Abstract
SOX genes are transcription factors with important roles in embryonic development and carcinogenesis. The SOX family of 20 genes is responsible for regulating lineage and tissue specific gene expression patterns, controlling numerous developmental processes including cell differentiation, sex determination, and organogenesis. As is the case with many genes involved in regulating development, SOX genes are frequently deregulated in cancer. In this perspective we provide a brief overview of how SOX proteins can promote or suppress cancer growth. We also present a pan-cancer analysis of aberrant SOX gene expression and highlight potential molecular mechanisms responsible for their disruption in cancer. Our analyses indicate the prominence of SOX deregulation in different cancer types and reveal potential roles for SOX genes not previously described in cancer. Finally, we summarize our recent identification of SOX15 as a candidate tumor suppressor in pancreatic cancer and propose several research avenues to pursue to further delineate the emerging role of SOX15 in development and carcinogenesis.
Keywords: SOX, SOX15, oncogene, tumor suppressor, development, cancer
INTRODUCTION
SOX genes (SRY-related high mobility group (HMG) box) encode a family of transcription factors containing the DNA binding domain of SRY, the first SOX gene identified [1-3]. The twenty different SOX proteins identified in mammals to date can be subdivided into 8 groups (A, B1, B2, C, D, E, F, G, H) based on similarities in HMG box domains, gene structure, and the presence of specific functional domains including coiled-coil, transactivation and transrepression domains [2, 4]. Depending on the domains present and their specific binding partners, SOX proteins can either activate or repress the expression of target genes in a tissue-specific manner [1, 2, 5, 6]. Through their lineage-specific modulation of gene expression, SOX proteins are involved in embryonic development, regulating processes such as cell differentiation, maintenance of stemness, sex determination, and development of the central nervous, haematopoietic and other organ systems [1, 2, 5]. As SOX members are critical regulators of cellular programming, it is not surprising that disruption of these genes has been implicated in several human diseases including cancer [1, 2, 7].
Roles of SOX proteins in cancer and cancer-associated pathways
Embryonic development is a tightly regulated process involving differentiation of cells into specialized cell types and rapid cell growth. It is well established that numerous genes and pathways with essential roles in development are frequently disrupted to promote carcinogenesis, which itself is characterized by aberrant cell proliferation and/or differentiation. The disruption of SOX proteins in various malignancies is a case in point.
Oncogenic and suppressive roles of SOX proteins in tumorigenesis
SOX family members may act as oncogenes, tumor suppressor genes, or both depending on the cellular context, and can be activated or inactivated through a variety of genetic and epigenetic mechanisms including DNA copy number alterations, DNA methylation changes and aberrant miRNA expression [1, 2, 8, 9]. For example, in squamous esophageal, non-small cell and small cell lung cancers, SOX2 acts as an oncogene and is activated through DNA amplification [10, 11]. SOX2 promotes cell proliferation, anchorage independent growth and is capable of transforming transbronchial epithelial cells [10, 11]. SOX9 is another example of an oncogenic SOX protein that is overexpressed in multiple cancer types including colorectal, glioma, and pancreatic cancers [12-14]. In contrast, SOX7 is downregulated via DNA deletions and methylation silencing, acting as a tumor suppressor gene in prostate, colon, lung, and breast cancers through its involvement in cell death, movement, invasion and proliferation [15, 16]. SOX4 appears to have a context-dependent role in cancer as it is upregulated and promotes growth of leukemia, colorectal, lung and breast cancers, but is underexpressed and suppresses growth of bladder and liver cancers [9]. SOX4 is disrupted through DNA copy gains, epigenetic changes involving DNA methylation and miRNAs, and sequence mutations [8, 9]. It mediates its oncogenic function via several mechanisms including suppression of apoptosis, promotion of metastasis, and maintenance of cancer-initiating cells. Similar to SOX4, SOX2 and SOX9 have also been shown to have tumor suppressive effects in specific cancer types (gastric cancer and melanoma, respectively), further emphasizing the context specific nature of SOX involvement in carcinogenesis [17, 18]. The differing actions of SOX proteins in cancer cells of various origins and genetic backgrounds likely underlie the disparate behaviors of SOX proteins in promoting or inhibiting tumor growth [9].
SOX gene disruption in various cancer types
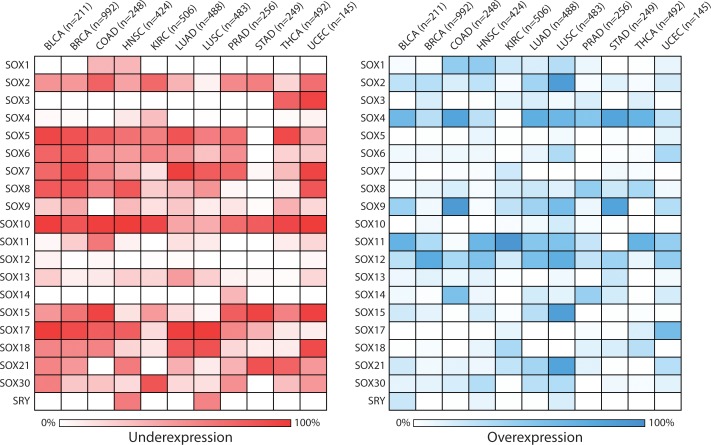
Not surprisingly given the known involvement of SOX members in cancer biology, a pan-cancer analysis of SOX expression using publically available RNA-sequencing data for 11 cancer types from The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov) revealed that SOX genes are frequently deregulated in several human malignancies (Figure 1, Supplemental Table 1). These results corroborate numerous reports of aberrant SOX gene disruption in various cancer types such as overexpression of SOX2 in lung squamous cell carcinoma [8], low expression of SOX10 in epithelial-derived carcinomas [19], as well as overexpression of SOX4 and SOX11 and underexpression of SOX7 and SOX17 in a variety of cancer types [1, 2, 9, 10, 16-18, 20-54]. Our analysis also identified deregulated SOX genes such as SOX12 and SOX30, which have not been well characterized in the context of cancer, suggesting these genes may be worthwhile candidates for further investigation. Many of these RNA expression changes have also been demonstrated at the protein level as evident in the Human Protein Atlas (e.g. SOX4, SOX7, SOX10, SOX17) [55].
Figure 1. Pan-cancer analysis of SOX gene expression levels.
Processed Cancer Genome Atlas (TCGA) SOX family gene expression data for tumor and non-malignant tissues from 11 different cancer types was downloaded from the UCSC Cancer Genomics Browser (https://genome-cancer.ucsc.edu/) [92]. The number of tumor samples for each cancer type is indicated in brackets. Gene expression was classified as over- or underexpressed in individual tumors if tumor expression was at least 2-fold more or less than the average expression of available tissue matched non-malignant samples, and the frequency of expression changes across tumors of each type was calculated. Darker coloration indicates a higher frequency of alteration as indicated in the legends below each heatmap, with underexpression depicted on the left in red and overexpression on the right in blue. Genes were considered recurrently, aberrantly expressed within a particular cancer type if they exhibited a 20% or greater frequency in disruption. Cancer types are annotated as follows: BLCA - bladder urothelial carcinoma; BRCA - breast invasive carcinoma; COAD - colon adenocarcinoma; HNSC - head and neck squamous cell carcinoma; KIRC - kidney clear cell carcinoma; LUAD - lung adenocarcinoma; LUSC - lung squamous cell carcinoma; PRAD - prostate adenocarcinoma; STAD - stomach adenocarcinoma; THCA - thyroid carcinoma; UCEC - uterine corpus endometrioid carcinoma.
The most recurrently deregulated SOX genes, arbitrarily defined here as having a minimum 20% frequency of deregulation in at least 9 of the 11 different cancer types, included underexpression of SOX2 (10/11), SOX5 (10/11), SOX6 (10/11), SOX7 (9/11), and SOX10 (11/11) and overexpression of SOX4 (10/11), SOX11 (9/11), and SOX12 (9/11) (Figure 1, Supplemental Table 1). Interestingly, although it has been suggested that SOX genes are predominantly oncogenic in cancer and we found that many were overexpressed [8], we also observed recurrent underexpression of SOX genes in tumors relative to matched non-malignant tissues. In the 11 TCGA cancer types we considered, 13/20 SOX genes showed transcriptional downregulation (≥20%) whereas only 5/20 showed recurrent upregulation (≥20%) in at least 5 cancer types. Of the 13 recurrently underexpressed SOX genes, SOX3, SOX5, SOX7 and SOX10 were exclusively underexpressed (i.e. they showed overexpression frequencies <20% in all cancer types). We speculate that the prominence of SOX underexpression may be due to the functional redundancy of individual SOX genes [5, 7], as loss of function could require inactivation of multiple SOX family members. For example, SOX5 and SOX6, members of the SOXD family, were frequently underexpressed concurrently in several cancer types (Figure 1). Most SOX genes were either over- or underexpressed within individual cancer types, though some exceptions were evident. SOX2 and SOX9 both exhibited frequent over- and underexpression within the same cancer type, potentially indicating their dual roles in cancer and that they could be differentially selected for in cells with different genetic backgrounds. The SOX genes least often disrupted at the expression level were SOX14, SOX3, and SRY.
As mentioned above, several genetic and epigenetic mechanisms have been associated with aberrant SOX expression in cancer. A similar pan-cancer investigation of TCGA genomics data for the same tumor types revealed that SOX genes are recurrently disrupted through copy number and methylation changes, and infrequently by sequence mutations (Supplemental Table 1). DNA amplifications were more frequent than deletions, while DNA hypermethylation was more frequent than hypomethylation at SOX gene loci. It is possible that these DNA-level changes underlie the prominent SOX gene deregulation we observed, although we acknowledge that additional mechanisms likely contribute to SOX expression as well. The prevalence of DNA and RNA alterations affecting SOX genes in various tumor types is strong evidence of their importance to cancer biology.
Malignant phenotypes and cellular pathways modulated by SOX members in cancer
As described above, SOX proteins can contribute to the malignant phenotype through their abilities to regulate numerous cancer hallmarks including cell proliferation, apoptosis, survival, invasion, migration, differentiation, stemness, senescence, and angiogenesis [1, 2, 8, 9, 56]. In the context of cancer biology, Wnt/β-catenin signaling is the most well documented cellular pathway affected by SOX proteins. This pathway plays important roles in the development of multiple organs and is aberrantly activated in several cancers, driving both cell proliferation and metastasis [57, 58]. Activation of the Wnt pathway results in liberation of β-catenin from a cytoplasmic inhibitory complex, enabling it to translocate to the nucleus and bind to TCF, recruit transcriptional co-activators and stimulate expression of Wnt target genes. Numerous reports have demonstrated that SOX proteins positively (e.g. SOX2 and SOX4 in breast and colon cancers, respectively) or negatively (e.g. SOX9, SOX7, and SOX17 in colorectal cancer) regulate Wnt-mediated transcriptional activity through a variety of mechanisms, including: binding with β-catenin to prevent TCF from interacting with β-catenin, interacting with TCFs directly to inhibit them from binding β-catenin, competitive binding with TCF proteins for DNA sites, recruitment of transcriptional repressors or activators, stabilization of TCF repression, or promotion of β-catenin degradation [59, 60]. Interestingly, some SOX members (e.g. SOX21 and SOX9) have also been implicated in non-canonical Wnt signaling due to their modulation of planar cell polarity signaling (PCP), which is known to contribute to tumor progression and metastasis [61-64]. Work exploring the involvement of SOX proteins in cancer through their effects on PCP may provide additional insights into how SOX disruption promotes aggressive tumor phenotypes.
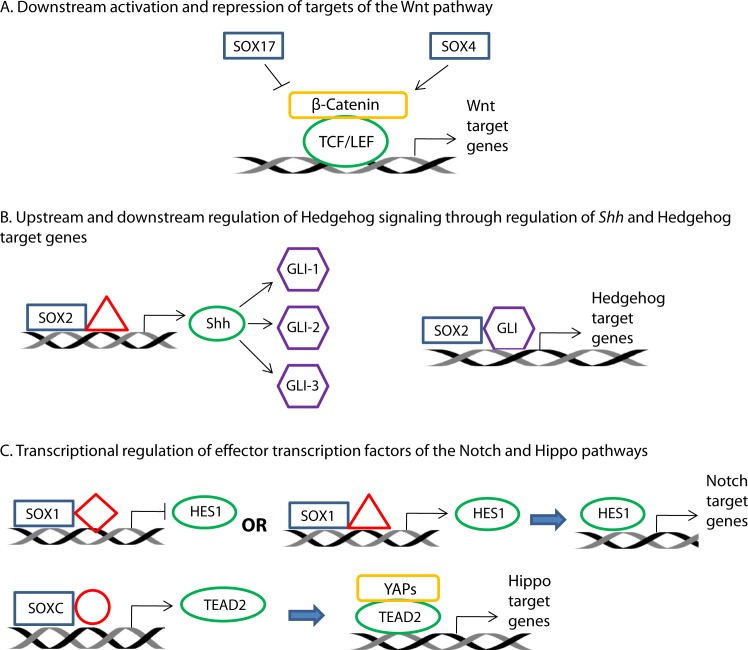
In addition to the Wnt/β-catenin pathway, SOX family members also have established roles in other developmental pathways including the Notch, Sonic Hedgehog, and Hippo pathways [5, 7, 65-68]. SOX genes can affect these pathways at both upstream (i.e. pathway stimulation) and downstream (i.e. transcriptional activity) levels, through transcriptional regulation of genes encoding pathway proteins or target genes, respectively. Figure 2 demonstrates known interactions between SOX proteins and developmental pathways implicated in carcinogenesis. SOX1 exhibits downstream regulation of the Notch pathway in neural progenitor cells by binding to the gene promoter of the HES1 transcription factor, repressing its transcription, thereby mitigating Notch signaling and promoting neuronal differentiation [69]. In contrast, through its transcriptional activation of HES1, SOX1 is involved in promoting the switch from neurogenesis to gliogenesis in the ventral spinal cord [70]. In the Hedgehog pathway, SOX9 and SOX2 partner with GLI transcription factors, the downstream effectors of Hedgehog signaling to activate expression of transcription factors required for cartilage development and spinal cord neural progenitor cells, respectively [65-67]. A similar example is evident in the regulation of organogenesis via the Hippo pathway, where SOXC factors (SOX4, SOX11, and SOX12) control the expression of the transcriptional mediator of the pathway, TEAD2 [71]. In the brain, SOX2 regulates the expression of sonic hedgehog (SHH), whose corresponding protein stimulates the Hedgehog signaling cascade; this example of upstream regulation is important for stem cell maintenance in brain development [72].
Figure 2. Involvement of SOX proteins in various developmental pathways that have been associated with tumorigenesis.
SOX members are involved in regulating several signaling pathways relevant to tumorigenesis, including the Wnt, Hedgehog, Notch, and Hippo pathways. (A) In the Wnt pathway, SOX proteins can bind β-catenin or TCF/LEF to either promote or suppress Wnt mediated transcriptional activity. (B) In the Hedgehog pathway, SOX proteins act upstream to control the expression of sonic hedgehog (Shh) and downstream through interaction with GLI to promote transcription of pathway target genes. (C) In the Notch and Hippo pathways, SOX proteins bind with other factors to control expression of their transcription factor effectors, HES1 and TEAD2, respectively.
Discovery of SOX15 as a potential tumor suppressor in pancreatic cancer
We recently identified SOX15 (also known as SOX20) as a potential tumor suppressor gene negatively associated with the Wnt/β-catenin pathway in pancreatic ductal adenocarcinoma (PDAC) [45]. A multi-dimensional, integrative genomic analysis of 20 PDAC cell lines revealed SOX15 was inactivated as a result of multiple molecular mechanisms. We observed recurrent two-hit inactivation, defined as concurrent copy number loss and DNA hypermethylation associated with underexpression within an individual sample, in 45% of the cell lines assessed. Our observation of SOX15 disruption was consistent with Knudson's two-hit hypothesis for tumor suppressor gene inactivation [73]. Following validation of SOX15 downregulation in clinical PDAC tumors, we performed experiments demonstrating that SOX15 exerts tumor suppressor properties in vitro and in vivo. Specifically, re-expression of SOX15 in PDAC lines with undetectable endogenous levels resulted in significantly reduced cell viability and tumor growth [45]. To deduce a potential mechanism through which SOX15 may exert its tumor suppressive effects, we turned our attention to the Wnt pathway since other SOX family members are known to regulate Wnt/β-catenin signaling and our pathway analysis suggested a potential role for SOX15 in this pathway. Multiple different assays revealed that SOX15 expression was associated with a modest but consistent reduction in Wnt pathway activity. Taken together, our findings provide novel evidence of the involvement of yet another SOX family member in the process of carcinogenesis.
The role of SOX15 in developmental and cancer biology
The role of SOX15 in cell biology and development is relatively understudied compared to other SOX family members, such as SOX2, SOX4 and SOX9. Early work demonstrated SOX15 expression in fetal brain, spinal cord, thymus, heart and adrenal tissues as well as in adult brain, lung, heart, liver, spleen, gut, small intestine, kidney, and testes tissues [74]. Knockout studies revealed that SOX15-null mice and embryonic stem cells are viable and grossly normal, perhaps suggesting functional redundancy with other SOX members [5, 7, 75, 76]. However, manipulation of SOX15 levels in mice results in muscular abnormalities, implicating SOX15 in the regulation of skeletal muscle development [75, 77-79]. More recent studies have suggested that SOX15 is a potential mediator of pluripotency and stemness due to its upregulation in induced pluripotent and embryonic stem cells and mesodermal progenitor cells [80, 81].
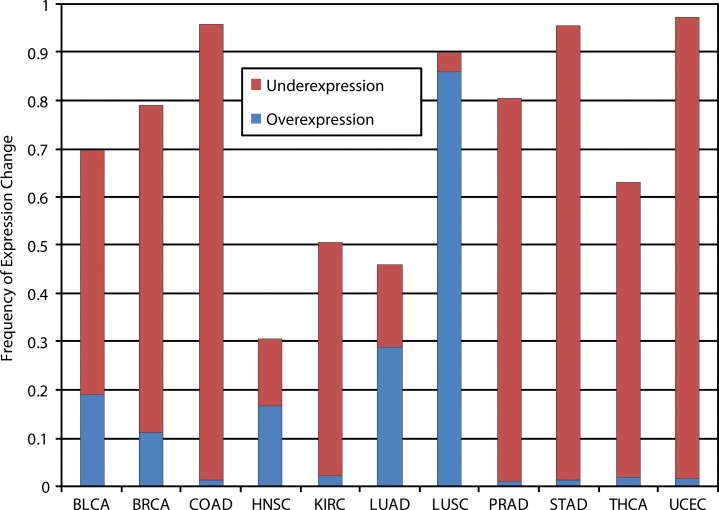
In cancer, SOX15 overexpression was found to inhibit the proliferation of human testicular embryonic carcinoma cells [82]. This negative regulation of cancer cell proliferation is consistent with our results of SOX15 expression reducing tumor growth, providing additional evidence of a potential suppressor role for SOX15 in tumorigenesis. Further supporting this concept, our pan-cancer analysis of SOX15 gene expression showed it was recurrently downregulated in multiple cancer types with high frequencies of disruption (≥ 80%), including colon, stomach, prostate, and uterine cancers (Figure 3). In contrast, SOX15 appeared to be frequently overexpressed in lung adenocarcinoma and squamous cell carcinoma, potentially exemplifying the tissue-specific dependency of SOX gene expression and function (Figure 3). Collectively, our pan-cancer expression analysis and functional validation of SOX15 in PDAC, suggests that SOX15 may be involved in multiple cancer types.
Figure 3. SOX15 expression status in various cancer types.
Frequency of SOX15 over- and underexpression in 11 cancer types, illustrating SOX15 is predominantly underexpressed in cancer with the exception of lung adenocarcinoma (LUAD) and squamous cell carcinoma (LUSC), in which it is frequently overexpressed. Cancer types are annotated as follows: BLCA - bladder urothelial carcinoma; BRCA - breast invasive carcinoma; COAD - colon adenocarcinoma; HNSC - head and neck squamous cell carcinoma; KIRC - kidney clear cell carcinoma; LUAD - lung adenocarcinoma; LUSC - lung squamous cell carcinoma; PRAD - prostate adenocarcinoma; STAD - stomach adenocarcinoma; THCA - thyroid carcinoma; UCEC - uterine corpus endometrioid carcinoma.
SOX15: Where do we go from here?
Much remains to be learned about SOX15 function in development, cell differentiation, and cancer. While we demonstrated a role for SOX15 in regulation of the Wnt/β-catenin pathway in PDAC, we acknowledge that the moderate suppression of Wnt activity does not reflect the large effect SOX15 expression had on inhibition of tumor growth, suggesting SOX15 may function through additional cellular pathways to mediate its inhibitory effect [45]. Moreover, since many SOX members have various roles in different cellular programs, it is possible that SOX15 is involved in normal biological processes other than just muscle development [5, 7]. We suggest three key avenues of research should be undertaken to elucidate novel roles of SOX15 in cell and developmental biology; these include identification of SOX15 transcriptional targets, understanding SOX15 expression patterns and transcriptional regulation, and identification of protein interacting partners.
Perhaps the most informative strategy for identifying novel SOX15 functions is to determine what genes it regulates (i.e. SOX15 target genes), appreciating that this may be cell-and tissue-dependent. A similar approach was recently used to infer the biological functions of SOX11 in mantle cell lymphoma and SOX2 in glioblastoma multiforme [83, 84]. SOX15 target genes could be revealed using a combination of approaches such as chromatin immunoprecipitation coupled with sequencing (ChIP-seq) to identify SOX15 DNA binding sites, and/or, genome wide expression profiling following manipulation of SOX15 levels to identify genes whose expression is strongly correlated to that of SOX15. We employed the latter approach to find cellular pathways associated with SOX15 in our PDAC study, and identified the Wnt and ERK5 signaling pathways as candidates for SOX15 regulation [45]. Although findings from genome wide approaches must be validated, they provide an excellent starting point for the identification of novel target genes.
Determining the spatial and temporal patterns of SOX15 expression throughout development will be extremely informative for identifying roles of SOX15 in developmental biology. Mapping of protein expression throughout mouse organogenesis provided insights into the role of SOX13 in multiple developmental processes, and this approach is a logical next step to further our understanding of SOX15 function [85]. Moreover, deciphering how SOX15 is transcriptionally regulated and understanding what cellular pathways or signals activate SOX15 gene expression could also reveal novel insights into its function. Evidence suggests that SOX gene expression can be controlled autonomously by other SOX factors, or regulated epigenetically, for example through DNA methylation or micro-RNA (miRNA) expression [5, 15, 17, 45, 80, 86, 87]; thus, knowing the biological roles of miRNAs governing SOX15 expression could possibly shed light on SOX15 function. We also do not overlook the possibility that post-translational modifications may play an important role in mediating SOX15 behavior, as has been observed for SOX2 [5].
Lastly, it is well documented that the activity of SOX transcription factors is highly dependent on the proteins they partner with to exert their effects [88, 89]. SOX proteins may bind with completely different proteins or other SOX members. SOXB1/C/F members bind to heterologous transcription factors; for example, SOX2 partners with OCT4 (also known as POU5F1) to maintain embryonic stem cell pluripotency [90]. On the other hand, SOX5 and SOX6 can dimerize and this binding enhances their ability to bind DNA [88, 91]. The finding of SOX proteins binding downstream protein components of the Wnt pathway (e.g. TCF or β-catenin) would implicate their involvement in Wnt signaling. Thus, clues about SOX15 function could come from discerning its protein binding partners using a variety of high throughput proteomic or immunoprecipitation strategies.
CONCLUSIONS
The reports of SOX transcription factors in the literature emphasize the critical roles SOX genes play in developmental and cancer biology. While some SOX proteins are well studied, we have barely scratched the surface in understanding the biological functions of many others, especially in the context of malignancy. Nevertheless, due to their regulation of stemness, pluripotency, developmental pathways, and numerous cancer processes, it is clear that SOX members are integral contributors to cancer biology. We have provided a snapshot of SOX deregulation in a variety of cancer types, revealing that SOX expression patterns are broadly, aberrantly expressed in cancer, heightening interest in several SOX members that are recurrently disrupted but have not yet been studied in carcinogenesis. We also summarized our recent finding of the relatively understudied SOX member, SOX15, as a potential tumor suppressor gene frequently inactivated in pancreatic cancer. Further work to study this gene in different cancer types and to elucidate additional mechanisms through which it may function is required to gain a better understanding of SOX15's physiological role in normal and diseased states, and could lead to the development of novel cancer therapeutic strategies [2].
SUPPLEMENTARY TABLE
Acknowledgments
The pan-cancer SOX gene expression results presented here are based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/). The authors would like to thank Dr. Victor Martinez for assisting with data acquisition and visualization. This work was supported by grants from the Canadian Institutes for Health Research, Canadian Cancer Society Research Institute, Terry Fox Foundation, PMH Foundation, Ontario Ministry of Health and Long Term Care, and Vanier Canada Graduate Scholarships to KLT, DDBS, NR and LAP. MST is the M Qasim Choksi Chair in Lung Cancer Translational Research.
Footnotes
Conflicts of Interest
The authors have no conflicts of interest to declare.
REFERENCES
- 1.Dong C, Wilhelm D, Koopman P. Sox genes and cancer. Cytogenetic and genome research. 2004;105(2-4):442–447. doi: 10.1159/000078217. [DOI] [PubMed] [Google Scholar]
- 2.Castillo SD M. The SOX family of genes in cancer development: biological relevance and opportunities for therapy. Expert opinion on therapeutic targets. 2012;16(9):903–919. doi: 10.1517/14728222.2012.709239. [DOI] [PubMed] [Google Scholar]
- 3.Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346(6281):245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- 4.Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Developmental biology. 2000;227(2):239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 5.Kamachi Y, Kondoh H. Sox proteins: regulators of cell fate specification and differentiation. Development. 2013;140(20):4129–4144. doi: 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- 6.Wegner M. All purpose Sox: The many roles of Sox proteins in gene expression. The international journal of biochemistry & cell biology. 2010;42(3):381–390. doi: 10.1016/j.biocel.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Sarkar A, Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell stem cell. 2013;12(1):15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Y, Li Y, Jun Wei JW, Liu X. The role of sox genes in lung morphogenesis and cancer. International journal of molecular sciences. 2012;13(12):15767–15783. doi: 10.3390/ijms131215767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vervoort SJ, van Boxtel R, Coffer PJ. The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe? Oncogene. 2013;32(29):3397–3409. doi: 10.1038/onc.2012.506. [DOI] [PubMed] [Google Scholar]
- 10.Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, Bergbower EA, Guan Y, Shin J, Guillory J, Rivers CS, Foo CK, Bhatt D, Stinson J, Gnad F, Haverty PM, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nature genetics. 2012;44(10):1111–1116. doi: 10.1038/ng.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, Ramos AH, Woo MS, Weir BA, Getz G, Beroukhim R, O'Kelly M, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nature genetics. 2009;41(11):1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L, He S, Yuan J, Mao X, Cao Y, Zong J, Tu Y, Zhang Y. Oncogenic role of SOX9 expression in human malignant glioma. Med Oncol. 2012;29(5):3484–3490. doi: 10.1007/s12032-012-0267-z. [DOI] [PubMed] [Google Scholar]
- 13.Kopp JL, von Figura G, Mayes E, Liu FF, Dubois CL, Morris JPt Pan, FC Akiyama, H Wright, CV Jensen, K Hebrok, M and Sander. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer cell. 2012;22(6):737–750. doi: 10.1016/j.ccr.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matheu A, Collado M, Wise C, Manterola L, Cekaite L, Tye AJ, Canamero M, Bujanda L, Schedl A, Cheah KS, Skotheim RI, Lothe RA, Lopez de Munain A, Briscoe J, Serrano M, Lovell-Badge R. Oncogenicity of the developmental transcription factor Sox9. Cancer research. 2012;72(5):1301–1315. doi: 10.1158/0008-5472.CAN-11-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stovall DB, Wan M, Miller LD, Cao P, Maglic D, Zhang Q, Stampfer MR, Liu W, Xu J, Sui G. The regulation of SOX7 and its tumor suppressive role in breast cancer. The American journal of pathology. 2013;183(5):1645–1653. doi: 10.1016/j.ajpath.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stovall DB, Cao P, Sui G. SOX7: From a developmental regulator to an emerging tumor suppressor. Histology and histopathology. 2014;29(4):439–445. doi: 10.14670/hh-29.10.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otsubo T, Akiyama Y, Yanagihara K, Yuasa Y. SOX2 is frequently downregulated in gastric cancers and inhibits cell growth through cell-cycle arrest and apoptosis. British journal of cancer. 2008;98(4):824–831. doi: 10.1038/sj.bjc.6604193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Passeron T, Valencia JC, Namiki T, Vieira WD, Passeron H, Miyamura Y, Hearing VJ. Upregulation of SOX9 inhibits the growth of human and mouse melanomas and restores their sensitivity to retinoic acid. The Journal of clinical investigation. 2009;119(4):954–963. doi: 10.1172/JCI34015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ordonez NG. Value of SOX10 immunostaining in tumor diagnosis. Advances in anatomic pathology. 2013;20(4):275–283. doi: 10.1097/PAP.0b013e318297a9d0. [DOI] [PubMed] [Google Scholar]
- 20.Chen QL, Zheng WL, Yao WJ, Nie LW, Cheng SH, Ma WL. Analysis of SOX4 gene mutation in non-small cell lung cancer tissues. Zhonghua yi xue yi chuan xue za zhi = Zhonghua yixue yichuanxue zazhi = Chinese journal of medical genetics. 2007;24(5):505–509. [PubMed] [Google Scholar]
- 21.Yin D, Jia Y, Yu Y, Brock MV, Herman JG, Han C, Su X, Liu Y, Guo M. SOX17 methylation inhibits its antagonism of Wnt signaling pathway in lung cancer. Discovery medicine. 2012;14(74):33–40. [PMC free article] [PubMed] [Google Scholar]
- 22.Sun M, Uozaki H, Hino R, Kunita A, Shinozaki A, Ushiku T, Hibiya T, Takeshita K, Isogai M, Takada K, Fukayama M. SOX9 expression and its methylation status in gastric cancer. Virchows Archiv: an international journal of pathology. 2012;460(3):271–279. doi: 10.1007/s00428-012-1201-7. [DOI] [PubMed] [Google Scholar]
- 23.Cox JL, Wilder PJ, Desler M, Rizzino A. Elevating SOX2 levels deleteriously affects the growth of medulloblastoma and glioblastoma cells. PloS one. 2012;7(8):e44087. doi: 10.1371/journal.pone.0044087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swartling FJ, Savov V, Persson AI, Chen J, Hackett CS, Northcott PA, Grimmer MR, Lau J, Chesler L, Perry A, Phillips JJ, Taylor MD, Weiss WA. Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer cell. 2012;21(5):601–613. doi: 10.1016/j.ccr.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgos-Ojeda D, Rueda BR, Buckanovich RJ. Ovarian cancer stem cell markers: prognostic and therapeutic implications. Cancer letters. 2012;322(1):1–7. doi: 10.1016/j.canlet.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang SS, Fang WT, Hou YH, Huang SF, Yen BL, Chang JL, Li SM, Liu HP, Liu YL, Huang CT, Li YW, Jang TH, Chan SH, Yang SJ, Hsiung CA, Wu CW, et al. Upregulation of SOX9 in lung adenocarcinoma and its involvement in the regulation of cell growth and tumorigenicity. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16(17):4363–4373. doi: 10.1158/1078-0432.CCR-10-0138. [DOI] [PubMed] [Google Scholar]
- 27.Li B, Ge Z, Song S, Zhang S, Yan H, Huang B, Zhang Y. Decreased expression of SOX7 is correlated with poor prognosis in lung adenocarcinoma patients. Pathology oncology research: POR. 2012;18(4):1039–1045. doi: 10.1007/s12253-012-9542-8. [DOI] [PubMed] [Google Scholar]
- 28.Sholl LM, Barletta JA, Yeap BY, Chirieac LR, Hornick JL. Sox2 protein expression is an independent poor prognostic indicator in stage I lung adenocarcinoma. The American journal of surgical pathology. 2010;34(8):1193–1198. doi: 10.1097/PAS.0b013e3181e5e024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yousem SA. Role of molecular studies in the diagnosis of lung adenocarcinoma. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2012;25(Suppl 1):S11–17. doi: 10.1038/modpathol.2011.156. [DOI] [PubMed] [Google Scholar]
- 30.Guo X, Xiong L, Sun T, Peng R, Zou L, Zhu H, Zhang J, Li H, Zhao J. Expression features of SOX9 associate with tumor progression and poor prognosis of hepatocellular carcinoma. Diagnostic pathology. 2012;7:44. doi: 10.1186/1746-1596-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Shi L, Zhang L, Li R, Liang J, Yu W, Sun L, Yang X, Wang Y, Zhang Y, Shang Y. The molecular mechanism governing the oncogenic potential of SOX2 in breast cancer. The Journal of biological chemistry. 2008;283(26):17969–17978. doi: 10.1074/jbc.M802917200. [DOI] [PubMed] [Google Scholar]
- 32.Fu DY, Wang ZM, Li C, Wang BL, Shen ZZ, Huang W, Shao ZM. Sox17, the canonical Wnt antagonist, is epigenetically inactivated by promoter methylation in human breast cancer. Breast cancer research and treatment. 2010;119(3):601–612. doi: 10.1007/s10549-009-0339-8. [DOI] [PubMed] [Google Scholar]
- 33.Acloque H, Ocana OH, Matheu A, Rizzoti K, Wise C, Lovell-Badge R, Nieto MA. Reciprocal repression between Sox3 and snail transcription factors defines embryonic territories at gastrulation. Developmental cell. 2011;21(3):546–558. doi: 10.1016/j.devcel.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu P, Ramachandran S, Ali Seyed M, Scharer CD, Laycock N, Dalton WB, Williams H, Karanam S, Datta MW, Jaye DL, Moreno CS. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer research. 2006;66(8):4011–4019. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- 35.Scharer CD, McCabe CD, Ali-Seyed M, Berger MF, Bulyk ML, Moreno CS. Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer research. 2009;69(2):709–717. doi: 10.1158/0008-5472.CAN-08-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, McKnight NC, Zhang T, Lu ML, Balk SP, Yuan X. SOX9 is expressed in normal prostate basal cells and regulates androgen receptor expression in prostate cancer cells. Cancer research. 2007;67(2):528–536. doi: 10.1158/0008-5472.CAN-06-1672. [DOI] [PubMed] [Google Scholar]
- 37.Zhong WD, Qin GQ, Dai QS, Han ZD, Chen SM, Ling XH, Fu X, Cai C, Chen JH, Chen XB, Lin ZY, Deng YH, Wu SL, He HC, Wu CL. SOXs in human prostate cancer: implication as progression and prognosis factors. BMC cancer. 2012;12:248. doi: 10.1186/1471-2407-12-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sinner D, Kordich JJ, Spence JR, Opoka R, Rankin S, Lin SC, Jonatan D, Zorn AM, Wells JM. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Molecular and cellular biology. 2007;27(22):7802–7815. doi: 10.1128/MCB.02179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azhikina T, Kozlova A, Skvortsov T, Sverdlov E. Heterogeneity and degree of TIMP4, GATA4, SOX18, and EGFL7 gene promoter methylation in non-small cell lung cancer and surrounding tissues. Cancer genetics. 2011;204(9):492–500. doi: 10.1016/j.cancergen.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 40.Castillo SD, Matheu A, Mariani N, Carretero J, Lopez-Rios F, Lovell-Badge R, Sanchez-Cespedes M. Novel transcriptional targets of the SRY-HMG box transcription factor SOX4 link its expression to the development of small cell lung cancer. Cancer research. 2012;72(1):176–186. doi: 10.1158/0008-5472.CAN-11-3506. [DOI] [PubMed] [Google Scholar]
- 41.Chen S, Xu Y, Chen Y, Li X, Mou W, Wang L, Liu Y, Reisfeld RA, Xiang R, Lv D, Li N. SOX2 gene regulates the transcriptional network of oncogenes and affects tumorigenesis of human lung cancer cells. PloS one. 2012;7(5):e36326. doi: 10.1371/journal.pone.0036326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, Wang J, Xu Z, Ahmad A, Li E, Wang Y, Qin S, Wang Q. Expression of sox2 and oct4 and their clinical significance in human non-small-cell lung cancer. International journal of molecular sciences. 2012;13(6):7663–7675. doi: 10.3390/ijms13067663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maddison P, Thorpe A, Silcocks P, Robertson JF, Chapman CJ. Autoimmunity to SOX2, clinical phenotype and survival in patients with small-cell lung cancer. Lung Cancer. 2010;70(3):335–339. doi: 10.1016/j.lungcan.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 44.Medina PP, Castillo SD, Blanco S, Sanz-Garcia M, Largo C, Alvarez S, Yokota J, Gonzalez-Neira A, Benitez J, Clevers HC, Cigudosa JC, Lazo PA, Sanchez-Cespedes M. The SRY-HMG box gene, SOX4, is a target of gene amplification at chromosome 6p in lung cancer. Human molecular genetics. 2009;18(7):1343–1352. doi: 10.1093/hmg/ddp034. [DOI] [PubMed] [Google Scholar]
- 45.Thu KL, Radulovich N, Becker-Santos DD, Pikor LA, Pusic A, Lockwood WW, Lam WL, Tsao MS. SOX15 is a candidate tumor suppressor in pancreatic cancer with a potential role in Wnt/beta-catenin signaling. Oncogene. 2014;33(3):279–288. doi: 10.1038/onc.2012.595. [DOI] [PubMed] [Google Scholar]
- 46.Titulaer MJ, Klooster R, Potman M, Sabater L, Graus F, Hegeman IM, Thijssen PE, Wirtz PW, Twijnstra A, Smitt PA, van der Maarel SM, Verschuuren JJ. SOX antibodies in small-cell lung cancer and Lambert-Eaton myasthenic syndrome: frequency and relation with survival. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009;27(26):4260–4267. doi: 10.1200/JCO.2008.20.6169. [DOI] [PubMed] [Google Scholar]
- 47.Wilbertz T, Wagner P, Petersen K, Stiedl AC, Scheble VJ, Maier S, Reischl M, Mikut R, Altorki NK, Moch H, Fend F, Staebler A, Bass AJ, Meyerson M, Rubin MA, Soltermann A, et al. SOX2 gene amplification and protein overexpression are associated with better outcome in squamous cell lung cancer. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2011;24(7):944–953. doi: 10.1038/modpathol.2011.49. [DOI] [PubMed] [Google Scholar]
- 48.Xiang R, Liao D, Cheng T, Zhou H, Shi Q, Chuang TS, Markowitz D, Reisfeld RA, Luo Y. Downregulation of transcription factor SOX2 in cancer stem cells suppresses growth and metastasis of lung cancer. British journal of cancer. 2011;104(9):1410–1417. doi: 10.1038/bjc.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin YW, Tsao CM, Yu PN, Shih YL, Lin CH, Yan MD. SOX1 suppresses cell growth and invasion in cervical cancer. Gynecologic oncology. 2013;131(1):174–181. doi: 10.1016/j.ygyno.2013.07.111. [DOI] [PubMed] [Google Scholar]
- 50.Choi YJ, Song JH, Yoon JH, Choi WS, Nam SW, Lee JY, Park WS. Aberrant expression of SOX9 is associated with gastrokine 1 inactivation in gastric cancers. Gastric cancer: official journal of the International Gastric Cancer Association and the Japanese Gastric Cancer Association. 2014;17(2):247–254. doi: 10.1007/s10120-013-0277-3. [DOI] [PubMed] [Google Scholar]
- 51.Hayano T, Garg M, Yin D, Sudo M, Kawamata N, Shi S, Chien W, Ding LW, Leong G, Mori S, Xie D, Tan P, Koeffler HP. SOX7 is down-regulated in lung cancer. Journal of experimental & clinical cancer research: CR. 2013;32:17. doi: 10.1186/1756-9966-32-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shivane A, Parkinson DB, Ammoun S, Hanemann CO. Expression of c-Jun and Sox-2 in human schwannomas and traumatic neuromas. Histopathology. 2013;62(4):651–656. doi: 10.1111/his.12062. [DOI] [PubMed] [Google Scholar]
- 53.Zhang S, Zhu C, Zhu L, Liu H, Liu S, Zhao N, Wu J, Huang X, Zhang Y, Jin J, Ji T, Ding X. Oncogenicity of the transcription factor SOX8 in hepatocellular carcinoma. Med Oncol. 2014;31(4):918. doi: 10.1007/s12032-014-0918-3. [DOI] [PubMed] [Google Scholar]
- 54.Naujokas A, Charli-Joseph Y, Ruben BS, Yeh I, LeBoit PE, McCalmont TH, Pincus LB. SOX-10 expression in cutaneous myoepitheliomas and mixed tumors. Journal of cutaneous pathology. 2014;41(4):353–363. doi: 10.1111/cup.12279. [DOI] [PubMed] [Google Scholar]
- 55.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, Wernerus H, Bjorling L, Ponten F. Towards a knowledge-based Human Protein Atlas. Nature biotechnology. 2010;28(12):1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 56.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 57.Polakis P. Wnt signaling in cancer. Cold Spring Harbor perspectives in biology. 2012;4(5) doi: 10.1101/cshperspect.a008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nature reviews Cancer. 2013;13(1):11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 59.Bernard P, Harley VR. Acquisition of SOX transcription factor specificity through protein-protein interaction, modulation of Wnt signalling and post-translational modification. The international journal of biochemistry & cell biology. 2010;42(3):400–410. doi: 10.1016/j.biocel.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 60.Kormish JD, Sinner D, Zorn AM. Interactions between SOX factors and Wnt/beta-catenin signaling in development and disease. Developmental dynamics: an official publication of the American Association of Anatomists. 2010;239(1):56–68. doi: 10.1002/dvdy.22046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jessen JR. Noncanonical Wnt signaling in tumor progression and metastasis. Zebrafish. 2009;6(1):21–28. doi: 10.1089/zeb.2008.0571. [DOI] [PubMed] [Google Scholar]
- 62.Wang Y. Wnt/Planar cell polarity signaling: a new paradigm for cancer therapy. Molecular cancer therapeutics. 2009;8(8):2103–2109. doi: 10.1158/1535-7163.MCT-09-0282. [DOI] [PubMed] [Google Scholar]
- 63.Hosoya M, Fujioka M, Matsuda S, Ohba H, Shibata S, Nakagawa F, Watabe T, Wakabayashi K, Saga Y, Ogawa K, Okano HJ, Okano H. Expression and function of Sox21 during mouse cochlea development. Neurochemical research. 2011;36(7):1261–1269. doi: 10.1007/s11064-011-0416-3. [DOI] [PubMed] [Google Scholar]
- 64.Darido C, Buchert M, Pannequin J, Bastide P, Zalzali H, Mantamadiotis T, Bourgaux JF, Garambois V, Jay P, Blache P, Joubert D, Holle F. Defective claudin-7 regulation by Tcf-4 and Sox-9 disrupts the polarity and increases the tumorigenicity of colorectal cancer cells. Cancer research. 2008;68(11):4258–4268. doi: 10.1158/0008-5472.CAN-07-5805. [DOI] [PubMed] [Google Scholar]
- 65.Leung VY, Gao B, Leung KK, Melhado IG, Wynn SL, Au TY, Dung NW, Lau JY, Mak AC, Chan D, Cheah KS. SOX9 governs differentiation stage-specific gene expression in growth plate chondrocytes via direct concomitant transactivation and repression. PLoS genetics. 2011;7(11):e1002356. doi: 10.1371/journal.pgen.1002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oosterveen T, Kurdija S, Alekseenko Z, Uhde CW, Bergsland M, Sandberg M, Andersson E, Dias JM, Muhr J, Ericson J. Mechanistic differences in the transcriptional interpretation of local and long-range Shh morphogen signaling. Developmental cell. 2012;23(5):1006–1019. doi: 10.1016/j.devcel.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 67.Peterson KA, Nishi Y, Ma W, Vedenko A, Shokri L, Zhang X, McFarlane M, Baizabal JM, Junker JP, van Oudenaarden A, Mikkelsen T, Bernstein BE, Bailey TL, Bulyk ML, Wong WH, McMahon AP. Neural-specific Sox2 input and differential Gli-binding affinity provide context and positional information in Shh-directed neural patterning. Genes & development. 2012;26(24):2802–2816. doi: 10.1101/gad.207142.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Corada M, Orsenigo F, Morini MF, Pitulescu ME, Bhat G, Nyqvist D, Breviario F, Conti V, Briot A, Iruela-Arispe ML, Adams RH, Dejana E. Sox17 is indispensable for acquisition and maintenance of arterial identity. Nature communications. 2013;4:2609. doi: 10.1038/ncomms3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kan L, Israsena N, Zhang Z, Hu M, Zhao LR, Jalali A, Sahni V, Kessler JA. Sox1 acts through multiple independent pathways to promote neurogenesis. Developmental biology. 2004;269(2):580–594. doi: 10.1016/j.ydbio.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 70.Genethliou N, Panayiotou E, Panayi H, Orford M, Mean R, Lapathitis G, Gill H, Raoof S, De Gasperi R, Elder G, Kessaris N, Richardson WD, Malas S. SOX1 links the function of neural patterning and Notch signalling in the ventral spinal cord during the neuron-glial fate switch. Biochemical and biophysical research communications. 2009;390(4):1114–1120. doi: 10.1016/j.bbrc.2009.08.154. [DOI] [PubMed] [Google Scholar]
- 71.Bhattaram P, Penzo-Mendez A, Sock E, Colmenares C, Kaneko KJ, Vassilev A, Depamphilis ML, Wegner M, Lefebvre V. Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nature communications. 2010;1:9. doi: 10.1038/ncomms1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Favaro R, Valotta M, Ferri AL, Latorre E, Mariani J, Giachino C, Lancini C, Tosetti V, Ottolenghi S, Taylor V, Nicolis SK. Hippocampal development and neural stem cell maintenance require Sox2-dependent regulation of Shh. Nature neuroscience. 2009;12(10):1248–1256. doi: 10.1038/nn.2397. [DOI] [PubMed] [Google Scholar]
- 73.Berger AH, Knudson AG, Polfi PP. A continuum model for tumour suppression. Nature. 2011;476(7359):163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vujic M, Rajic T, Goodfellow PN, Stevanovic M. cDNA characterization and high resolution mapping of the human SOX20 gene. Mammalian genome: official journal of the International Mammalian Genome Society. 1998;9(12):1059–1061. doi: 10.1007/s003359900925. [DOI] [PubMed] [Google Scholar]
- 75.Lee HJ, Goring W, Ochs M, Muhlfeld C, Steding G, Paprotta I, Engel W, Adham IM. Sox15 is required for skeletal muscle regeneration. Molecular and cellular biology. 2004;24(19):8428–8436. doi: 10.1128/MCB.24.19.8428-8436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maruyama M, Ichisaka T, Nakagawa M, Yamanaka S. Differential roles for Sox15 and Sox2 in transcriptional control in mouse embryonic stem cells. The Journal of biological chemistry. 2005;280(26):24371–24379. doi: 10.1074/jbc.M501423200. [DOI] [PubMed] [Google Scholar]
- 77.Beranger F, Mejean C, Moniot B, Berta P, Vromme M. Muscle differentiation is antagonized by SOX15, a new member of the SOX protein family. The Journal of biological chemistry. 2000;275(21):16103–16109. doi: 10.1074/jbc.275.21.16103. [DOI] [PubMed] [Google Scholar]
- 78.Meeson AP, Shi X, Alexander MS, Williams RS, Allen RE, Jiang N, Adham IM, Goetsch SC, Hammer RE, Garry DJ. Sox15 and Fhl3 transcriptionally coactivate Foxk1 and regulate myogenic progenitor cells. The EMBO journal. 2007;26(7):1902–1912. doi: 10.1038/sj.emboj.7601635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Savage J, Conley AJ, Blais A, Skerjanc IS. SOX15 and SOX7 differentially regulate the myogenic program in P19 cells. Stem Cells. 2009;27(6):1231–1243. doi: 10.1002/stem.57. [DOI] [PubMed] [Google Scholar]
- 80.Nishino K, Toyoda M, Yamazaki-Inoue M, Makino H, Fukawatase Y, Chikazawa E, Takahashi Y, Miyagawa Y, Okita H, Kiyokawa N, Akutsu H, Umezawa A. Defining hypo-methylated regions of stem cell-specific promoters in human iPS cells derived from extra-embryonic amnions and lung fibroblasts. PloS one. 2010;5(9):e13017. doi: 10.1371/journal.pone.0013017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pacini S, Carnicelli V, Trombi L, Montali M, Fazzi R, Lazzarini E, Giannotti S, Petrini M. Constitutive expression of pluripotency-associated genes in mesodermal progenitor cells (MPCs) PloS one. 2010;5(3):e9861. doi: 10.1371/journal.pone.0009861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yan HT, Shinka T, Sato Y, Yang XJ, Chen G, Sakamoto K, Kinoshita K, Aburatani H, Nakahori Y. Overexpression of SOX15 inhibits proliferation of NT2/D1 cells derived from a testicular embryonal cell carcinoma. Molecules and cells. 2007;24(3):323–328. [PubMed] [Google Scholar]
- 83.Vegliante MC, Palomero J, Perez-Galan P, Roue G, Castellano G, Navarro A, Clot G, Moros A, Suarez-Cisneros H, Bea S, Hernandez L, Enjuanes A, Jares P, Villamor N, Colomer D, Martin-Subero JI, et al. SOX11 regulates PAX5 expression and blocks terminal B-cell differentiation in aggressive mantle cell lymphoma. Blood. 2013;121(12):2175–2185. doi: 10.1182/blood-2012-06-438937. [DOI] [PubMed] [Google Scholar]
- 84.Fang X, Yoon JG, Li L, Yu W, Shao J, Hua D, Zheng S, Hood L, Goodlett DR, Foltz G, Lin B. The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC genomics. 2011;12:11. doi: 10.1186/1471-2164-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Y, Ristevski S, Harley VR. SOX13 exhibits a distinct spatial and temporal expression pattern during chondrogenesis, neurogenesis, and limb development. The journal of histochemistry and cytochemistry: official journal of the Histochemistry Society. 2006;54(12):1327–1333. doi: 10.1369/jhc.6A6923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Biswal BK, Beyrouthy MJ, Hever-Jardine MP, Armstrong D, Tomlinson CR, Christensen BC, Marsit CJ, Spinella MJ. Acute hypersensitivity of pluripotent testicular cancer-derived embryonal carcinoma to low-dose 5-aza deoxycytidine is associated with global DNA Damage-associated p53 activation, anti-pluripotency and DNA demethylation. PloS one. 2012;7(12):e53003. doi: 10.1371/journal.pone.0053003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137(4):647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 88.Kamachi Y, Uchikawa M, Kondoh H. Pairing SOX off: with partners in the regulation of embryonic development. Trends in genetics: TIG. 2000;16(4):182–187. doi: 10.1016/s0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- 89.Kondoh H, Kamachi Y. SOX-partner code for cell specification: Regulatory target selection and underlying molecular mechanisms. The international journal of biochemistry & cell biology. 2010;42(3):391–399. doi: 10.1016/j.biocel.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 90.Chambers I, Tomlinson SR. The transcriptional foundation of pluripotency. Development. 2009;136(14):2311–2322. doi: 10.1242/dev.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lefebvre V, Li P, de Crombrugghe B. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. The EMBO journal. 1998;17(19):5718–5733. doi: 10.1093/emboj/17.19.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goldman M, Craft B, Swatloski T, Ellrott K, Cline M, Diekhans M, Ma S, Wilks C, Stuart J, Haussler D, Zhu J. The UCSC Cancer Genomics Browser: update 2013. Nucleic acids research. 2013;41:D949–954. doi: 10.1093/nar/gks1008. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.