Abstract
Microphthalmia-associated transcription factor (MITF) acts via pigment epithelium-derived factor (PEDF), an antiangiogenic protein, to regulate retinal pigment epithelium migration. PEDF expression and/or regulation during melanoma development have not been investigated previously. Using immunohistochemistry, we determined expression of PEDF in common and dysplastic melanocytic nevi, melanoma in situ, invasive melanoma, and metastatic melanoma (n = 102). PEDF expression was consistently decreased in invasive and metastatic melanoma, compared with nevi and melanoma in situ (P < 0.0001). PEDF was lost in thicker melanomas (P = 0.003), and correlated with depth of invasion (P = 0.003) and distant metastasis (P = 0.0331), but only marginally with mitotic index, AJCC stage, nodal metastasis, or blood vascular density (0.05 < P < 0.10). Quantitative real-time PCR and microarray analyses confirmed PEDF down-regulation at the mRNA level in several melanoma lines, compared with melanocytes. MITF positively correlated with PEDF expression in invasive melanomas (P = 0.0003). Searching for PEDF regulatory mechanisms revealed two occupied conserved E-boxes (DNA recognition elements) in the first intron of the human and mouse PEDF promoter regions, confirmed by binding assays. Dominant-negative and siRNA approaches in vivo demonstrated direct transcriptional influence of MITF on PEDF, establishing the PEDF gene (SERPINF1) as a MITF target in melanocytes and melanoma cells. These findings suggest that loss of PEDF expression promotes early invasive melanoma growth.
The incidence and mortality of melanoma have continually increased for decades in the United States, and the difficulties of clinical management remain a significant challenge.1 Genetically, melanoma is a highly heterogeneous disease exemplified by numerous recurrent chromosomal rearrangements, gene amplifications, point mutations, and a highly complex miRNA transcriptome,2 as well as by interactions with the environment.3,4 Characterization of these genetic events has defined several key signaling pathways involved in melanoma development, leading to rational targeted therapies5,6 and immunotherapy breakthroughs.7
Cutaneous melanoma has a propensity for early microscopic dissemination to sentinel lymph nodes. Several investigators have independently demonstrated that nodal metastasis is associated with lymphangiogenesis (ie, formation of new lymphatic vessels) in human melanoma8–12 and in mouse xenograft models.13,14 Importantly, lymphatic invasion, defined as the presence of tumor cells within intraluminal lymphatic spaces, may represent an early step whereby melanoma cells gain entry into lymphatic vessels by interacting with newly formed vessels in the dermis.15 Indeed, lymphatic invasion was strongly associated with melanoma metastasis to sentinel lymph nodes in a cohort of 94 patients16 and with poorer survival in a cohort of 251 patients.17 By contrast, the role of tumor angiogenesis (ie, formation of new blood vessels) in melanoma progression and as a prognostic marker has been shown to be less significant. A recent systematic and meta-analysis of angiogenesis, lymphangiogenesis, and lymphatic invasion in melanoma, based on 26 published studies in 1695 patients, demonstrated that lymphangiogenesis and lymphatic invasion (but not angiogenesis) had prognostic values in these patients.18
Although lymphatic dissemination may reflect the metastatic propensity of melanoma, tumor angiogenesis has a major influence on melanoma growth, but the balance of secreted factors promoting or inhibiting angiogenesis in melanoma is yet to be defined.19 Pigment epithelium-derived factor (PEDF), a serine protease inhibitor (serpin) encoded by SERPINF1 (alias PEDF), is a potent inhibitor of angiogenesis,20 one that can antagonize vascular endothelial growth factor (VEGF).21 Originally, the PEDF protein, isolated from cultured retinal pigment epithelial (RPE) cells, was shown to have neurotrophic activity on retinoblastoma tumor cells.22 Given that PEDF is a ligand for four diverse receptors, receptor binding of dysregulated PEDF can cause a wide range of pathological conditions, including eye neovascularization, lipid metabolism, cardiovascular disease, and cancer (reviewed by Craword et al23). PEDF has been shown to have antitumorigenic and antimetastatic activities in a number of human malignancies, including melanoma, and in mouse xenograft models (reviewed by Becerra and Notario24). For example, overexpression of PEDF decreases angiogenesis and can inhibit melanoma growth25 and metastasis26 in mouse xenograft models. Moreover, it has been shown by immunohistochemistry (IHC) that PEDF is expressed in human melanocytic tumors (melanocytic nevi and melanoma), but not in nonmelanocytic tumors (eg, carcinomas of various organs).27 No previous study has investigated the expression of PEDF during the various stages of melanoma development or the mechanism of its cell-specific regulated expression.
Melanocytes express multiple lineage markers and microphthalmia-associated transcription factor (MITF), which encodes a Myc-related transcription factor essential for melanocyte development.28,29 In humans, mutations of MITF produce Waardenburg syndrome type 2A, a condition associated with melanocyte deficiencies in the hair follicle, skin, and inner ear.29 MITF is part of the MiT family of transcription factors that includes TFEB, TFEC, and TFE3, which are able to homodimerize and heterodimerize with each other; MITF appears to be the dominant family member within melanocytes. MITF regulates target gene expression through the binding of the canonical E-box promoter sequence CACGTG or the nonpalindromic sequence CACATG.28,29 Mutational analyses suggest that MITF mediates several discrete functions in the melanocyte lineage, including regulation of differentiation as well as proliferation and survival.30 Among the transcriptional targets believed to be regulated by MITF are survival factors (BCL2, BCL2A1), pigmentation enzyme-related genes, the metabolic regulator PGC1α, DNA repair machinery, and factors involved in cellular migration or invasion.
Despite the established transcription factor–target gene relationship between MITF and PEDF in RPE cells,31 this relationship has not previously been explored in melanocytes or in the various stages of melanoma development. In the present study, we identified PEDF as a direct transcriptional target of MITF in melanocytes and in melanoma cell lines using genomic and biochemical analyses. Our IHC analysis of a series of human clinical specimens (n = 102) spanning the spectrum of benign melanocytic nevi to metastatic melanoma identified PEDF down-regulation during melanoma progression and metastasis. PEDF expression correlated inversely with tumor thickness, a known prognostic indicator of survival.
Materials and Methods
Immunofluorescence and IHC
Cultured primary human melanocytes were double-stained using a mouse monoclonal antibody against PEDF (dilution 1:100; Chemicon; EMD Millipore, Billerica, MA), followed by incubation with the respective secondary antibodies labeled with either Texas Red or fluorescein isothiocyanate (both at dilution 1:50; Jackson ImmunoResearch, West Grove, PA), as described previously.32 Cell nuclei were counterstained with Hoechst bisbenzimide (Sigma-Aldrich, St. Louis, MO) at 20 μg/mL.
We used 102 formalin-fixed, paraffin-embedded specimens from 102 patients having undergone a biopsy and/or excision of a melanocytic lesion from the Pathology Departments of Brigham and Women’s Hospital, Massachusetts General Hospital, or Stanford University Medical Center. These specimens comprised common melanocytic nevi (CN) (n = 13), dysplastic melanocytic nevi (n = 3), melanoma in situ (MIS) (n = 13), primary cutaneous melanomas (PCM, n = 61), and metastatic melanomas to skin or lymph node (MM) (n = 12). The diagnoses were confirmed by two board-certified pathologists or dermatopathologists. The institutional review boards of the three hospitals approved this protocol. Formalin-fixed, paraffin-embedded sections (4 μm thick) were cut and mounted on Superfrost Plus slides (Thermo Fisher Scientific, Waltham, MA). The sections were then deparaffinized and hydrated by passing through xylene and a graded series of ethanol. Antigen retrieval was performed for 30 minutes at 37°C by trypsinization, and then endogenous peroxidase activity was inhibited in 0.3% hydrogen peroxide for 10 minutes. After blocking for 10 minutes in 0.75% normal horse serum, the sections were incubated with mouse monoclonal PEDF antibody (dilution 1:100; Chemicon; EMD Millipore) for 1 hour at room temperature or MITF antibody as previous described.33 We developed the immunohistochemical stain using an ABC Elite kit (Dako, Carpinteria, CA) at 1:200 dilution for 1.5 hours, according to the manufacturer’s instructions. Hematoxylin was used for counterstaining. During antibody optimization, a negative control (no primary antibody) was included; this was completely negative (data not shown). The final slides were examined using a Nikon E-600 microscope (Nikon, Melville, NY); digital images were captured using a SPOT Imaging Solutions digital camera (Diagnostic Instruments, Sterling Heights, MI).
Immunoscoring and Statistical Analysis
A semi-quantitative, four-point ordinal immunoreactivity score was established. A PEDF signal detected in <10% of tumor cells was scored as negative reactivity (0); positive reactivity was defined as a PEDF signal detected in ≥11% of tumor cells and was scored as 1 for weakly positive staining in epidermal or follicular keratinocytes, 2 for moderately positive staining in epidermal or follicular keratinocytes, and 3 (strongly positive) for diffuse and homogeneous staining in the cytoplasm and nucleolus. Immunostaining of tumor cells was scored in comparison with adjacent keratinocytes (internal positive control). Melanophages (specialized macrophages containing dark, brown coarse melanin pigment) were not scored. Two investigators (S.S.D. and G.R.) scored the stained slides independently. Individual and repeated scores were averaged, and final PEDF values were categorized into a three-level grouping of negative, low (>0 and ≤1.5), or high (≥1.6) for analyses that included all 102 lesions (Table 1).
Table 1.
PEDF Expression among Melanoma Diagnostic Groups
| Diagnostic group | Sample size | PEDF immunoreactivity, no. (%) |
|||
|---|---|---|---|---|---|
| Negative | Low, >0 and ≤1.5 | High, ≥1.6 | Means ± SD | ||
| Common melanocytic nevus | n = 13 | 0 (0) | 9 (69) | 4 (31) | 1.63 ± 0.63 |
| Dysplastic melanocytic nevus | n = 3 | 0 (0) | 3 (100) | 0 (0) | 0.5 ± 0 |
| Melanoma in situ | n = 13 | 2 (15) | 2 (15) | 9 (70) | 2.19 ± 1.82 |
| Primary cutaneous melanoma | n = 61 | 16 (26) | 36 (59) | 9 (15) | 0.93 ± 0.87 |
| Metastatic melanoma | n = 12 | 6 (50) | 6 (50) | 0 (0) | 0.33 ± 0.44 |
| Total | 102 | 24 (100) | 56 (100) | 22 (100) | |
Percentages have been rounded to add up to 100.
Boxplots were generated and Kruskal–Wallis testing was performed for overall comparison among all diagnostic groups. Bonferroni procedure, based on the ranks of the observations, was used for multiple pairwise comparisons (family level of significance α = 0.10). The relationship between PEDF expression and clinicopathological variables was examined by using both a three-level grouping (negative, ≤1.5, or ≥1.6) and raw PEDF immunoreactivity scores (0 to 3). These variables included age, sex, anatomic location, histological subtype, tumor thickness, ulceration, mitotic index (/mm2), Clark’s level, regression, AJCC stage (I, II, III, IV), nodal metastases (sentinel and nonsentinel lymph nodes), organ metastasis, distant metastasis (to skin and soft tissue), death of disease, and angiogenic parameters, as well as MITF immunoreactivity. We used Kendall’s τB correlation coefficient to evaluate the association between PEDF expression and ordinal variables and used Mantel–Haenszel χ2 test for nonordinal variables (sex, anatomic location, and histological subtype). The unpaired Student’s t-test was used to determine statistical significance (P value) of the association between mean tumor thickness and PEDF expression. The correlation between PEDF and MITF immunoreactivity was measured by both Kendall’s τB and the CORREL function of Microsoft Excel 2011 with two-tailed Student’s t-test.
Cell Culture and Media
Human melanoma cell lines were obtained from the National Cancer Institute (NIH–NCI, Rockville, MD) or were purchased from ATCC (Manassas, VA). The ARPE19 human RPE cell line (ATCC) and UACC62 cell line (NCI) were cultured in Ham’s F10 with 10% fetal bovine serum. Human foreskin primary melanocytes were purchased from Lonza–Clonetics (Walkersville, MD) and maintained in TICVA medium (Ham’s F10 with penicillin–streptomycin–glutamine, 7.5% fetal bovine serum, 50 ng/mL tetradecanoyl phorbol acetate, 225 mol/L 3-isobutyl-1-methylxanthine, 1 mol/L Na3VO4, and 1 mmol/L dibutyryl cAMP) as described previously.34 For expression microarray analysis, melanocytes were harbored from different individuals, purchased from ATCC, and cultured as above.
Adenovirus Infection and RNA Preparation
Adenoviruses encoding wild-type (WT) MITF, dominant-negative (DN) mutant MITF, or vector control encoding a random polypeptide were generated as described previously.34 In brief, 106 cells were plated per 100-mm plate. On the second day, cells were overlaid with 2 mL serum-free Ham’s F10 medium containing 10 mmol/L MgCl2, and concentrated adenovirus was added at multiplicity of infection MOI = 100. The cells were incubated at 37°C for 30 minutes, after which virus was removed and fresh complete medium was added. Total RNA was isolated with an RNAqueous-4PCR kit (Ambion; Life Technologies, Carlsbad, CA) at 48 or 72 hours after infection.
siRNA Gene Silencing
Synthetic siRNAs against MITF and nontargeting control oligo (siGENOME) were obtained from Thermo Fisher Scientific. Cells were transfected with siRNA using lipidoid reagent C12-113 as described previously35 and were assayed at 72 hours after siRNA transfection.
Microarray and Gene Correlation Analyses
Expression profiling data on human primary melanocytes and primary melanoma specimens were collected using high-density oligonucleotide microarrays (Hu6800–Hu35KsubA or HG-U95A; Affymetrix, Santa Clara, CA) as described previously.32 For correlation studies, the expression level of MITF or PEDF in each melanoma specimen was shown as the individual variance from the mean for all melanoma specimens in the data set (n = 10).
ChIP Assay
Chromatin immunoprecipitation (ChIP) assays were performed as described previously.32 In brief, 2 × 106 primary human melanocytes were grown to logarithmic phase, harvested by scraping, and homogenized in a hypotonic buffer [10 mmol/L Tris-HCl (pH 7.4), 15 mmol/L NaCl, 60 mmol/L KCl, 1 mmol/L EDTA, 0.1% Nonidet P40, 5% sucrose, 1× complete proteinase inhibitor cocktail (Roche Diagnostics, Indianapolis, IN)] on ice using a Dounce homogenizer. Nuclei were isolated by centrifugation onto a 10% sucrose pad and then were cross-linked with 1% formaldehyde in phosphate-buffered saline for 20 minutes at room temperature with gentle shaking. Nuclei were then spun down and resuspended in ChIP buffer [10 mmol/L Tris-HCl (pH 7.4), 100 mmol/L NaCl, 60 mmol/L KCl, 0.1% Nonidet P40, 1× complete proteinase inhibitor cocktail (Roche Diagnostics)], then sonicated by two 1-minute pulses on ice using a dismembranator (Thermo Fisher Scientific) fitted with a microtip. Antibodies [rabbit anti-MITF, rabbit anti–glutathione S-transferase, rabbit anti–acetylated histone H4 (all from EMD Millipore)] were then added to a 10-fold ChIP buffer diluted sample and incubated on a nutator for 3 hours at room temperature. Ultralink protein-A/G-beads (Pierce; Thermo Fisher Scientific) were added to the sample (and to control samples), which was incubated for an additional hour. Immunoprecipitates were then washed twice with ChIP buffer, twice with 500 mmol/L NaCl ChIP buffer, and once with Tris–EDTA (pH 8). The immunoprecipitates were released from the beads by incubation at 65°C for 20 minutes in 1% SDS–Tris–EDTA and protein was digested by proteinase K treatment side by side with an additional unprecipitated sample as input control. Crosslinks were released by heating at 70°C for 10 hours, and DNAs were recovered by extraction with phenol and chloroform at high salt concentration [0.6 mol/L NaOAc (pH 8)] and then were ethanol-precipitated. Next, semiquantitative PCR was performed on samples to amplify a fragment containing E-boxes E1/E2/E3 (approximately 250 bp) or the E-box E4 (approximately 300 bp), as well as an upstream control region in the human SERPINF1 (alias PEDF) genomic locus (2.7 kb upstream of the transcription start site). The forward and reverse primers for the E1/E2/E3 region were 5′-CTCAATCCCCTCCGGAGAGCCAGG-3′ and 5′-GCCCACATCAAGAAACACAGAAGC-3′, respectively. The forward and reverse primers for the E4 region were 5′-TGGCTCACGCCTGTAGTCCCAGCA-3′ and 5′-AGTACAGAGGTACCATCTCGGCTC-3′, respectively.
qPCR
The human primers used for quantitative real-time PCR (qPCR) were as follows. PEDF primers were 5′-ATTCCCGATGAGATCAGCATTC-3′, 5′-AACTTTGTTACCCACTGCCCC-3′; probe was 5′-6-FAM ATCGGAGAGGGCACGTACGGAGTTGT-TAMRA-3′ (IDT). β-Actin primers were 5′-ATTGCCGACAGGATGCAGAA-3′ and 5′-GCTGATCCACATCTGCTGGAA-3′; probe was 5′-6-FAM-CAAGATCATTGCTCCTCCTGAGCGCA-3′ (IDT). Additional primers were used as follows: SERPINF1 (PEDF), forward: 5′-AAAAAGCTCTGTGCTGGCTG-3′; reverse: 5′-TCCAATGCAGAGGAGTAGCAC-3′. MITF, forward: 5′-CATTGTTATGCTGGAAATGCTAGAA-3′; reverse: 5′-GGCTTGCTGTATGTGGTACTTGG-3′. RPL11, forward: 5′-GTTGGGGAGAGTGGAGACAG-3′, reverse: 5′-TGCCAAAGGATCTGACAGTG-3′. Reverse transcription proceeded at 48°C, 30 minutes; then 40 cycles of PCR at 95°C, 15 seconds and at 60°C for 1 minute. The total volume of each reaction was 25 μL. PCR was performed using an ABI PRISM 7700 sequence detection system (Life Technologies) and the results were analyzed using the manufacturer’s integrated Sequence Detection System software version 1.7. Standard curves were generated for all primer sets to confirm linearity of signals over the experimentally measured ranges. All reaction assays were performed in triplicate, using β-actin as an endogenous control. The qPCR was repeated, with the same results. CT values for each mRNA were normalized to β-actin (ΔCT) and calculated as relative quantification, RQ = 2−ΔCt.
Results
PEDF Expression Is Down-Regulated during Melanoma Progression
Using a monoclonal anti-human PEDF antibody and immunohistochemistry, we examined the expression of PEDF in 102 formalin-fixed, paraffin-embedded, well-characterized melanocytic lesions (n = 102): 13 CN, 3 dysplastic melanocytic nevi, 13 MIS, 61 PCM, and 12 MM (to skin or lymph node), as well as normal skin (n = 5). Before scoring immunoreactivity in the lesions, we examined and established the expression pattern for PEDF in normal skin. Weak to moderate signal was detected in epidermal and follicular keratinocytes, capillary-size blood vessels in the venus plexuses, eccrine apparatus, and the intimal layer of large caliber blood vessels (data not shown). These finding were consistent with the low but predictable expression of PEDF in normal skin and thus served as a positive control for the study. We established a semiquantitative immunoreactivity score categorized into a three-level grouping as negative, low (>0 and ≤1.5), or high (≥1.6) (Table 1).
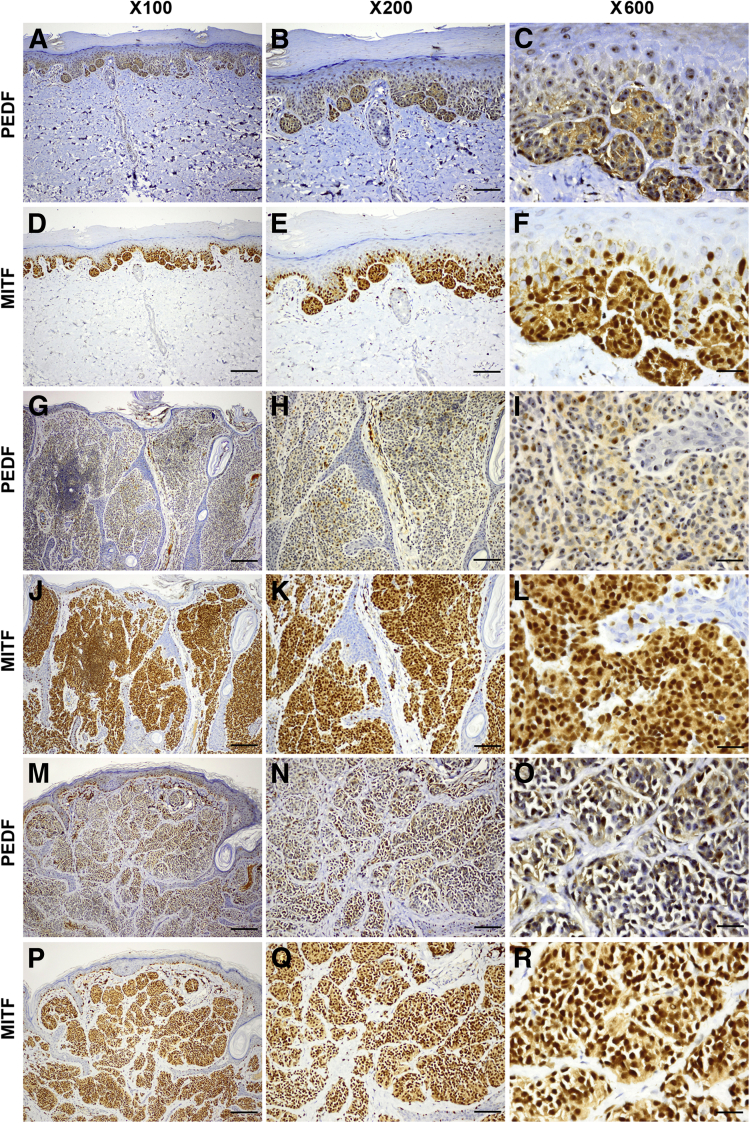
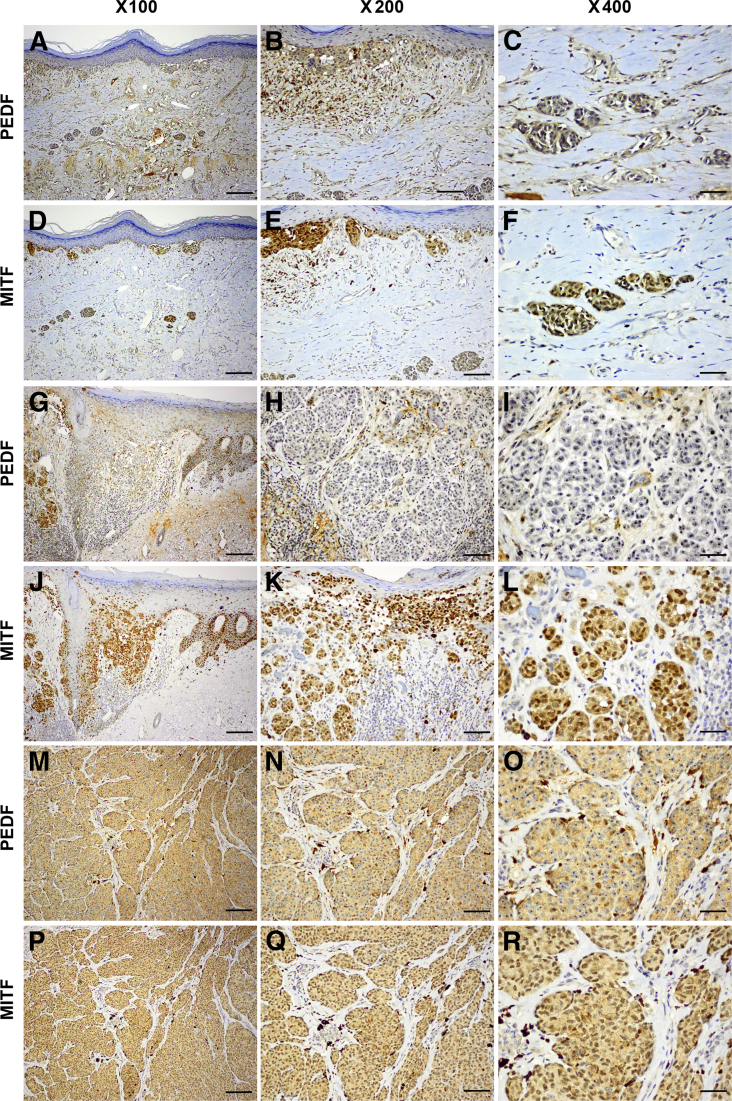
All CN specimens exhibited diffusely positive homogeneous immunoreactivity (9/13 low and 4/13 high) in the cytoplasm and nucleolus of junctional and intradermal nevus cells (Figure 1, A–C, G–I, and M–O). We compared PEDF and MITF immunoreactivities in serial sections of the same CN specimens and found that PEDF was coexpressed in the MITF+ nevus cells (Figure 1, D–F, J–L, and P–R). All three specimens of dysplastic melanocytic nevi exhibited consistently low PEDF expression (Table 1). Most of the 13 MIS specimens (9/13 with high and 2/13 with low immunoreactivity) retained expression of PEDF; however, in the remaining two specimens (15%) no PEDF was detected. By contrast, a majority (52/61) of PCM specimens (16/61 negative and 36/61 low) exhibited lost to low levels of PEDF immunoreactivity (Table 1). PCM lesions consistently exhibited strong and diffuse expression of PEDF in the junctional (in situ) cells, but exhibited decreased to lost expression in the invasive (dermal component) cells (Figure 2, A–C and G–I). Interestingly, 9/61 (15%) PCM specimens exhibited high PEDF immunoreactivity (Figure 2, M–O and Table 1). MITF was consistently expressed in the serial sections of PCM specimens, even in PEDF− melanoma cells (Figure 2, D–F, J–L, and P–R). Overall, we detected significantly less PEDF expression in melanoma cells in vertical growth phase, whereas epidermal keratinocytes and MIS cells (junctional component) retained PEDF expression (Figure 2, A–C and G–I).
Figure 1.
Pigment epithelium-derived factor (PEDF) is strongly expressed in the nevus cells of common melanocytic nevi (CN). A–C, G–I, and M–O: Three representative specimens of CN exhibit diffuse expression of PEDF in both junctional and intradermal nevus cells. PEDF expression is localized to the cytoplasm and nucleoli (C, I, and O). D–F, J–L, and P–R: Serial sections from the same three specimens show diffuse nuclear and cytoplasmic expression of microphthalmia-associated transcription factor (MITF). Scale bars: 50 μm (left column); 25 μm (middle column); 6 μm (right column).
Figure 2.
PEDF is significantly decreased or lost in invasive melanoma cells. A–C and G–I: Two invasive primary cutaneous melanomas (PCMs), representative of a majority [52/61 (85%)] of specimens, exhibited strong expression of PEDF in the junctional (in situ) cells, but expression was decreased to lost in the invasive cells. M–O: A third PCM specimen is representative of the minority [9/16 (15%)] that retained high levels of PEDF expression. D–F, J–L, and P–R: Serial sections from the same three specimens show diffuse nuclear and cytoplasmic expression of MITF. Scale bars: 50 μm (left column); 25 μm (middle column); 12.5 μm (right column).
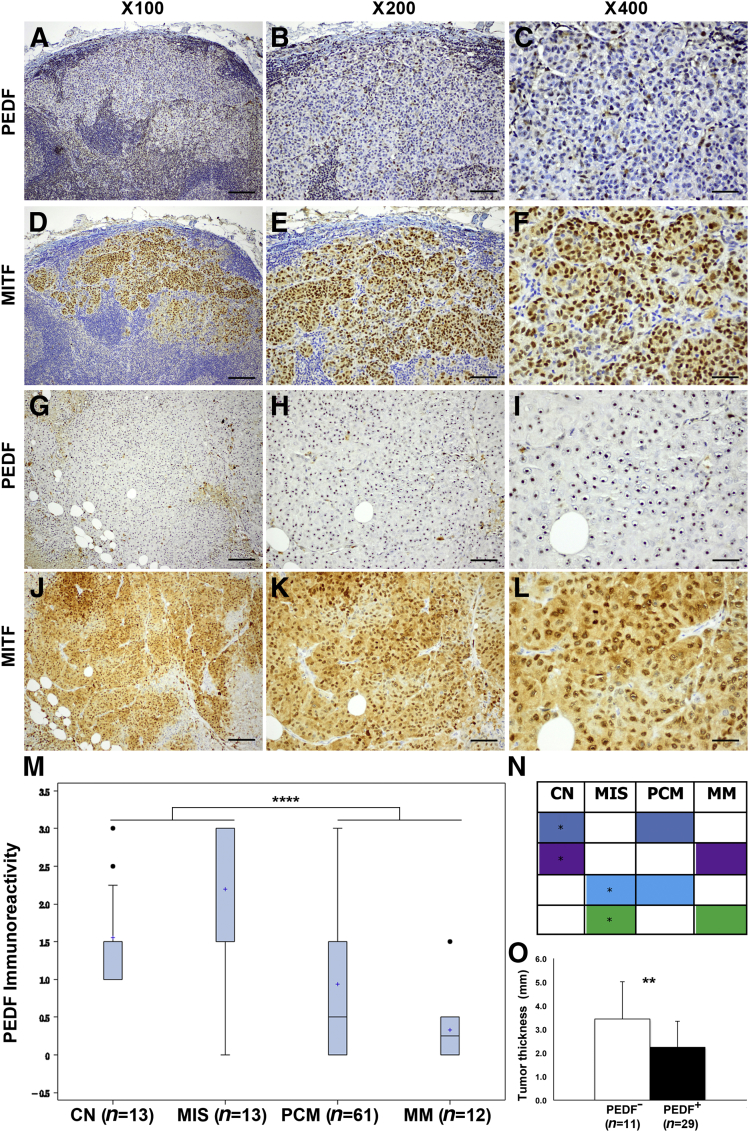
PEDF was dramatically lost in MM specimens (Figure 3, A–C and G–I), in which expression of MITF was consistently retained in serial sections (Figure 3, D–F and J–L). The high endothelial venules (specialized blood vessels in lymph nodes) stained positive for PEDF, which served as internal positive control for metastatic lesions to lymph node (data not shown). Boxplots and Kruskal–Wallis testing both confirmed significant reduction of PEDF expression in primary invasive melanoma (PCM) and MM, compared with CN and MIS (P < 0.0001) (Figure 3M). These results were independently confirmed by Bonferroni procedure (Figure 3N). Interestingly, a single MM specimen exhibited strong PEDF staining, whereas all others were negative; this specimen was the only one that was separately found to contain strong amplification of MITF (>100 copies using quantitative genomic PCR).45
Figure 3.
Pigment epithelium-derived factor (PEDF) is significantly decreased or lost from metastatic melanoma (MM) cells. A–C and G–I: Two representative specimens of metastasis to lymph node (A–C) and to subcutaneous adipose tissue (G–I) exhibit a dramatic loss of PEDF expression. D–F and J–L: Serial sections from the same two specimens show diffuse nuclear and cytoplasmic expression of MITF. M: PEDF immunoreactivity indicates a significant loss of expression in PCMs and MM, compared with CN and melanoma in situ (MIS) by Kruskal–Wallis test (P < 0.0001). Immunoreactivity data are expressed as boxplots; the box includes the first through third quartiles, a plus sign indicates the median, whiskers indicate the minimum and maximum (excluding outliers), and black dots indicate outliers. N: Based on multiple pairwise comparisons, Bonferroni procedure confirmed the boxplot analysis and showed a significant difference in PEDF with same color. Within a color, the diagnostic group with the higher level of PEDF is marked with an asterisk. O: In PCM specimens, tumor thickness was inversely correlated with PEDF expression (P = 0.003). Data are presented as means ± SD. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗∗P < 0.0001. Scale bars: 50 μm (left column); 25 μm (middle column); 12.5 μm (right column).
Comparison of mean tumor thickness (mm) in PEDF+ and PEDF− melanomas revealed a significant inverse correlation (P = 0.003) (Figure 3O). The mean thickness for PEDF− melanomas was 3.5 mm and for PEDF+ melanomas was 2.0 mm (Table 2), a finding consistent with the low to lost PEDF expression observed in vertical growth phase melanomas. Our results from examination of 102 melanocytic tumors spanning several stages of melanoma development demonstrated a consistent down-regulation of PEDF expression during tumor progression. Comparison of immunoreactivity of MITF and PEDF in serial sections of 51 specimens (11 CN, 11 MIS, 7 MM, and 22 PCM) revealed a significantly positive correlation (τB = 0.35, P = 0.0003) (Table 3).
Table 2.
Inverse Correlation between Tumor Thickness and PEDF Immunoreactivity According to Positive and Negative Expression in Primary Cutaneous Melanoma (n = 40)
| Expression | No. of specimens | Mean thickness, mm | P value |
|---|---|---|---|
| PEDF− | 11 | 3.5 | 0.003 |
| PEDF+ | 29 | 2.0 |
Table 3.
Positive Correlation between PEDF and MITF Immunoreactivities According to Positive and Negative Expression in Melanocytic Lesions (n = 51)
| Diagnostic group | Sample size | PEDF, no. (%) |
MITF, no. (%) |
||
|---|---|---|---|---|---|
| + | − | + | − | ||
| Common melanocytic nevus | 11 | 11 (100) | 0 (0) | 11 (100) | 0 (0) |
| Melanoma in situ | 11 | 11 (100) | 0 (0) | 11 (100) | 0 (0) |
| Primary cutaneous melanoma | 22 | 11 (50) | 11 (50) | 20 (91) | 2 (9) |
| Metastatic melanoma | 7 | 1 (14) | 6 (86) | 6 (86) | 1 (14) |
Kendall’s τB = 0.35; P = 0.0003.
Detailed clinicopathological data with clinical follow-up were available for 41 patients (Table 4). We examined the relationship between PEDF expression and clinicopathological characteristics by both a three-level grouping [negative, low (>0 and ≤1.5), or high (≥1.6)] and raw PEDF immunoreactivity scores (0 to 3). Both methods yielded nearly identical results, and we report only the three-level grouping results (Table 4). Higher PEDF expression was significantly associated with distant metastases to skin and soft tissue (P = 0.0331). The correlations between PEDF expression and mitotic index, AJCC stage, nodal metastases, and blood vascular density were marginally significant (0.05 < P < 0.10).
Table 4.
Correlations between PEDF Expression and Clinicopathological Characteristics of Melanoma Patients (n = 41)
| Characteristic | Value | P value, H:0 τ = 0 | Kendall’s τB∗ |
|---|---|---|---|
| Age (years) | 0.6546 | −0.05642 | |
| Mean | 54 | ||
| Median | 54 | ||
| Range | 21–84 | ||
| Sex | 0.34† | ||
| Male | 26 | ||
| Female | 15 | ||
| Anatomic location (no.) | 0.1471† | ||
| Head & neck | 7 | ||
| Upper extremities | 9 | ||
| Trunk | 17 | ||
| Lower extremities | 8 | ||
| Histological subtype (no.) | 0.8008† | ||
| Superficial spreading | 33 | ||
| Nodular | 6 | ||
| Lentigo maligna | 1 | ||
| Other | 1 | ||
| Tumor thickness (mm) | 0.1413 | −0.18608 | |
| Mean | 2.67 | ||
| Median | 1.41 | ||
| Ulceration (no.) | 0.1252 | 0.23179 | |
| Present | 9 | ||
| Absent | 32 | ||
| Mitotic index (/mm2) | 0.053 | 0.25852 | |
| Mean | 4.9 | ||
| Median | 3 | ||
| Clark’s level | 0.6077 | −0.07342 | |
| II | 3 | ||
| III | 11 | ||
| IV | 23 | ||
| V | 4 | ||
| Regression | 40 | 0.7638 | 0.04597 |
| Present | 7 | ||
| Absent | 33 | ||
| Clinical follow-up (months) | 0.1076 | 0.20138 | |
| Mean | 47.6 | ||
| Median | 34.5 | ||
| Range | 3.9–120.7 | ||
| AJCC stage | 0.0796 | 0.24762 | |
| I | 7 | ||
| II | 12 | ||
| III | 3 | ||
| IV | 19 | ||
| Nodal metastasis | 0.0868 | 0.25889 | |
| Positive | 21 | ||
| Negative | 20 | ||
| Organ metastasis | 0.3225 | 0.14956 | |
| Positive | 12 | ||
| Negative | 29 | ||
| Distant metastasis | 0.0331 | 0.32215 | |
| Positive | 19 | ||
| Negative | 22 | ||
| Death of disease | 0.1273 | 0.23055 | |
| Positive | 14 | ||
| Negative | 27 | ||
| Angiogenic parameters | 32 | ||
| Blood vascular density (/mm2) | 0.0675 | 0.25894 | |
| Mean | 52.6 | ||
| Median | 47 | ||
| Blood vessel size (mm2) | 0.3653 | −0.12821 | |
| Mean | 662.7 | ||
| Median | 634.82 | ||
| Blood vascular area (%) | 0.3031 | 0.14596 | |
| Mean | 3.3 | ||
| Median | 3.2 |
P values in bold are statistically significant (<0.05).
Positive values of τB denote a positive correlation; negative values, an inverse correlation.
Mantel–Haenszel χ2 test.
The PEDF Gene Is a MITF Transcriptional Target in Vitro
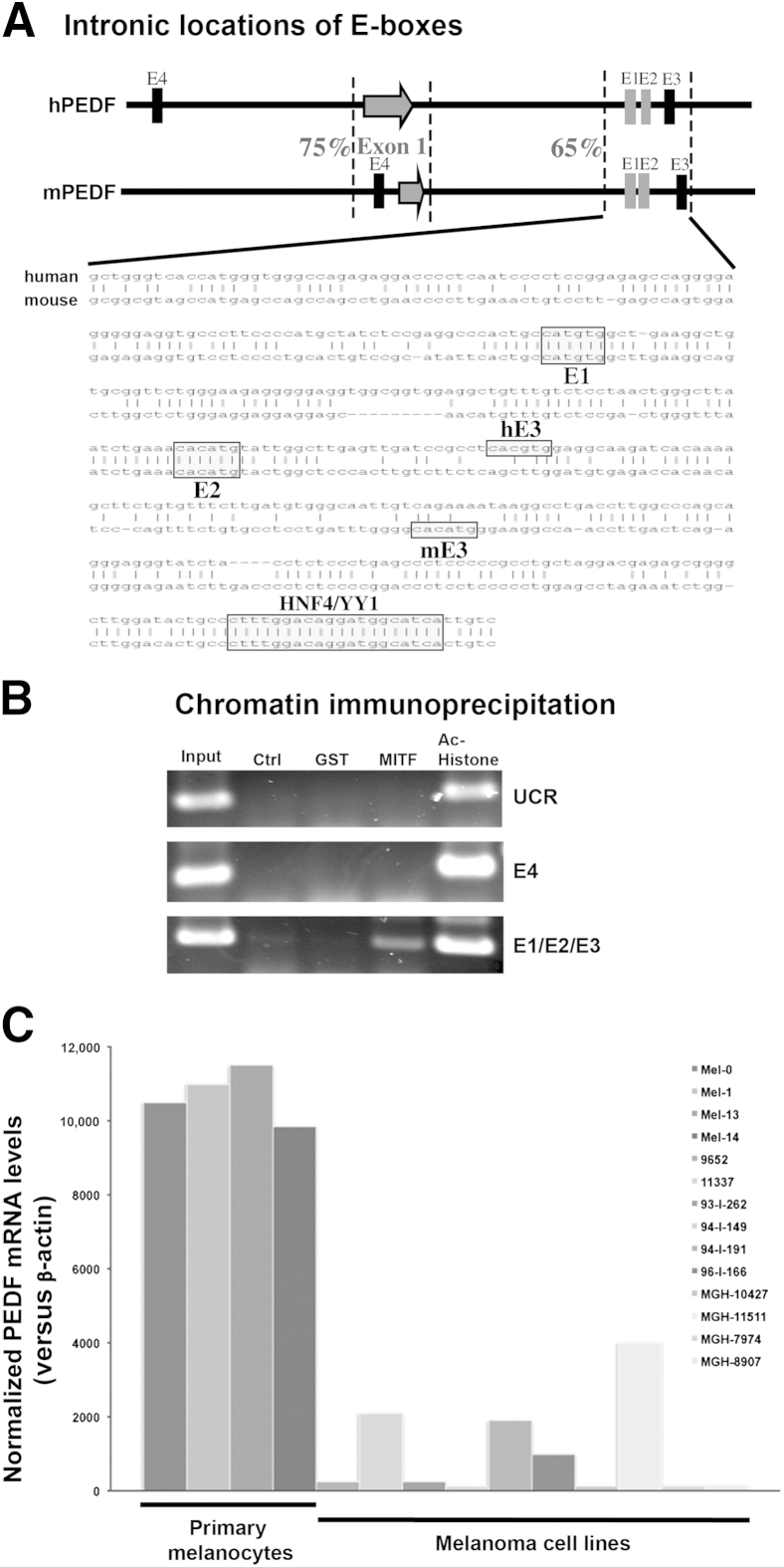
Given the observed coexpression of PEDF and MITF in benign and malignant melanocytic tumors, we investigated the mechanism of transcriptional control of PEDF expression by MITF in vitro. Previous experiments using a microarray-based strategy identified the PEDF gene as a potential MITF target in human primary melanocytes.34 Alignment of the human and mouse PEDF genomic loci, including 5 kb upstream and downstream of the transcription start site and the first exon, revealed relatively little sequence conservation, with overall homology of <10%. Only two stretches of conserved sequence were found, one located within approximately 380 bp surrounding the transcription start site and the first exon (75% identical) and another region of approximately 400 bp located approximately 1 kb into the first intron (68% identical) (Figure 4A). The human and mouse PEDF gene promoter regions up to 1.5 kb upstream of transcription start site shared little sequence homology (18%), suggesting that these regions might not be important in regulating PEDF gene expression. The E-box E4 was located 1.3 kb upstream of the transcription start site in the human PEDF promoter and 176 bp upstream in the mouse PEDF promoter (Figure 4A). The locations and sequences of these two E-boxes were not conserved across species. In addition, luciferase reporter constructs containing either of these two E-boxes failed to respond to MITF in transient transfection assays (data not shown), suggesting that these E-boxes were not MITF regulatory elements in vivo. However, examination of the conserved intronic sequences between the human and mouse PEDF genes revealed the presence of three E-boxes, the closest of which was approximately 1.3 kb from the transcriptional start sites (Figure 4A). Importantly, the first two E-boxes (E1 and E2) were highly conserved between human and mouse; E-box E3, although very close to E1 and E2, was not conserved.
Figure 4.
Identification of the PEDF gene as a target MITF in human primary melanocytes. A: Alignment of human and mouse PEDF genomic loci revealed conserved MITF binding sites (E-boxes) in the first intron. Dashed lines bracket two highly conserved regions of the PEDF genome. Arrows mark the transcription start site and the first exon; vertical bars represent E-boxes. E1 and E2 (gray bars), are conserved between human (h) and mouse (m) sequence; E3 and E4 (black bars) are not conserved between the two species. The sequence alignment of the conserved intronic region is shown, E-boxes and a conserved HNF4/YY1 site are labeled. B:In vivo occupancy of the intronic E-boxes by MITF is shown by PCR amplification of immunoprecipitated chromatin DNA. Chromatin from human primary melanocytes was sonicated and precipitated by control, anti–glutathione S-transferase (GST), anti-MITF, or anti–acetylated histone (Ac-Histone) antibodies. The input DNA represents 5% of the amount of the total DNA used in the ChIP assay. The primers for the upstream control region (UCR) were approximately 2.7 kb upstream of the transcription start site; the E4 and E1/E2/E3 primers span E-boxes in the human PEDF gene promoter and the first intron. C: The qPCR analysis shows markedly decreased PEDF mRNA levels in melanoma cell lines, compared with human primary melanocytes. All reaction assays were performed in triplicate, using β-actin as an endogenous control. CT values for each mRNA were normalized to β-actin and calculated as relative quantification RQ = 2−ΔCt.
We investigated the significance of these conserved E-boxes and their occupancy by MITF in vivo through a ChIP assay. Protein–DNA complexes in primary human melanocytes were cross-linked by formaldehyde, sonicated, and immunoprecipitated with antibodies against control (glutathione S-transferase), MITF, or acetylated histone H4 (Figure 4B). PCR primers were designed to span E-boxes E1 and E2 (and including E3, because it was physically too close to be reliably discriminated), E4, and a negative upstream control region 2.7 kb from the transcription start site. This assay revealed that MITF is bound to the intronic conserved E-boxes, as shown by the amplification of the E1/E2/E3 DNA fragment (Figure 4B). Neither PCR amplification product was seen from control or anti–glutathione S-transferase antibodies, nor any amplification of either upstream nonconserved E-box E4 or upstream control region primers. To identify other potential conserved transcription factor binding sites in the intronic region of the PEDF gene, we conducted an extensive search of the TRANSFAC database (2006.1 release) using MatInspector (release 7.4; Genomatix Software, Munich, Germany) and TESS software (version 2006; http://membres-timc.imag.fr/Olivier.Francois/tess.html).36 This analysis revealed a conserved HNF4/YY1 binding site in the conserved intronic sequence (Figure 4A), as well as several potential binding sites for nuclear receptor superfamily members (data not shown). Importantly, similar analysis of the conserved promoter region of the PEDF gene spanning the first exon revealed only a conserved CCAAT box and possible binding sites for c-ETS and the myeloid zinc finger protein-1, again suggesting that this region of the PEDF gene is unlikely to be directly regulated in vivo by MITF. Overall, these results showed that MITF occupies the endogenous conserved intronic E-boxes in the human PEDF gene in primary human melanocytes.
To further characterize the relationship between MITF and PEDF, using anti-PEDF and anti-MITF antibodies, immunofluorescence localized the PEDF signal to cytoplasm and nucleus, whereas the MITF signal was localized only to the nucleus of human primary melanocytes (data not shown). qPCR analyses confirmed high levels of PEDF mRNA in human primary melanocytes, which were on average decreased 10-fold in all of the melanoma cell lines examined (Figure 4C). As an independent means for validation of the MITF–PEDF transcription factor–target relationship, we examined the correlation of their microarray expression profiles in a series of primary melanomas. MITF expression correlated with PEDF gene expression in the majority of primary melanomas, which independently corroborated our IHC results (Table 3).
MITF Regulates the Expression of PEDF in Melanocytes and Melanomas
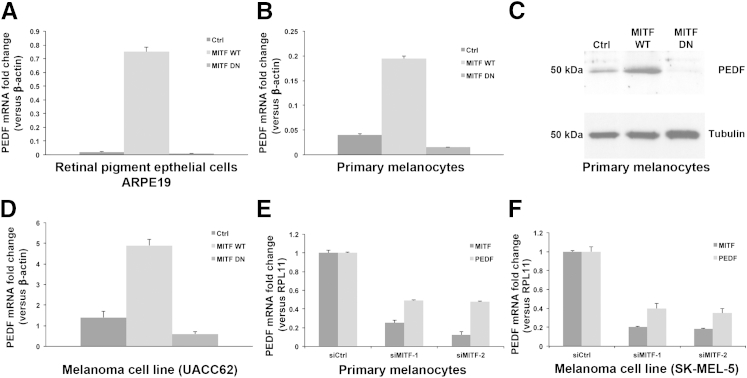
We next investigated whether PEDF expression can be regulated by MITF by using an early-passage RPE cell line (ARPE19), human primary melanocytes, and melanoma cell lines (UACC62 and SK-MEL-5) and by using two different gene silencing approaches: DN and siRNA (Figure 5). MITF up-regulation was achieved by overexpression of WT MITF (Figure 5, A–D). MITF down-regulation was achieved in two ways: expression of a DN mutant of MITF (Figure 5, A–D) or use of two independent siRNAs against MITF (Figure 5, E and F). We graphed relative mRNA levels of MITF (suppressed by both siRNAs in both cell types) and of PEDF (also suppressed by both siRNAs against MITF in both of these cells (Figure 5, E and F).
Figure 5.
MITF regulates PEDF gene expression in vivo, as demonstrated by both DN and siRNA gene silencing methods. A and B: qPCR analysis of PEDF mRNA levels in RPE cells (ARPE19) (A) and primary melanocytes (B) after infection with adenovirus expressing a random polypeptide control, WT MITF, or DN MITF shows a marked increase in endogenous PEDF mRNA levels by functional MITF and a decrease by nonfunctional MITF in RPE cells and melanocytes, compared with control. C: In accord, a marked increase in PEDF protein levels was demonstrated in melanocytes expressing WT MITF, and a marked decrease in DN MITF melanocytes, compared with control. D: Similarly, marked increase and decrease in PEDF mRNA levels were demonstrated in melanoma cell line (UACC62) expressing WT and DN MITF, respectively, compared with control. E and F: In two independent experiments, siMITF markedly decreased both MITF and PEDF mRNA levels in melanocytes (E) and in the melanoma cell line SK-MEL-5 (F). Ctrl, control; DN, dominant negative; WT, wild type.
ARPE19 cells, primary melanocytes, and UACC62 cells were infected with adenovirus expressing a random polypeptide (control), WT MITF, or DN MITF. We then measured the endogenous PEDF mRNA and protein fold changes by qPCR and Western blotting. Compared with control, WT MITF significantly increased (37-fold), whereas DN MITF reduced (twofold) the endogenous levels of PEDF mRNA in the RPE cell line ARPE19 (Figure 5A). In human melanocytes, WT MITF increased the endogenous levels of PEDF mRNA 4.8-fold, whereas DN MITF reduced levels 2.6-fold, compared with control (Figure 5B). PEDF protein levels mirrored the changes in PEDF mRNA levels in primary melanocytes (Figure 5C). Similar changes were observed in the melanoma cell line UACC62 for PEDF mRNA (Figure 5D) and protein levels (data not shown). Using another approach, we found that siMITF markedly suppressed PEDF mRNA levels in primary melanocytes (Figure 5E) and in the SK-MEL-5 melanoma cell line (Figure 5F). Two other melanoma cell lines (MALME-3M and UACC257) did not respond to siMITF, because of unusually low endogenous PEDF mRNA levels, despite relatively high MITF expression (data not shown). These results suggested that MITF regulates expression of the endogenous PEDF gene in both melanocytes and melanomas. Taken together with results of the ChIP assay, these data are consistent with MITF being a direct transcriptional regulator of the PEDF gene in the cell lines examined.
Discussion
Although the transcription factor–target gene relationship between MITF and PEDF has been established in RPE cells, no previous study has investigated this relationship in melanocytes or the various stages of melanoma development. We not only examined the expression of PEDF in 102 melanocytic tumors spanning several stages of melanoma development (including benign and dysplastic melanocytic nevi, early-stage radial growth phase to vertical growth phase to metastatic melanomas), but we also investigated the correlation of PEDF expression with melanoma clinicopathological prognostic and tumor angiogenic parameters, as well as the mechanism of transcriptional regulation by MITF in melanocytes and melanoma cells. Expression of PEDF protein, as determined by semiquantitative IHC, consistently decreased in PCM and MM specimens, compared with CN and MIS. Moreover, comparison of PEDF mRNA and protein level in cultured primary human melanocytes and established melanoma cell lines revealed that PEDF was indeed expressed at much higher level in primary melanocytes than melanomas, supporting the IHC results. MITF positively correlated with PEDF protein expression; serial sections clearly showed that MITF+ melanoma cells indeed expressed PEDF. Comparison of MITF and PEDF mRNA levels by microarray expression values confirmed the positive correlation between MITF and PEDF expression in primary melanomas. These data are consistent with a model in which PEDF expression is down-regulated in the majority of non–MITF-amplified melanomas but is retained in the minority (15%) of MITF-amplified lesions examined here.
MITF is a melanocyte-specific transcription factor whose targets are essential for melanocyte differentiation, as well as for survival.29 We have now identified the gene for PEDF (which is an antiangiogenic factor in retinal homeostasis37) as another MITF target gene in melanocytes and melanoma cells. Comparison of retinal epithelium and cutaneous epidermis may help explain this novel role of MITF as a regulator of PEDF expression in human skin. The epidermis is an avascular epithelium, in this way resembling the retina. Only during injury or wound healing does the skin becomes extensively vascularized as a physiological response.38 The concentration of PEDF in the epidermis and cutaneous vasculature supports the notion that it may contribute to maintenance of an antiangiogenic state in the skin, possibly through MITF-driven PEDF expression in melanocytes, along with either low-level expression within keratinocytes or, conceivably, through transferring to nearby keratinocytes from melanocytes via melanosomes.39 Thus, MITF regulation of PEDF expression in epidermal keratinocytes and melanocytes may contribute to the maintenance of an antiangiogenic balance in skin under normal physiological conditions.
Previous studies have implicated the tumor suppressive role of PEDF in pancreatic and prostate carcinoma40,41 and in mouse melanoma xenograft models.25,26 If melanoma growth in a mouse xenograft model were a surrogate for the depth of invasion in human melanoma, then PEDF− melanomas should be thicker. Indeed, in the present study PEDF levels were absent to low in the majority [52/61 (85%)] of PCM specimens (Table 1), and PEDF expression was lost in thicker invasive melanomas (3.5 mm), compared with thinner lesions (2.0 mm) (Table 2). Moreover, PEDF expression was retained in MIS specimens and in the in situ component of invasive specimens but was lost in invasive dermal cells (vertical growth phase), supporting the notion that loss of PEDF expression enhances tumor growth. However, a study examining the relationship between PEDF expression and tumor angiogenesis in 32 primary invasive melanomas revealed only a marginal significance for blood vascular density and no significance for blood vessel size or blood vascular area,8 three important determinants of angiogenesis in tumor tissue sections. In the present study, comparison of PEDF expression with ulceration and mitotic index (two other major AJCC prognostic parameters in melanoma) revealed no significant correlation. Except for distant metastases, PEDF expression did not show any significant relationship to other parameters of disease progression (including death of disease, organ metastasis, nodal metastasis, or AJCC stage) during an average of 47.6 months of clinical follow up in 41 melanoma patients. These results indicate that tumor angiogenesis can influence melanoma growth (invasive depth) but not survival outcome, consistent with the notion that other factors (eg, tumor lymphangiogenesis) are significantly associated with melanoma prognosis, as evidenced by a number of published studies and a recent meta-analysis.18 Alternatively, the number of patients in our present analysis may be too small to detect a statistically significant difference.
To further scrutinize the intimate correlation between MITF and PEDF tissue expression, we investigated the mechanism of transcriptional control of MITF on PEDF in melanocytes and melanoma cells. We identified two conserved E-boxes in the first intron of the human and mouse PEDF genes using binding assays in vivo and demonstrating direct influence of MITF on PEDF by using DN and siRNA approaches in vivo. The high conservation of both sequence and positions for these E-boxes, together with the absence of conserved E-boxes in the proximal promoter region, suggests a mechanism of intronic gene regulation through a MITF-dependent enhancer element. Indeed, the ChIP assay confirmed the specific binding of MITF to the conserved E-boxes, but not to upstream, nonconserved E-box sequences. The ability of MITF to modulate PEDF gene expression through an intronic enhancer is plausible, given the prior demonstration that MITF regulates the melanocyte-lineage gene PMEL through a similarly positioned, species-conserved enhancer element located within the first intron.42 Given that the E-boxes were located at an intronic location (and not within the promoter region), the use of luciferase reporter assays would have been artificial, because the intron would have been removed from its natural location to reside in a usable plasmid reporter. Instead, the ChIP assay provided direct evidence for the interaction of MITF with the E-boxes of PEDF. Taken together, these results are consistent with a MITF–PEDF transcription factor–target relationship.
To validate PEDF as a MITF target, melanocytes and melanoma cells were enforced to express DN MITF or siMITF. In both approaches, PEDF endogenous mRNA (and protein) levels directly increased in response to WT MITF, but decreased on suppression of MITF (by DN or siRNA) in melanocytes and melanoma cell lines. Therefore, given binding of endogenous MITF to a conserved consensus element within the first intron of PEDF, it is likely that it regulates the PEDF gene directly, not indirectly. PEDF is also expressed within nonmelanocytic cell types (eg, keratinocytes, prostate, and pancreas) that are not known to express MITF. It is possible that the closely related MITF family members TFEB, TFE3, or TFEC may modulate PEDF in these nonmelanocytic cell types (as has been seen for cathepsin K expression in osteoclasts43) or, alternatively, distinct transcriptional mechanisms may regulate PEDF in these cell types. Despite the involvement of MITF in regulating PEDF expression, it remains incompletely understood exactly which mechanism or mechanisms are responsible for PEDF down-regulation during progression of melanocytic neoplasm. One possibility is that MITF expression is largely responsible, because its expression tends to diminish,44 although in a minority of specimens MITF is amplified.45 Similarly, in the present study PEDF down-regulation was measured within intermediate-thickness cutaneous melanomas, and the diminished expression was seen to correlate with metastatic propensity. Therefore, down-regulation of MITF should similarly affect PEDF expression. Although it is possible that selection for MITF down-regulation could arise from growth benefits associated with diminished differentiation and/or pigmentation, it is also possible that loss of PEDF expression may contribute (and perhaps even more importantly) to this selection.
Finally, PEDF has been observed to harbor potent endothelial killing activity, as well as antineoplastic activity against multiple solid tumors in animal models.40,41 Moreover, enforced overexpression of PEDF in melanoma cells can lead to growth inhibition and tumor necrosis in xenografted mouse models and to prevention of lung metastases in mice injected with melanoma cells.26 These reports appear to be highly complemented by our findings of a MITF–PEDF transcription factor–target relationship and down-regulation of PEDF expression as a potential angiogenic factor during early melanoma invasive growth. A thorough understanding of the mechanisms regulating PEDF expression in melanoma may lead to novel therapeutic strategies aimed at restoring its expression in advanced disease.46 A proof of principle using such a strategy has recently been shown in pancreatic cancer models, in which adenoviral-mediated PEDF gene transfer inhibited tumor growth and intraperitoneal metastasis.47 Because suppression of PEDF-mediated antiangiogenesis may confer advantageous tumor growth, aiming to reactivate PEDF expression might be a plausible therapeutic strategy for melanoma.
Acknowledgments
We thank Dr. Rong Wu (Connecticut Institute for Clinical and Translational Science) for statistical analyses, members of D.E.F.’s laboratory for useful discussions and comments, and Nandita Bhattacharya of M.F.L.’s laboratory for help with immunohistochemistry.
Footnotes
Supported by NIH grants 1-P01-CA163222 and R01-AR043369-17 (D.E.F.), the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (D.E.F.), and the American Cancer Society New England Division (R.J.L.).
S.S.D. and R.J.L. contributed equally to this work.
Disclosures: E.F. is a full-time employee and stockholder of VBL Therapeutics. VBL Therapeutics has no research or commercial interest in MITF or PEDF and does not have any active clinical program in melanoma. The data reported here were generated before E.F. joined VBL Therapeutics.
Current address of E.F., VBL Therapeutics, Or Yehuda, Israel; of M.D., Swiss Federal Institute of Technology (ETH) Zurich, Zurich, Switzerland; of R.J.L., NYU Langone Medical Center, New York, NY
References
- 1.Tsao H., Atkins M.B., Sober A.J. Management of cutaneous melanoma. N Engl J Med. 2004;351:998–1012. doi: 10.1056/NEJMra041245. [Erratum appeared in N Engl J Med 2004, 351:2461] [DOI] [PubMed] [Google Scholar]
- 2.Kozubek J., Ma Z., Fleming E., Duggan T., Wu R., Shin D.G., Dadras S.S. In-depth characterization of microRNA transcriptome in melanoma. PLoS One. 2013;8:e72699. doi: 10.1371/journal.pone.0072699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chudnovsky Y., Khavari P.A., Adams A.E. Melanoma genetics and the development of rational therapeutics. J Clin Invest. 2005;115:813–824. doi: 10.1172/JCI24808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kabbarah O., Chin L. Revealing the genomic heterogeneity of melanoma. Cancer Cell. 2005;8:439–441. doi: 10.1016/j.ccr.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 5.Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 6.Flaherty K.T., Yasothan U., Kirkpatrick P. Vemurafenib. Nat Rev Drug Discov. 2011;10:811–812. doi: 10.1038/nrd3579. [DOI] [PubMed] [Google Scholar]
- 7.Flaherty K.T., Hodi F.S., Fisher D.E. From genes to drugs: targeted strategies for melanoma. Nat Rev Cancer. 2012;12:349–361. doi: 10.1038/nrc3218. [DOI] [PubMed] [Google Scholar]
- 8.Dadras S.S., Paul T., Bertoncini J., Brown L.F., Muzikansky A., Jackson D.G., Ellwanger U., Garbe C., Mihm M.C., Detmar M. Tumor lymphangiogenesis: a novel prognostic indicator for cutaneous melanoma metastasis and survival. Am J Pathol. 2003;162:1951–1960. doi: 10.1016/S0002-9440(10)64328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shields J.D., Borsetti M., Rigby H., Harper S.J., Mortimer P.S., Levick J.R., Orlando A., Bates D.O. Lymphatic density and metastatic spread in human malignant melanoma. Br J Cancer. 2004;90:693–700. doi: 10.1038/sj.bjc.6601571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dadras S.S., Lange-Asschenfeldt B., Velasco P., Nguyen L., Vora A., Muzikansky A., Jahnke K., Hauschild A., Hirakawa S., Mihm M.C., Detmar M. Tumor lymphangiogenesis predicts melanoma metastasis to sentinel lymph nodes. Mod Pathol. 2005;18:1232–1242. doi: 10.1038/modpathol.3800410. [DOI] [PubMed] [Google Scholar]
- 11.Massi D., Puig S., Franchi A., Malvehy J., Vidal-Sicart S., González-Cao M., Baroni G., Ketabchi S., Palou J., Santucci M. Tumour lymphangiogenesis is a possible predictor of sentinel lymph node status in cutaneous melanoma: a case-control study. J Clin Pathol. 2006;59:166–173. doi: 10.1136/jcp.2005.028431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shayan R., Karnezis T., Murali R., Wilmott J.S., Ashton M.W., Taylor G.I., Thompson J.F., Hersey P., Achen M.G., Scolyer R.A., Stacker S.A. Lymphatic vessel density in primary melanomas predicts sentinel lymph node status and risk of metastasis. Histopathology. 2012;61:702–710. doi: 10.1111/j.1365-2559.2012.04310.x. [DOI] [PubMed] [Google Scholar]
- 13.Harrell M.I., Iritani B.M., Ruddell A. Tumor-induced sentinel lymph node lymphangiogenesis and increased lymph flow precede melanoma metastasis. Am J Pathol. 2007;170:774–786. doi: 10.2353/ajpath.2007.060761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bracher A., Cardona A.S., Tauber S., Fink A.M., Steiner A., Pehamberger H., Niederleithner H., Petzelbauer P., Gröger M., Loewe R. Epidermal growth factor facilitates melanoma lymph node metastasis by influencing tumor lymphangiogenesis. J Invest Dermatol. 2012;133:230–238. doi: 10.1038/jid.2012.272. [DOI] [PubMed] [Google Scholar]
- 15.Dadras S.S. An unexpected role for EGF in lymphangiogenesis-mediated melanoma metastasis to sentinel lymph nodes. J Invest Dermatol. 2013;133:14–16. doi: 10.1038/jid.2012.436. [DOI] [PubMed] [Google Scholar]
- 16.Doeden K., Ma Z., Narasimhan B., Swetter S.M., Detmar M., Dadras S.S. Lymphatic invasion in cutaneous melanoma is associated with sentinel lymph node metastasis. J Cutan Pathol. 2009;36:772–780. doi: 10.1111/j.1600-0560.2008.01166.x. [DOI] [PubMed] [Google Scholar]
- 17.Xu X., Chen L., Guerry D., Dawson P.R., Hwang W.T., VanBelle P., Elder D.E., Zhang P.J., Ming M.E., Schuchter L., Gimotty P.A. Lymphatic invasion is independently prognostic of metastasis in primary cutaneous melanoma. Clin Cancer Res. 2012;18:229–237. doi: 10.1158/1078-0432.CCR-11-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pastushenko I., Vermeulen P.B., Carapeto F.J., Van den Eynden G., Rutten A., Ara M., Dirix L.Y., Van Laere S. Blood microvessel density, lymphatic microvessel density and lymphatic invasion in predicting melanoma metastases: systematic review and meta-analysis. Br J Dermatol. 2014;170:66–77. doi: 10.1111/bjd.12688. [DOI] [PubMed] [Google Scholar]
- 19.Dadras S.S., Detmar M. Angiogenesis and lymphangiogenesis of skin cancers. Hematol Oncol Clin North Am. 2004;18:1059–1070. doi: 10.1016/j.hoc.2004.06.009. viii. [DOI] [PubMed] [Google Scholar]
- 20.Dawson D.W., Volpert O.V., Gillis P., Crawford S.E., Xu H., Benedict W., Bouck N.P. Pigment epithelium-derived factor: a potent inhibitor of angiogenesis. Science. 1999;285:245–248. doi: 10.1126/science.285.5425.245. [DOI] [PubMed] [Google Scholar]
- 21.Browne M., Stellmach V., Cornwell M., Chung C., Doll J.A., Lee E.J., Jameson J.L., Reynolds M., Superina R.A., Abramson L.P., Crawford S.E. Gene transfer of pigment epithelium-derived factor suppresses tumor growth and angiogenesis in a hepatoblastoma xenograft model. Pediatr Res. 2006;60:282–287. doi: 10.1203/01.pdr.0000232789.86632.91. [DOI] [PubMed] [Google Scholar]
- 22.Steele F.R., Chader G.J., Johnson L.V., Tombran-Tink J. Pigment epithelium-derived factor: neurotrophic activity and identification as a member of the serine protease inhibitor gene family. Proc Natl Acad Sci USA. 1993;90:1526–1530. doi: 10.1073/pnas.90.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craword S.E., Fitchev P., Veliceasa D., Volpert O.V. The many facets of PEDF in drug discovery and disease: a diamond in the rough or split personality disorder? Expert Opin Drug Discov. 2013;8:769–792. doi: 10.1517/17460441.2013.794781. [DOI] [PubMed] [Google Scholar]
- 24.Becerra S.P., Notario V. The effects of PEDF on cancer biology: mechanisms of action and therapeutic potential. Nat Rev Cancer. 2013;13:258–271. doi: 10.1038/nrc3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abe R., Shimizu T., Yamagishi S., Shibaki A., Amano S., Inagaki Y., Watanabe H., Sugawara H., Nakamura H., Takeuchi M., Imaizumi T., Shimizu H. Overexpression of pigment epithelium-derived factor decreases angiogenesis and inhibits the growth of human malignant melanoma cells in vivo. Am J Pathol. 2004;164:1225–1232. doi: 10.1016/s0002-9440(10)63210-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia M., Fernandez-Garcia N.I., Rivas V., Carretero M., Escamez M.J., Gonzalez-Martin A., Medrano E.E., Volpert O., Jorcano J.L., Jimenez B., Larcher F., Del Rio M. Inhibition of xenografted human melanoma growth and prevention of metastasis development by dual antiangiogenic/antitumor activities of pigment epithelium-derived factor. Cancer Res. 2004;64:5632–5642. doi: 10.1158/0008-5472.CAN-04-0230. [DOI] [PubMed] [Google Scholar]
- 27.Tsuru M., Arima N., Toyozumi Y., Kato S. Pigment epithelium-derived factor as a new diagnostic marker for melanocytic tumors. Kurume Med J. 2005;52:81–87. doi: 10.2739/kurumemedj.52.81. [DOI] [PubMed] [Google Scholar]
- 28.Steingrimsson E., Copeland N.G., Jenkins N.A. Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- 29.Widlund H.R., Fisher D.E. Microphthalamia-associated transcription factor: a critical regulator of pigment cell development and survival. Oncogene. 2003;22:3035–3041. doi: 10.1038/sj.onc.1206443. [DOI] [PubMed] [Google Scholar]
- 30.Haq R., Fisher D.E. Biology and clinical relevance of the micropthalmia family of transcription factors in human cancer. J Clin Oncol. 2011;29:3474–3482. doi: 10.1200/JCO.2010.32.6223. [DOI] [PubMed] [Google Scholar]
- 31.Ma X., Pan L., Jin X., Dai X., Li H., Wen B., Chen Y., Ma A., Qu J., Hou L. Microphthalmia-associated transcription factor acts through PEDF to regulate RPE cell migration. Exp Cell Res. 2012;318:251–261. doi: 10.1016/j.yexcr.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 32.Du J., Widlund H.R., Horstmann M.A., Ramaswamy S., Ross K., Huber W.E., Nishimura E.K., Golub T.R., Fisher D.E. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell. 2004;6:565–576. doi: 10.1016/j.ccr.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 33.King R., Weilbaecher K.N., McGill G., Cooley E., Mihm M., Fisher D.E. Microphthalmia transcription factor. A sensitive and specific melanocyte marker for melanoma diagnosis. Am J Pathol. 1999;155:731–738. doi: 10.1016/S0002-9440(10)65172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGill G.G., Horstmann M., Widlund H.R., Du J., Motyckova G., Nishimura E.K., Lin Y.L., Ramaswamy S., Avery W., Ding H.F., Jordan S.A., Jackson I.J., Korsmeyer S.J., Golub T.R., Fisher D.E. BCL2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109:707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 35.Brock A., Krause S., Li H., Kowalski M., Goldberg M.S., Collins J.J., Ingber D.E. Silencing HoxA1 by intraductal injection of siRNA lipidoid nanoparticles prevents mammary tumor progression in mice. Sci Transl Med. 2014;6:217ra2. doi: 10.1126/scitranslmed.3007048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 37.King G.L., Suzuma K. Pigment-epithelium-derived factor–a key coordinator of retinal neuronal and vascular functions. N Engl J Med. 2000;342:349–351. doi: 10.1056/NEJM200002033420511. [DOI] [PubMed] [Google Scholar]
- 38.Velasco P., Lange-Asschenfeldt B. Dermatological aspects of angiogenesis. Br J Dermatol. 2002;147:841–852. doi: 10.1046/j.1365-2133.2002.05073.x. [DOI] [PubMed] [Google Scholar]
- 39.Setaluri V. The melanosome: dark pigment granule shines bright light on vesicle biogenesis and more. J Invest Dermatol. 2003;121:650–660. doi: 10.1046/j.1523-1747.2003.12500.x. [DOI] [PubMed] [Google Scholar]
- 40.Uehara H., Miyamoto M., Kato K., Ebihara Y., Kaneko H., Hashimoto H., Murakami Y., Hase R., Takahashi R., Mega S., Shichinohe T., Kawarada Y., Itoh T., Okushiba S., Kondo S., Katoh H. Expression of pigment epithelium-derived factor decreases liver metastasis and correlates with favorable prognosis for patients with ductal pancreatic adenocarcinoma. Cancer Res. 2004;64:3533–3537. doi: 10.1158/0008-5472.CAN-03-3725. [DOI] [PubMed] [Google Scholar]
- 41.Halin S., Wikström P., Rudolfsson S.H., Stattin P., Doll J.A., Crawford S.E., Bergh A. Decreased pigment epithelium-derived factor is associated with metastatic phenotype in human and rat prostate tumors. Cancer Res. 2004;64:5664–5671. doi: 10.1158/0008-5472.CAN-04-0835. [DOI] [PubMed] [Google Scholar]
- 42.Du J., Miller A.J., Widlund H.R., Horstmann M.A., Ramaswamy S., Fisher D.E. MLANA/MART1 and SILV/PMEL17/GP100 are transcriptionally regulated by MITF in melanocytes and melanoma. Am J Pathol. 2003;163:333–343. doi: 10.1016/S0002-9440(10)63657-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Motyckova G., Weilbaecher K.N., Horstmann M., Rieman D.J., Fisher D.Z., Fisher D.E. Linking osteopetrosis and pycnodysostosis: regulation of cathepsin K expression by the microphthalmia transcription factor family. Proc Natl Acad Sci USA. 2001;98:5798–5803. doi: 10.1073/pnas.091479298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salti G.I., Manougian T., Farolan M., Shilkaitis A., Majumdar D., Das Gupta T.K. Micropthalmia transcription factor: a new prognostic marker in intermediate-thickness cutaneous malignant melanoma. Cancer Res. 2000;60:5012–5016. [PubMed] [Google Scholar]
- 45.Garraway L.A., Widlund H.R., Rubin M.A., Getz G., Berger A.J., Ramaswamy S., Beroukhim R., Milner D.A., Granter S.R., Du J., Lee C., Wagner S.N., Li C., Golub T.R., Rimm D.L., Meyerson M.L., Fisher D.E., Sellers W.R. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 46.Tombran-Tink J., Barnstable C.J. Therapeutic prospects for PEDF: more than a promising angiogenesis inhibitor. Trends Mol Med. 2003;9:244–250. doi: 10.1016/s1471-4914(03)00074-1. [DOI] [PubMed] [Google Scholar]
- 47.Hase R., Miyamoto M., Uehara H., Kadoya M., Ebihara Y., Murakami Y., Takahashi R., Mega S., Li L., Shichinohe T., Kawarada Y., Kondo S. Pigment epithelium-derived factor gene therapy inhibits human pancreatic cancer in mice. Clin Cancer Res. 2005;11:8737–8744. doi: 10.1158/1078-0432.CCR-05-1323. [DOI] [PubMed] [Google Scholar]