R. I. Sastroewignjo, M. Imai, Y. Miyakawa, and M. Mayumi. 1988. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J. Gen. Virol. 69:2575-2583. [DOI] [PubMed] [Google Scholar]
- 39.Okamoto, H., S. Yotsumoto, Y. Akahane, T. Tamanaka, Y. Miyazaki, Y. Sugai, F. Tsuda, T. Tanaka, Y. Miyakawa, and M. Mayumi. 1990. Hepatitis B viruses with precore regions defects prevail in persistently infected hosts along with seroconversion to the antibody against e antigen. J. Virol. 64:1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okamoto, H., K. Yano, Y. Nozaki, A. Matsui, H. Miyazaki, K. Yamamoto, F. Tsuda, A. Machida, and S. Mishiro. 1992. Mutations within the S gene of hepatitis B virus transmitted form mothers to babies immunized with hepatitis-B immune globulin and vaccine. Pediatr. Res. 32:264-268. [DOI] [PubMed] [Google Scholar]
- 41.Oon, C. J., K. L. Tan, T. Harrison, and A. Zuckerman. 1996. Natural history of hepatitis B surface antigen mutants in children. Lancet 348:1524.. [DOI] [PubMed] [Google Scholar]
- 42.Oon, C. J., and W. N. Chen. 1998. Current aspects of hepatitis B surface antigen mutants in Singapore. J. Viral Hepatitis 5(Suppl. 2):17-23. [DOI] [PubMed] [Google Scholar]
- 43.Orito, E., T. Ichida, H. Sakugawa, M. Sata, N. Horiike, K. Hino, K. Okita, T. Okanoue, S. Iino, E. Tanaka, K. Suzuki, H. Watanabe, S. Hige, and M. Mizokami. 2001. Geographic distribution of hepatitis B virus (HBV) genotype in patients with chronic HBV infection in Japan. Hepatology 34:590-594. [DOI] [PubMed] [Google Scholar]
- 44.Orito, E., M. Mizokami, H. Sakugawa, K. Michitaka, K. Ishikawa, T. Ichida, T. Okanoue, H. Yotsuyanagi, S. Lino, et al. 2001. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Hepatology 33:218-223. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez-Frias, F., M. Buti, R. Jardi, M. Cotrina, L. Viladomiu, R. Esteban, and J. Guardia. 1995. Hepatitis B virus infection: precore mutants and its relation to viral genotypes and core mutations. Hepatology 22:1641-1647. [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez-Frias, F., M. Buti, R. Jardi, V. Vargas, J. Quer, M. Cotrina, M. Martell, R. Esteban, and J. Guardia. 1999. Genetic alterations in the S gene of hepatitis B virus in patients with acute hepatitis B, chronic hepatitis B, and hepatitis B liver cirrhosis before and after liver transplantation. Liver 19:177-182. [DOI] [PubMed] [Google Scholar]
- 47.Sanchez-Tapias, J. M., J. Costa, A. Mas, M. Bruguera, and J. Rodes. 2003. Influence of hepatitis B virus genotype on the long-term outcome of chronic hepatitis B in western patients. Gastroenterology 125:1559-1560. [DOI] [PubMed] [Google Scholar]
- 48.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stuyver, L., S. De Gendt, C. Van Geyt, F. Zoulim, M. Fried, R. F. Schinazi, and R. Rossau. 2000. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J. Gen. Virol. 81:67-74. [DOI] [PubMed] [Google Scholar]
- 50.Su, H., and J. K. Yee. 1992. Regulation of hepatitis B virus gene expression by its two enhancers. Proc. Natl. Acad. Sci. USA 89:2708-2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahashi, K., K. Aoyama, N. Ohno, K. Iwata, Y. Akahane, K. Baba, H. Yoshizawa, and S. Mishiro. 1995. The pre core/core promoter mutant (T1762A1764) of hepatitis B virus: clinical significance and easy method for detection. J. Gen. Virol. 76:3159-3164. [DOI] [PubMed] [Google Scholar]
- 52.Teles, S. A., R. M. Martins, B. Vanderborght, L. Stuyver, A. M. Gaspar, and C. F. Yoshida. 1999. Hepatitis B virus: genotypes and subtypes in Brazilian hemodialysis patients. Artif. Organs 23:1074-1078. [DOI] [PubMed] [Google Scholar]
- 53.Theamboonlers, A., V. Chongsrisawat, P. Jantaradsamee, and Y. Poovorawan. 2001. Variants within the “a” determinant of HBs gene in children and adolescents with and without hepatitis B vaccination as part of Thailand's Expanded Program on Immunization (EPI). Tohoku J. Exp. Med. 193:197-205. [DOI] [PubMed] [Google Scholar]
- 54.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tong, C. Y. W. 2000. Genetic variations of hepatitis B virus. Curr. Opin. Infect. Dis. 13:481-487. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, X., F. Zoulim, F. Habersetzer, S. Xiong, and C. Trepo. 1996. Analysis of hepatitis B virus genotypes and pre-core region variability during interferon treatment of HBe antigen negative chronic hepatitis B. J. Med. Virol. 48:8-16. [DOI] [PubMed] [Google Scholar]