Abstract
Three bovine group B rotavirus strains were detected from diarrheic calves during a surveillance study of rotaviral diarrhea in West Bengal, India. The sequence analysis of VP7 and NSP5 genes of these strains demonstrates a high degree of sequence variation from other group B rotavirus strains, indicating the emergence of a new genotype.
Group B rotaviruses (GBRs) are associated with severe gastroenteritis in humans, pigs, cattle, lambs, and rats (2-4, 6, 10, 12-15, 17, 20-24, 26). Bovine GBRs have been reported only from the United States, the United Kingdom, and Japan (2, 3, 6, 24). Recently, an extensive review on molecular characterization and epidemiology of GBRs has been written by Sen et al. (20). However, studies on animal rotaviruses are very limited in India (8). Here we report the detection of three bovine GBRs, collectively referred to as Kolkata strains (DB101, DB176, and DB180), from calves for which the VP7 and NSP5 gene sequences showed variations from other GBR sequences.
A total of 175 diarrheic stool samples were collected during the study period (November 2001 to May 2002) from calves between 1 and 6 months of age. All the samples were screened for rotaviruses by RNA electrophoresis as described by Herring et al. (9). Extraction of double-stranded RNA (dsRNA) from fecal samples, reverse transcription (RT), and amplification of the resultant cDNA were carried out according to the methods described by Das et al. and Sen et al. (7, 18, 19). Oligonucleotide primers MEV7-1 (5′-GGAAATAATCAGAGATGCCGTT-3′, nucleotides [nt] 1 to 20) and MEV7-3 (5′-CTACTCGTTTGGCTCCCTCC-3′, nt 795 to 776) were designed based on the VP7 gene of bovine GBR strain Mebus for amplification of gene 9 coding for VP7, and BBR11-3F (5′-GGAATAAAAGAGACAGGTAG-3′, nt 1 to 20) and BBR11-3R (5′-GGGTATTATTCCAGCACTAA-3′, nt 625 to 606) were designed from the ovine KB63 strain for amplification of gene 11 coding for NSP5. The amplified PCR products were cloned and sequenced as described earlier (7, 25). Multiple alignments were carried out using the program CLUSTAL W (version DDBJ, www.ddbj.nig.ac.jp/E-mail/homology.html) with default parameters. Amino acid identity and pairwise alignment were carried out as described by Varghese et al. (25). The phylogenetic tree was constructed using the DNASIS (version 2.1) software program.
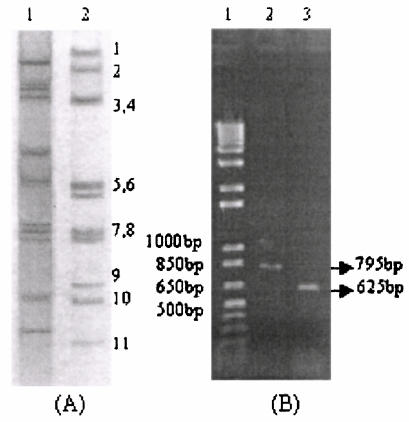
A total of 18 (10.3%) out of 175 samples were positive for group A rotavirus, and three strains (1.7%) showed the specific migration pattern of dsRNA (4-2-2-3) (15) for bovine GBRs in RNA polyacrylamide gel electrophoresis (Fig. 1A). The presence of GBRs in all three samples was confirmed by RT-PCR assays using bovine GBR VP7 gene-specific primers. The amplified PCR products were 795 bp long (partial) for VP7 and 625 bp long (full length) for NSP5 in all three strains (Fig. 1B).
FIG. 1.
(A) Electropherotyping of rotavirus dsRNA in 10% polyacrylamide gel. Lanes 1 and 2 represent the electrophoretic migration patterns of bovine group A (pattern 4-2-3-2) and group B (pattern 4-2-2-3) rotavirus strains detected during this study. (B) The RT-PCR products of the bovine GBR isolates. Lanes 2 and 3 represent the amplified VP7 gene (795 bp, partial length) and the NSP5 gene (625 bp, full length). Lane 1, 1 kb plus molecular weight marker (GIBCO BRL, Grand Island, N.Y.).
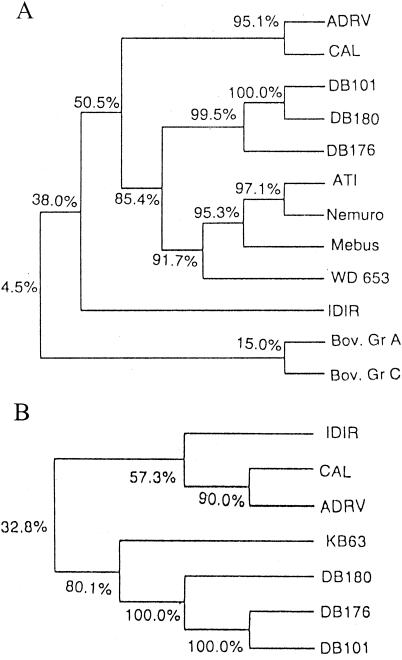
Due to lack of tissue culture adaptation of GBRs, except for one porcine strain (16), no serotyping scheme has been defined for GBRs at present (13). Moreover, only a few GBR VP7 genes have been analyzed and compared (2, 4, 11, 14, 20, 24) to develop a systemic genotyping scheme for GBRs. The VP7 gene sequences of all four published bovine GBR strains showed significantly high identity (92 to 97%) among themselves, and they may belong to the same genotype (24). In phylogenetic analysis, these bovine GBRs formed a distinct cluster away from other GBRs, indicating that they are closely related to each other (Fig. 2A). The VP7 gene sequence of Kolkata strains had an open reading frame (nt 15 to 756), which encoded a 247-amino-acid-long protein. Although the basic structural features of this protein are similar to those of other bovine GBRs, comparison of VP7 sequences of Kolkata strains with other bovine GBRs showed a high level of diversity between them (78.4 to 80.2% identity at the nucleotide and 85.4 to 88.7% at the amino acid level) (Table 1), and amino acid substitution occurred in 27 positions. These data indicated that the strains might be genetically distinct from other bovine GBRs. When the VP7 sequences of three Kolkata strains were compared with other GBR sequences (ADRV, CAL, and IDIR), they showed greater diversity both at the amino acid (51.2 to 62.2%) and the nucleotide (60.9 to 66.5%) level (Table 1). This divergence of the sequences clusters in several discrete regions (11). The amino acid substitutions of Kolkata strain VP7 sequences from other bovine GBRs are also confined to these clusters of discrete regions. These data suggest that this clustering of diverse regions may be the putative basis for genotype differentiation of GBRs.
FIG. 2.
(A) Phylogenetic tree of VP7 amino acid sequences of all the GBRs. Numbers at nodes represent the homology percentages. (B) Phylogenetic tree of NSP5 amino acid sequences of all the GBRs. Numbers at nodes represent the homology percentages.
TABLE 1.
Percentages of identities of nucleotides (upper right) and deduced amino acids (lower left) between the VP7 gene of the three Kolkata strains (DB101, DB176, and DB180) and those of other GBRs
| Strain | DB101 | DB176 | DB180 | NEMURO | MEBUS | ATI | WD653 | ADRV | CAL | IDIR |
|---|---|---|---|---|---|---|---|---|---|---|
| DB101 | 99.6 | 99.9 | 80.2 | 78.9 | 79.3 | 78.5 | 66.4 | 66.8 | 61.0 | |
| DB176 | 99.6 | 99.5 | 80.2 | 78.9 | 79.3 | 78.5 | 66.5 | 67.1 | 61.0 | |
| DB180 | 100 | 99.6 | 80.0 | 78.7 | 79.1 | 78.4 | 66.3 | 66.7 | 60.9 | |
| NEMURO | 88.7 | 88.3 | 88.7 | 92.9 | 94.4 | 94.4 | 64.9 | 64.4 | 62.8 | |
| MEBUS | 85.8 | 85.4 | 85.8 | 96.0 | 94.2 | 91.7 | 63.7 | 63.4 | 61.4 | |
| ATI | 87.4 | 87.0 | 87.4 | 97.6 | 95.1 | 93.1 | 64.7 | 64.3 | 61.5 | |
| WD653 | 87.4 | 87.0 | 87.4 | 96.4 | 92.3 | 93.9 | 63.9 | 64.4 | 62.6 | |
| ADRV | 61.4 | 61.4 | 61.4 | 62.2 | 59.4 | 60.6 | 62.2 | 92.1 | 61.5 | |
| CAL | 62.2 | 62.2 | 62.2 | 63.1 | 60.2 | 61.4 | 63.1 | 95.2 | 59.9 | |
| IDIR | 51.2 | 51.2 | 51.2 | 52.6 | 51.0 | 52.2 | 52.2 | 51.2 | 50.0 |
Sequence analysis of NSP5 genes further supports the diverse nature of Kolkata strains. To the best of our knowledge, no other bovine GBR NSP5 sequence is deposited in the GenBank database. The NSP5 protein of the Kolkata strains was 166 amino acids long, like that of the KB63 strain, but 4 and 8 amino acids shorter than the human (ADRV and CAL) and murine (IDIR) strains (5, 14, 20, 21). In group A rotaviruses, NSP5 sequences are generally conserved from different sources (>78% identity) (1), whereas the NSP5 gene of Kolkata strains showed 54.9 to 60.9% and 49.1 to 50.6% identity with ADRV, CAL, and IDIR and 80.5 and 82.5% with KB63 at the nucleotide and the amino acid level, respectively (Table 2). Amino acid sequence alignment of NSP5 of Kolkata strains with those of ADRV, CAL, IDIR, and KB63 revealed three conserved regions from 1 to 7, 48 to 77, and 121 to 166 (numbers according to IDIR) which is identical with the KB63 strain. On the other hand, protein alignment of NSP5 of Kolkata strains with KB63 showed three variable regions from 22 to 47, 83 to 107, and 133 to 138 (numbers according to DB176) within which 29 amino acids were different from KB63. The NSP5 protein of Kolkata strains contains 16.8% acidic and 16.2% basic amino acids. It also contains high proportions of serine, threonine, lysine, and glutamic acid, which are typical of NSP5 of group A rotaviruses. Similar to KB63, it contains only one potential glycosylation site at amino acid positions 76 to 78.
TABLE 2.
Percentages of identities of nucleotide and deduced amino acid sequences of the NSP5 gene of Kolkata strains and the equivalent genes from ADRV, CAL, IDIR, and KB63
| Strain | Kolkata | KB63 | ADRV | CAL | IDIR |
|---|---|---|---|---|---|
| Kolkata | 80.5 | 59.5 | 54.9 | 60.9 | |
| KB63 | 82.5 | 59.0 | 54.4 | 59.1 | |
| ADRV | 50.6 | 43.4 | 82.9 | 72.0 | |
| CAL | 50.0 | 44.3 | 90.6 | 64.2 | |
| IDIR | 49.1 | 43.1 | 67.8 | 68.4 |
Phylogenetic analysis of the VP7 and NSP5 proteins of Kolkata strains revealed a different cluster away from other GBRs (Fig. 2A and B). The number of amino acid substitutions in both VP7 and NSP5 sequences of GBR that were higher than in group A or C supports the hypothesis that GBR strains of different species origin may have had a longer period of time to diverge from one another than did group A and C rotaviruses (18, 20, 24). Based on the differences in sequences of the VP7 gene, one may postulate that Kolkata strains represent a new genotype of bovine GBRs. Phylogenetic analysis of all GBR VP7 and NSP5 protein sequences further supports the distinct nature of these strains.
Nucleotide sequence accession numbers.
GenBank accession numbers of DB101, DB176, and DB180 are AY158155, AF531910, and AF529214 for the VP7 genes and AY347930, AY347929, and AY347928 for the NSP5 genes, respectively.
Acknowledgments
We are thankful to S. C. Bhunia for excellent technical assistance.
The research was funded by the Indian Council of Medical Research (ICMR), Government of India. S. Das and S. Ghosh were supported by Senior Research Fellowships from the Council of Scientific and Industrial Research, Government of India. V. Varghese, P. Barman, and S. Chaudhuri were supported by Senior and Junior Research Fellowships from ICMR, Government of India.
REFERENCES
- 1.Blackhall, J., A. Fuentes, K. Hansen, and G. Magnusson. 1997. Serine protein kinase activity associated with rotavirus phosphoprotein NSP5. J. Virol. 71:138-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, K. O., A. V. Parwani, D. Smith, and L. J. Saif. 1997. Detection of group B rotavirus in fecal samples from diarrheic calves and adult cows and characterization of their VP7 genes. J. Clin. Microbiol. 35:2107-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chasey, D., and P. Davies. 1984. Atypical rotaviruses in pigs and cattle. Vet. Rec. 114:16-17. [DOI] [PubMed] [Google Scholar]
- 4.Chen, G. M., T. Hung, and E. R. Mackow. 1990. Identification of the gene encoding the group B rotavirus VP7 equivalent: primary characterization of the ADRV segment 9 RNA. Virology 178:311-315. [DOI] [PubMed] [Google Scholar]
- 5.Chen, G. M., T. Hung, and E. R. Mackow. 1990. cDNA cloning of each genomic segment of the group B rotavirus ADRV: molecular characterization of the 11th RNA segment. Virology 175:605-609. [DOI] [PubMed] [Google Scholar]
- 6.Chinsangaram, J., G. Y. Akita, and B. I. Osburn. 1994. Detection of bovine group B rotaviruses in feces by polymerase chain reaction. J. Vet. Diagn. Investig. 6:302-307. [DOI] [PubMed] [Google Scholar]
- 7.Das, S., A. Sen, G. Uma, V. Varghese, S. Chaudhury, S. K. Bhattacharya, T. Krishnan, P. Dutta, D. Dutta, M. K. Bhattacharya, U. Mitra, N. Kobayashi, and T. N. Naik. 2002. Genomic diversity of group A rotavirus strains infecting humans in eastern India. J. Clin. Microbiol. 40:146-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gulati, B. R., D. P. Patnayak, R. Singh, and R. Pandey. 1998. Rotavirus research in man and animals in India. Ind. J. Anim. Sci. 68:781-792. [Google Scholar]
- 9.Herring, A. J., N. F. Inglis, C. K. Ojeh, D. R. Snodgrass, and J. D. Menzies. 1982. Rapid diagnosis of rotavirus infection by direct detection of viral nucleic acid in silver-stained polyacrylamide gels. J. Clin. Microbiol. 16:473-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hung, T., G. Chen, C. Wang, Z. Chou, T. Chao, W. Ye, H. Yao, and K. Meng. 1983. Rotavirus-like agent in adult non-bacterial diarrhoea in China. Lancet 2:1078-1079. [PubMed] [Google Scholar]
- 11.Kobayashi, N., T. N. Naik, Y. Kusuhara, T. Krishnan, A. Sen, S. K. Bhattacharya, K. Taniguchi, M. Alam, T. Urasawa, and S. Urasawa. 2001. Sequence analysis of genes encoding structural and nonstructural proteins of a human group B rotavirus detected in Calcutta, India. J. Med. Virol. 64:583-588. [DOI] [PubMed] [Google Scholar]
- 12.Krishnan, T., A. Sen, J. S. Choudhury, S. Das, T. N. Naik, and S. K. Bhattacharya. 1999. Emergence of adult diarrhoea rotavirus in Calcutta, India. Lancet 353:380-381. [DOI] [PubMed] [Google Scholar]
- 13.Mackow, E. R. 1995. Group B and C rotaviruses, p. 983-1008. In M. J. Blaser, P. D. Smith, J. I. Ravdin, H. B. Greenberg, and R. L. Guerrant (ed.), Infections of the gastrointestinal tract. Raven Press Ltd., New York, N.Y.
- 14.Patric, M., K. Mayur, S. Vondefecht, and J. J. Eiden. 1991. Comparison of group B rotavirus genes 9 and 11. J. Gen. Virol. 72:2801-2804. [DOI] [PubMed] [Google Scholar]
- 15.Saif, L. J., and B. Jiang. 1994. Non group A rotaviruses of humans and animals, p. 339-371. In R. F. Ramig (ed.), Rotaviruses. Springer-Verlag, New York, N.Y.
- 16.Sanekata, T., Y. Kuwamoto, S. Akamatsu, N. Sakon, M. Oseto, K. Taniguchi, S. Nakata, and M. K. Estes. 1996. Isolation of group B porcine rotavirus in cell culture. J. Clin. Microbiol. 34:759-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanekata, T., M. U. Ahmed, A. Kader, K. Taniguchi, and N. Kobayashi. 2003. Human group B rotavirus infections cause severe diarrhea in children and adults in Bangladesh. J. Clin. Microbiol. 41:2187-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sen, A., N. Kobayashi, S. Das, T. Krishnan, S. K. Bhattacharya, and T. N. Naik. 2001. The evolution of human group B rotaviruses. Lancet 357:198-199. [DOI] [PubMed] [Google Scholar]
- 19.Sen, A., N. Kobayashi, S. Das, T. Krishnan, S. K. Bhattacharya, S. Urasawa, and T. N. Naik. 2000. Amplification of various genes of human group B rotavirus from stool specimens by RT-PCR. J. Clin. Virol. 17:177-181. [DOI] [PubMed] [Google Scholar]
- 20.Sen, A., N. Kobayashi, S. K. Bhattacharya, and T. N. Naik. 2003. Molecular characterization and epidemiology of group B rotaviruses, p. 137-178. In D. Raghunath and R. Nayak (ed.), Diarrhoeal diseases: current status, research trends and field studies. Tata McGraw-Hill Publishing Company Limited, New Delhi, India.
- 21.Shen, S., T. A. McKee, Z. D. Wang, U. Desselberger, and D. X. Liu. 1999. Sequence analysis and in vitro expression of genes 6 and 11 of an ovine group B rotavirus isolate, KB63: evidence for a non-defective, C-terminally truncated NSP1 and a phosphorylated NSP5. J. Gen. Virol. 80:2077-2085. [DOI] [PubMed] [Google Scholar]
- 22.Snodgrass, D. R., A. J. Herring, I. Campbell, J. M. Inglis, and F. D. Hargreaves. 1984. Comparison of atypical rotaviruses from calves, piglets, lambs and man. J. Gen. Virol. 65:909-914. [DOI] [PubMed] [Google Scholar]
- 23.Theil, K. W., D. L. Grooms, C. M. McCloskey, and D. R. Redman. 1995. Group B rotavirus associated with an outbreak of neonatal lamb diarrhea. J. Vet. Diagn. Investig. 7:148-150. [DOI] [PubMed] [Google Scholar]
- 24.Tsunemitsu, H., D. Morita, H. Takaku, T. Nishimori, K. Imai, and L. J. Saif. 1999. First detection of bovine group B rotavirus in Japan and sequence of its VP7 gene. Arch. Virol. 144:805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varghese, V., S. Das, N. B. Singh, K. Kojima, S. K. Bhattacharya, T. Krishnan, N. Kobayashi, and T. N. Naik. 2004. Molecular characterization of a human rotavirus reveals porcine characteristics in most of the genes including VP6 and NSP4. Arch. Virol. 149:155-172. [DOI] [PubMed] [Google Scholar]
- 26.Vonderfecht, S. L., D. A. Lindsay, and J. J. Eiden. 1994. Detection of rat, porcine, and bovine group B rotavirus in fecal specimens by solid-phase enzyme immunoassay. J. Clin. Microbiol. 32:1107-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]