Introduction
Many patients on antiretroviral therapy experience episodes of low-level viremia (LLV), commonly defined as viral loads between 50 and 1000 HIV-RNA copies/mL [1]. Since many treatment guidelines define virologic success as maintaining viral loads below the limits of assay detection [2–5], LLV can be a concern for both physicians and patients.
Resistance testing has been shown to be an effective predictor of future virologic failure in a number of studies [6–9]. However, most commercial resistance assays can only be performed on samples with viral loads above a minimum of 500-2000 copies/mL [10,11]. Despite this, in-house resistance assays can be performed on samples with low-level viraemia below 1000 copies/mL [12–14], and the success rate of such testing has increased over time in some settings [15]. Indeed, several studies have found that LLV is associated with subsequent virologic failure, immune activation, inadequate CD4 recovery, and development of drug resistance [16–21], and that resistance can be detected at LLV [22–24]. However, there is limited evidence that risk of virologic failure after LLV can be further elevated by the presence of resistance. Intriguingly, however, two recent studies on a modest number of individuals indicated that LLV resistance may be associated with virologic failure [25,26].
In British Columbia, Canada, resistance testing on LLV samples has been performed since approximately 2000. Starting in 2004, the results of resistance testing on LLV samples were made available to the ordering physician prospectively. We undertook the present analysis to evaluate the impact of emergent HIV drug resistance at LLV on the risk of subsequent virologic failure.
Methods
Resistance testing methods
Samples with viral loads below 1000 copies/mL underwent standard population-based sequencing using methodology identical to that performed on higher viral load samples. However these methods evolved over the years with successive generations of various laboratory technologies. For instance, viral load values were obtained using the Roche COBAS Amplicor HIV-1 Monitor Test v1.5 until 2009 and the Roche COBAS TaqMan HIV-1 v1.0 assay after 2009. HIV RNA was extracted from 500 uL of plasma using either manual or automated methodologies depending on the testing year. The protease and reverse transcriptase regions were amplified using nested RT-PCR, with a product spanning from the beginning of protease to codon 400 of RT. Bidirectional sequencing was performed using one of several ABI sequencers (3100, 3130, 3700, 3730), followed by sequence analysis using Sequencher (Genecodes) or RECall [27]. Samples which failed this process were re-extracted and reamplified with primers spanning a smaller region of pol (to codon 250 of RT), with the proportion of such cases increasing as viral loads decreased (Gonzalez-Serna 2013, Accepted, Clinical Infectious Diseases). In total, there were 4915 LLV samples tested for drug resistance from a total of 2492 patients.
Patient selection
Of these 2492 patients, we selected the 2176 patients (87%) who experienced their first documented LLV episode while on antiretroviral therapy. Low-level viremia was defined as an HIV RNA result <1000 copies/mL, consistent with the U.S. Department of Health and Human Services definition [28]. This definition includes patients experiencing “blips” [18,19,29] as well as patients with higher and less-transient episodes of elevated viremia below 1000 copies/mL. Resistance testing was successful in 1965 of these patients (90%) and unsuccessful in 211 (10%), consistent with the approximate 90% success rate of our resistance assay at LLV [24].
To determine the extent of resistance at LLV, the sequences obtained from these patients were interpreted separately using the Stanford HIV Drug Resistance Database [30] or Virco/Janssen VirtualPhenotype [31,32]. For each patient, at the time of first LLV, a score was generated based on the number of active drugs in their antiretroviral regimen. We estimated the scores, called genotypic susceptibility scores (GSS) using the Stanford HIV Drug Resistance Database [30] and separately, we estimated virtual phenotypic susceptibility scores (vPSS) using the Virco/Janssen VirtualPhenotype [31–33]. The GSS and vPSS were used to stratify patients into 4 categories based on the residual antiviral activity of the ARV regimen at the time of LLV. For each drug, a GSS or vPSS value of 1 was assigned if resistance interpretation identified no resistance to low-level resistance. A GSS or vPSS of 0.5 was assigned to drugs with intermediate resistance, and a value of 0 was assigned to drugs with high-level resistance. The GSS or vPSS values for all drugs in a regimen were then totaled, and patients were grouped corresponding to the number of active drugs prescribed: <1; 1-1.5; 2-2.5; and ≥3. Thus, a value of ≥3 or more indicates a fully-active regimen, and a value of <3 indicates increasingly higher drug resistance and secondarily, increasingly less residual antiviral potency of the ARV regimen. For simplicity, the GSS is reported for all analyses, unless otherwise indicated. The results of these analyses remain virtually unchanged when the GSS was replaced with the vPSS.
Patient follow-up and statistical analyses
Ethical approval for this study was granted by the University of British Columbia–Providence Health Care Research Ethics Board. Study observation comprised the period between August 1996 and May 2013. Patients were evaluated for their long-term risk of subsequent virologic failure ≥1000 copies/mL following their LLV episode. A sensitivity analysis was also performed using a more stringent failure definition of ≥5000 copies/mL. Patient adherence to therapy was estimated from the percentage of prescription refills obtained over the 12 months following their LLV episode regardless of whether they changed therapies. Patients with fewer than 6 months of follow-up after LLV (N=23) were not given an estimation of their adherence level.
Risk was determined through Kaplan-Meier analyses of time to virologic failure, and through Cox proportional hazards models. For clarity, Kaplan-Meier curves are displayed up to five years, though longer-term follow-up was obtained for some patients and was included in the Cox models. Patients who did not experience virologic failure were classified by whether they maintained LLV, changed therapy, had subsequent virologic suppression, or were lost to follow- up, and were censored at that point. Statistical tests for the dependence between categorical variables and categorical outcomes were performed using Fisher's exact test or the Chi-square test. The Kruskall-Wallis test was used to compare viral loads across GSS groups. The log-rank test was used to compare survival curves.
Following univariate analyses, two multivariate Cox proportional hazards models were built (one for GSS and another for vPSS) in order to estimate the effects of a number of variables and their influence on risk of virologic failure. In addition to GSS, the variables included were viral load at LLV, year at LLV, regimen type at LLV, presence of nucleotide mixtures in the sequence, whether testing was retrospective, patient gender, patient treatment experience, and patient adherence. Patients with missing data for these variables were excluded, leaving 1904 of 1965 (97%) observations used to fit the model. The exact method was used to handle ties in the failure times. Regression coefficients were estimated by maximizing the partial likelihood.
Stepwise selection with a 5% significance level for entry and removal was used to choose the best model. Both forward selection and backward elimination yielded the same final model. Martingale residuals were used to assess the assumption of proportional hazards for these variables using a supremum test of 1,000 simulated residual patterns and the observed values of the covariates. If certain variables violated this assumption, new models were made with stratification by these variables.
Results
Characteristics at baseline and at low-level viremia
The median pre-therapy plasma viral load of these patients (where available; N=1301) was 4.8 log copies/mL (Interquartile range: 4.3-5.0 log copies/mL). Patients were on treatment a median of 4.0 years (IQR: 1.0–7.9 years) prior to their first low-level viraemia episode, and were permitted to change therapies over this period. At LLV, the median plasma viral load was 481 copies/mL (Interquartile range: 331-696 copies/mL). At the time of their first low-level viraemia episode, patients in the study were on a variety of antiretroviral regimens, with 69% having changed regimens from their initial therapies (N=1365). Characteristics prior to therapy (baseline) and at low-level viremia are listed in Table 1.
Table 1.
Characteristics prior to therapy and at low-level viremia
| Baseline Characteristics | Characteristics at Low-Level Viremia | ||
|---|---|---|---|
| Sex - % Male (N male/total) | 78% (1514/1942) | ||
| Plasma viral load: Median (Interquartile Range) (No.) | 4.83 log copies/mL (4.27-5.04 log copies/mL) (N=1301) | 2.68 log copies/mL (2.52-2.84 log copies/mL) (N=1965) | |
| Treatment era of therapy initiation, % of patients | Pre-1996 | 20% (386/1965) | |
| 1996-1999 | 34% (661/1965) | ||
| 2000-2003 | 16% (317/1965) | ||
| 2004-2007 | 17% (333/1965) | ||
| 2008-2012 | 14% (268/1965) | ||
| Regimen type, % of patients | NNRTI-based | 20% (388/1965) | 20% (391/1965) |
| PI-based | 23% (459/1965) | 24% (472/1965) | |
| Boosted PI-based | 17% (329/1965) | 27% (531/1965) | |
| Other | 40% (789/1965) | 29% (571/1965) | |
| Percentage with resistance | 14% (103/736) | 30% (594/1965) | |
Patient characteristics were observed at two different time points, and they are recorded in two columns in Table 1 The column titled Baseline Characteristics represents observations for patients prior to beginning antiretroviral therapy (where available), and the column titled Characteristics at Low-Level Viremia represents the corresponding characteristics at LLV if they have changed. At baseline, resistance is defined as having drug-resistance associated mutations for any class of medications as observed prior to initiating antiretroviral therapy. At LLV, resistance is defined as having a GSS <3. Some baseline characteristic data was not available for all patients. A total of 23 patients did not have gender information available, and 664 patients did not have a documented pre-therapy (baseline) viral load. Neither the sex nor treatment era of therapy initiation changed between baseline and LLV.
HIV drug resistance was detected in 30% of patients during their first episode of low-level viraemia. Resistance to the nucleoside reverse transcriptase inhibitor (NRTI) drug class was most common at 28% (N=541), followed by the non-nucleoside reverse transcriptase inhibitors (NNRTIs) at 16% (N=305) and the protease inhibitors (PIs) at 7% (N=146). Multiclass resistance at LLV ranged from 3% for resistance to both PIs and NNRTIs to 8% for resistance to both NNRTIs and NRTIs.
Of the patients in this study, those on boosted-PI-based regimens were significantly less likely to have resistance at LLV (GSS <3) compared to all other regimens, with 15% (79/531) having resistance (p<0.0001). The other regimens were all associated with higher rates of resistance at LLV: 28% (134/472) for the unboosted PIs, 26% (101/391) for the NNRTIs and 30% (127/418) for other regimens of ≥3 agents. Of the patients prescribed <3 ARVs, 71% (110/153) had a GSS <2.
While the overall median viral load at LLV was 481 copies/mL, the different GSS groups had slightly different median viral loads. Samples with GSS ≥3 had significantly lower median viral loads at LLV (456 copies/mL) compared to samples with GSS 2-2.5 (514 copies/mL), 1-1.5 (580 copies/mL) or <1 (574 copies/mL) (p<0.01). Patients on regimens with <3 ARVs had the highest viral loads at LLV (median 635 copies/mL), significantly higher (p<0.001) than those on other regimens, whose median viral loads ranged from 436-494 copies/mL. There was no significant difference in viral load between samples which did or did not generate successful genotyping results: median 481 copies/mL (IQR: 331-696); versus 459 copies/mL (IQR: 318-676), respectively (p=0.4).
Excluded participants
Of the 1965 patients, 263 either changed therapy (N=238) or were lost to follow-up (N=25) before they were observed again following LLV, and were thus excluded from our Kaplan-Meier analyses at study baseline. The median plasma viral loads of these excluded patients did not differ significantly from those patients with additional observations (median 510 versus 480 copies/mL, p=0.07). The excluded patients did, however, have significantly more results with GSS <3 (i.e., resistant), with 46% having a GSS <3 (120/263) at LLV compared to 28% of those who had follow-up observations available (474/1702), p<0.0001. The groups were not significantly different in terms of their regimen makeup, with two exceptions; where the excluded patients had ~10% fewer patients on boosted-PI based regimens and ~10% more patients on “other” regimens compared to the included patients. The excluded patients also had 8% more males than the included participants.
Resistance at LLV and subsequent virologic failure
Included patients (N=1702) were then followed longitudinally following their LLV episode, with a mean of 1.2 years of follow-up, and a median of 7 months (Interquartile range [IQR]: 3 months – 1.5 years). They were assessed for their risk of subsequent virologic failure above 1000 copies/mL while remaining on the same antiretroviral regimen. Patients were followed as long as they continued on the same antiretroviral regimen until reaching one of five pre-specified endpoints: virologic failure, virologic suppression, sustained low-level-viremia, changing therapy, or loss-to-follow-up.
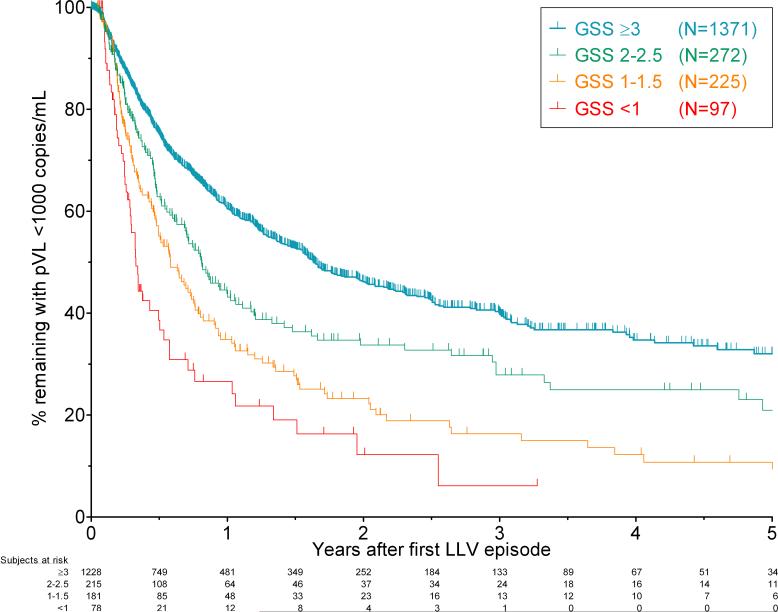
Overall, 50% of patients experienced virologic failure during study follow-up (855/1702 patients). Patients with resistance at LLV had a significantly higher risk of virologic failure compared to those who did not (Figure 1). Results were similar by vPSS rather than GSS (Supplementary Figure 1). The hazard ratio for risk of virologic failure increased as the extent of resistance increased. Relative to a GSS of ≥3, the hazard ratios were: 1.4 (95% confidence interval [CI]: 1.2-1.7) for patients with GSS 2-2.5. This increased to 2.0 (1.7-2.5) for patients with GSS 1-1.5, and 3.0 (2.2-4.0) for patients with GSS <1. Similarly, the proportion of patients who went on to achieve virologic suppression on the same therapy following their LLV episode fell as resistance level increased, as did the proportion who maintained LLV without virologic failure (Table 2).
Figure 1. Virologic failure was faster and more common in patients with lower genotypic susceptibility scores during low-level viraemia.
Kaplan-Meier curves for the proportion of patients remaining on the same therapy with viral loads <1000 copies/mL following their first low-level viraemia (LLV) episode. Patients are divided into 4 groups according to their GSS, and followed for up to five years while remaining on constant therapy. Patients with GSS ≥3 had the best outcomes following LLV, while patients with GSS <1 had the worst outcomes. The survival curves were all significantly different by the log-rank test (p<0.001). The numbers of patients remaining at risk at each six month interval are shown below the figure. Ticks represent censoring of patients suppressing on the same therapy, changing therapy, lost to follow-up, or at their last available time point.
Table 2.
Outcomes after low-level viremia varied with genotypic susceptibility scores
| GSS <1 (N=97) | GSS 1-1.5 (N=225) | GSS 2-2.5 (N=272) | GSS ≥3 (N=1371) | Failed testing (N=211) | p value | |
|---|---|---|---|---|---|---|
| Virologic Failure, % (N) | 54% (52) | 58% (130) | 45% (123) | 40% (550) | 32% (67) | <0.001 |
| Suppressed <50 copies, % (N) | 1% (1) | 4% (9) | 14% (39) | 26% (362) | 36% (77) | <0.001 |
| Maintained LLV, % (N) | 1% (1) | 0% (1) | 3% (8) | 5% (72) | 9% (20) | <0.001 |
| Changed therapy, % (N) | 43% (42) | 38% (85) | 36% (97) | 26% (352) | 20% (43) | <0.001 |
| Lost to follow-up, % (N) | 1% (1) | 0% (0) | 2% (5) | 3% (35) | 2% (4) | 0.14 |
Lower genotypic susceptibility scores (GSS) indicate more-compromised regimen activity (i.e., higher levels of resistance). Patients are grouped by their GSS values at low-level viraemia (LLV), ordered by decreasing extent of resistance (i.e. increasing GSS). A group of patients with failed resistance assays at LLV are also included. Statistical significance of differences amongst the GSS groups was tested using the Chi-square test. As resistance decreased (GSS increased): rates of virologic failure decreased, rates of virologic suppression increased, rates of maintaining LLV increased, and rates of changing therapy decreased. There was not a significant difference in the proportions of patients who were lost to follow-up amongst the groups. Patients whose resistance assays failed at LLV tended to have the most favourable responses. All proportions are excluding patients who were excluded at baseline due to changing therapy or loss to follow-up.
As a sensitivity analysis, we used a more conservative definition, with virologic failure defined as having a viral load ≥5000 instead of ≥1000 copies/mL. The results were broadly similar for both definitions of virologic failure. Slightly fewer patients experienced virologic failure under this more stringent definition of 5000 copies, at 42% of patients (N=714), compared to 50% under the 1000 copies definition (N=855). Hazard ratios in this sensitivity analysis were similar but smaller compared to the original analysis. For patients with GSS 2-2.5 at LLV, the hazard ratio was 1.3 (p=0.007). For those with 1-1.5, the hazard ratio was 1.7 (p<0.0001), and those with GSS <1 had a hazard ratio of 2.1 (p<0.0001). Kaplan-Meier plots appeared largely similar for either definition of virologic failure.
LLV resistance predicts subsequent response regardless of patient subgroup
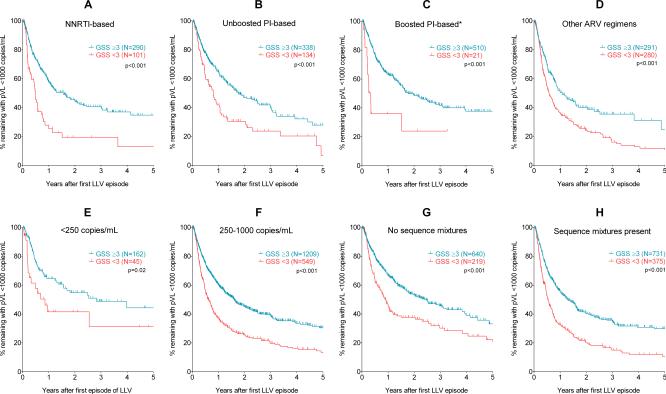
In additional sensitivity analyses, we tested whether resistance testing results were robust to various patient or sample characteristics (Figures 2 & 3). For these subanalyses, a patient was considered to have resistance if their GSS was <3 and was considered not to have resistance if their GSS was ≥3. Resistance testing was considered to be predictive of subsequent response if the “resistant” and “not resistant” groups were significantly different by a log-rank test on their Kaplan-Meier survival curves. Resistance was predictive of response for all patients regardless of the regimen they were taking at LLV (Figures 2A-2D), with p-values <0.01 for patients on NNRTI-based regimens, PI-based regimens, and other antiretroviral regimens (comprising <3 or ≥3 drugs). Boosted PI-based regimens were borderline-significant (p<0.05), but when ritonavir was included in the GSS scoring, the statistical significance increased (p<0.001).
Figure 2. Prediction of virologic failure by low-level viraemia resistance genotyping was robust to antiretroviral regimen, viral load, and presence of nucleotide mixtures.
All figure parts contain Kaplan-Meier plots with patients divided by GSS ≥3 (“Susceptible” – black lines) or GSS <3 (“Resistant” – grey dashed lines). Figures 2A-2D show patients stratified by antiretroviral regimen in use at the time of LLV. Figure 2C shows the outcomes including ritonavir in the GSS scoring. Figures 2E and 2F show patients with very low viral loads (<250 copies/mL) or higher viral loads. Figures 2G and 2H stratify responses by whether or not nucleotide mixtures were observed in the sequence detected at LLV. The log-rank test was used to test for statistically significant differences between the survival curves.
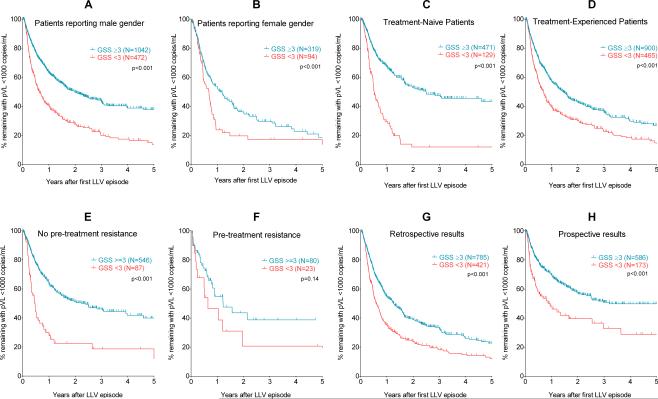
Figure 3. Prediction of virologic failure by low-level viraemia resistance genotyping was robust to gender, treatment-experience, pre-therapy resistance, or time of testing.
All figure parts contain Kaplan-Meier plots with patients divided by GSS ≥3 (“Susceptible” – black lines) or GSS <3 (“Resistant” – grey dashed lines). Figures 3A and 3B show patients reporting male or female gender. Figures 3C and 3D show treatment-naïve or treatment-experienced groups. Figures 3E and 3F show patients according to whether they had pre-treatment (i.e., transmitted) drug resistance. Figures 3G and 3H show patients tested retrospectively or prospectively. The log-rank test was used to test for statistically significant differences between the survival curves.
Resistance testing was also robust to the level of LLV (Figures 2E & 2F), even in patients with extremely low viral loads below 250 copies/mL (p=0.02). Patients with higher viral loads during LLV (250-1000 copies/mL) also had significantly different responses depending on whether resistance was detected during LLV (p<0.001). Patients with successively higher viral loads had worse outcomes than those with lower viral loads, but resistance profile at LLV remained a significant predictor of virologic failure (Supplementary Figure 2).
There were also 859 patients whose sequences obtained at LLV did not contain any nucleotide mixtures – a potential marker that only a few viral copies were amplified and a possible source of bias. However, LLV resistance was also predictive of failure even in these patients with no nucleoside mixtures, similar to those where nucleotide mixtures were detected (Figures 2G & 2H). Results were also robust to male or female gender (Figures 3A & 3B), and whether patients were taking their first antiretroviral regimen or were previously treatment- experienced (Figures 3C & 3D). Of the 736 patients with pre-treatment resistance data available, 102 (14%) had pre-treatment mutations which would have resulted in a reduced GSS. Detection of resistance at LLV was predictive of virologic failure regardless of pre-existing resistance (Figures 3E & 3F), though this did not reach statistical significance in the group with pre-treatment resistance, potentially due to the smaller number of patients.
LLV resistance results are acted on by physicians and result in better patient outcomes
Approximately 45% (N=759) of genotypic testing results were obtained within 30 days of the draw date and made available to the ordering physician. Amongst those patients for whom their resistance results were available within 30 days (the “prospective” group), overall outcomes seemed to be better than those for whom the resistance result was made available to the physician more than 30 days after the draw date (the “retrospective” group).
As with the above analyses, resistance at LLV remained a significant predictor of subsequent virologic failure regardless of whether the results were prospective or retrospectively collected (Figures 3G & 3H). There were also more therapy switches in the prospective group (53%) versus the retrospective group (31%) when patients had a GSS <3 (p<0.001), but in patients without resistance (GSS ≥3), therapy changes were lower and similar in both prospective (26%) and retrospective (26%) groups (p=n.s.), since treatment switches due to resistance were likely not necessary.
Outcomes after therapy changes
We then examined a group of 576 patients who changed therapy following their low-level viraemia episode. Comparing their GSS at LLV to the GSS of their next regimen, 244 patients (42%) changed to regimens with higher GSS values, 258 (45%) changed regimens but had the same GSS, and only 74 patients (13%) changed to regimens with lower GSS values.
Of those who maintained the same GSS, 204 (79%) already had regimens with GSS values of 3 or more. Similarly, a majority (N=151, 62%) of the patients who changed to regimens with higher GSS values had former GSS values of less than 3, indicating switches to more potent regimens. Finally, in terms of virologic outcomes on these new regimens, patients whose regimen changes resulted in higher GSS values had better outcomes than those with lower GSS values for their new regimens. Additionally, we again observed a “dose-dependent” effect of GSS on the virologic outcomes of patients on their new regimens (p=0.002) (Supplementary Figure 3).
Patients who maintained low-level viraemia
There were a total of 441 patients (26%) who had at least 2 LLV episodes prior to changing therapy, experiencing virologic failure, suppressing on the same therapy, or being lost-to-follow-up. These patients were followed longitudinally while maintaining LLV, and were assessed for their risk of accumulating resistance, as indicated by a decrease in the GSS between their first and last LLV genotype. Of these 441 patients, 12% (52 patients) accumulated resistance to the agents in their regimen.
Outcomes in patients with failed resistance testing results
Resistance testing failed for 211 patients. Interestingly, these patients had generally better outcomes to those who had successful LLV genotypes, including those whose genotypes indicated a fully susceptible regimen (Supplementary Figure 4). As previously noted, there were no significant differences in viral loads between those with successful and failed resistance testing.
Impact of adherence
Overall patient adherence was high in this study, according to their percentage of prescription refills obtained over the first 12 months following LLV. The median level of adherence was 93.4% (IQR: 66.0% - 99.7%). Patients were divided into quartiles according to their adherence levels: 0-65% adherent, 66-93% adherent, 93-99% adherent, and 100% adherent. Times to virologic failure were evaluated for each of these adherence groups (Supplementary Figure 5). GSS was a strong predictor of virologic failure for patients with good adherence (p<0.0001; Supplementary Figure 5C & 5D), a marginal predictor for patients with moderate adherence (p=0.06; Supplementary Figure 5B), and a poor predictor of virologic failure for patients with the lowest adherence levels (p=0.28, Supplementary Figure 5A).
Significance of resistance at low-level viremia confirmed by multivariate analysis
In a multivariate Cox proportional hazards model, the effect of GSS remained highly significant after controlling for a number of other variables. After exclusion of non-statistically significant variables, the resulting hazards model included the following variables: GSS, patient treatment experience, patient adherence, year at LLV, whether the sample was retrospectively tested, and whether the sequence contained nucleotide mixtures.
Having a GSS <3 at LLV was associated with a significantly increased risk of virologic failure, with a hazard ratio of 1.34 (95% confidence interval: 1.14 – 1.57, p<0.001). Other variables increasing the risk of future virologic failure were, in order of decreasing magnitude: the results not being available to the physician due to retrospective testing (hazard ratio: 1.28, p=0.007), and whether the sequence contained nucleotide mixtures (hazard ratio: 1.28, p <0.001). Three variables significantly decreased the risk for treatment failure: being treatment-naïve (hazard ratio: 0.80, p=0.005), experiencing LLV in later calendar years (hazard ratio: 0.98, p=0.013), and higher patient adherence (hazard ratio: 0.98, p<0.001).
The proportional hazards assumption was found to be violated for three variables: whether the sample was retrospectively tested (p<0.0001), year at LLV (p=0.01), and degree of adherence (p=0.001). A new Cox proportional hazards model was built which was stratified by the retrospective testing variable. After this stratification, year at LLV no longer violated the proportional hazards assumption (p=0.27) and adherence only violated it to a moderate extent (p=0.02). Furthermore, this stratified model yielded similar results and hazard ratios as before, suggesting that the model was robust to violations in the proportional hazards assumption. Finally, models generated using vPSS also had similar results to the GSS models.
Discussion
We have presented a large-scale study of the impact of antiretroviral resistance on virologic outcomes following low-level viraemia (LLV). Our results demonstrate that patients experiencing their first episode of LLV while on ARV therapy were up to three times more likely to experience subsequent virologic failure if they had emergent drug resistance at the time of LLV. Virologic failure followed a “dose-dependent” response in relationship to the GSS values, with progressively decreasing GSS associated with increasing risk of subsequent virologic failure. Our observations were robust in a diverse set of sensitivity analyses, focusing on a more stringent failure definition, antiretroviral class, viral load, treatment-experience, gender, baseline resistance, and patient adherence. GSS remained significant predictor of subsequent virologic failure even when controlling for these other variables. Finally, resistance testing at LLV was acted on by the ordering physicians and resulted in better outcomes.
This study builds on a growing body of literature surrounding low-level viraemia [1,13,16,17,20–22,26,34–42]. Current guidelines suggest that “[i]n persons with HIV RNA levels >500 but <1000 copies/mL, [resistance] testing may be unsuccessful but should still be considered”, and give it a moderate strength of recommendation (BII) [28]. Furthermore, these guidelines specifically do not recommend testing for patients with viral loads <500 copies/mL, with a strong recommendation against such testing (AIII). In addition, resistance assay kits are only approved by the FDA to test samples with viral loads above 1000 or 2000 copies/mL [10]. However, our results indicate that resistance testing of samples with viral loads below 1000 copies/mL provides clinically relevant information. These findings provide a strong rationale for the reevaluation of current drug resistance testing guidelines.
Some strengths of this study are the large number of patients examined, long-term patient follow-up of up to five years, and the fact that various sensitivity analyses yielded similar results to our original analyses. We also observed that physicians use this information and act on it by changing therapies where patients are found to have resistance at LLV. As with other studies of resistance [43,44], LLV resistance tended to be most common for the NRTI drug class, and least common for the PI drug class, especially for patients on boosted-PI-based regimens. However, there is some evidence that low-level viraemia itself may be more common in patients receiving boosted-PI-based regimens [29].
Along with the genotype susceptibility scores, the viral load at LLV influenced the likelihood of virologic failure over the study period, with higher levels of viraemia associated with higher risk. Similar to GSS, virologic outcomes stratified by viral load stratum followed a “dose-dependent” association. Thus, both the level of viraemia and the presence of resistance should be taken into account when assessing a patient's risk of future treatment failure.
Interestingly, we found that patients whose resistance test failed to produce usable sequences actually tended to have better virologic outcomes compared to those who had results – even those who did not have regimens compromised by resistance (GSS ≥3). These patients had lower rates of virologic failure, and were more likely to suppress below 50 copies/mL or maintain LLV during follow-up. While these patients had slightly lower viral load levels at LLV, they were not significantly lower. Thus, the improved outcomes over patients with LLV resistance results may be driven by something other than viraemia – or that these viral loads may have been due to assay false-positives [45]. Another observation in this study was the fact that resistance remained a significant predictor of virologic failure even adjusting for adherence. GSS was a significant predictor of virologic failure for a majority of patients, regardless of adherence level. However, in lower strata of adherence levels, the effect of resistance was reduced.
The retrospective, observational nature of this study represents a possible limitation, as this may lead to bias in the distribution and treatment of patients. The patients excluded from our analysis due to lack of useable follow-up differed slightly from those patients who were included in our analyses, and this may have impacted our results. However, these excluded patients were actually more likely to have a GSS <3, meaning that our observations may actually be conservative estimates of the impact of resistance at LLV. The consistent results observed when virologic failure was defined at 1000 or 5000 copies/mL are reassuring. However, as in all observational studies, unknown confounding factors, which were not distributed evenly throughout the population cannot be adjusted for or excluded.
Data for this study were collected on samples dating back to 1996. Various resistance testing and viral load monitoring methods have been in place over the years in British Columbia since that time, and while they are largely similar, there may have been changes in the sensitivity or accuracy of these methods as they were gradually updated and implemented. Furthermore, only a small number of patients remained in the study after five years without experiencing an event or changing therapy. This study had an overall rate of virologic failure of 44%, which is very high – even compared to other studies in British Columbia [46]. However, this is likely due to the fact that the patients in the current study were already experiencing virologic failure at LLV using a strict definition of a viral load ≥50 copies/mL, implying they were likely already at an elevated risk of future higher virologic failure compared to a more general treatment population.
In conclusion, our results demonstrate that emergent HIV drug resistance at LLV is strongly associated with subsequent virologic failure. Furthermore, we uncovered a “dose-dependent” increase in the hazard ratio for virologic failure with decreasing GSS estimated at the time of LLV. Our results were robust in a number of sensitivity analyses. Based on these findings we propose that resistance genotyping be encouraged among HIV infected individuals on ARV therapy who experience their first viral rebound with viral loads between 50 and 1000 copies/mL.
Supplementary Material
Acknowledgments
The authors would like to thank Guillaume Colley for assistance in obtaining the adherence data. LCS is supported by a Canadian Institutes of Health Research Doctoral Award. PRH holds a GSK-CIHR Research Chair in HIV/AIDS. JSGM holds a grant from the National Institutes of Drug Abuse (NIDA) 1R01DA036307-01: “Seek and Treat for Optimal Prevention of HIV & AIDS (STOP HIV/AIDS) in BC”.
Footnotes
Presented in part at: The International Workshop on HIV & Hepatitis Virus Drug Resistance and Curative Strategies. June 4-8, 2013. Toronto, Ontario.
References
- 1.Cohen C. Low-level viremia in HIV-1 infection: consequences and implications for switching to a new regimen. HIV clinical trials. 2009;10:116–24. doi: 10.1310/hct1002-116. [DOI] [PubMed] [Google Scholar]
- 2.BC Centre for Excellence in HIV/AIDS Therapeutic Guidelines Committee Therapeutic Guidelines: Antiretroviral Treatment (ARV) of Adult HIV Infection. 2011:1–48. [Google Scholar]
- 3.European AIDS Clinical Society European AIDS Clinical Society Guidelines: Clinical Management and Treatment of HIV-infected Adults in Europe. 2011 [Google Scholar]
- 4.EACS European AIDS Clinical Society (EACS) Guidelines version 6.1. 2012 [Google Scholar]
- 5.Williams I, Churchill D, Anderson J, Boffito M, Bower M, Cairns G, et al. British HIV Association guidelines for the treatment of HIV-1-positive adults with antiretroviral therapy 2012 British HIV Association guidelines for the treatment of HIV-1-positive adults with antiretroviral therapy 2012. HIV Medicine. 2012;13:1–85. [Google Scholar]
- 6.Harrigan PR, Côté HC. Clinical utility of testing human immunodeficiency virus for drug resistance. Clinical infectious diseases. 2000;30(Suppl 2):S117–22. doi: 10.1086/313861. [DOI] [PubMed] [Google Scholar]
- 7.Clevenbergh P, Durant J, Halfon P, del Giudice P, Mondain V, Montagne N, et al. Persisting long-term benefit of genotype-guided treatment for HIV-infected patients failing HAART. The Viradapt Study: week 48 follow-up. Antiviral therapy. 2000;5:65–70. [PubMed] [Google Scholar]
- 8.Baxter JD, Mayers DL, Wentworth DN, Neaton JD, Hoover ML, Winters MA, et al. A randomized study of antiretroviral management based on plasma genotypic antiretroviral resistance testing in patients failing therapy. CPCRA 046 Study Team for the Terry Beirn Community Programs for Clinical Research on AIDS. AIDS. 2000;14:F83–93. doi: 10.1097/00002030-200006160-00001. [DOI] [PubMed] [Google Scholar]
- 9.DeGruttola V, Dix L, D'Aquila R, Holder D, Phillips A, Ait-Khaled M, et al. The relation between baseline HIV drug resistance and response to antiretroviral therapy: re-analysis of retrospective and prospective studies using a standardized data analysis plan. Antiviral therapy. 2000;5:41–8. doi: 10.1177/135965350000500112. [DOI] [PubMed] [Google Scholar]
- 10.Shafer RW, Dupnik K, Winters MA, Eshleman SH. A Guide to HIV-1 Reverse Transcriptase and Protease Sequencing for Drug Resistance Studies. Los Alamos, NM: 2001. [PMC free article] [PubMed] [Google Scholar]
- 11.Monogram Biosciences. Features of GenoSure PRIme. GenoSure PRIme. 2013:1. [Google Scholar]
- 12.Stelzl E, Troppan KT, Winkler M, Korn K, Kessler HH. Optimized protocol for detection of HIV-1 drug mutations in patients with low viral load. Journal of virological methods. 2010;168:152–4. doi: 10.1016/j.jviromet.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Tobin NH, Learn GH, Holte SE, Wang Y, Melvin AJ, McKernan JL, et al. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: expression of archival virus and replication of virus. Journal of virology. 2005;79:9625–34. doi: 10.1128/JVI.79.15.9625-9634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallien S, Delaugerre C, Charreau I, Braun J, Boulet T, Barrail-Tran A, et al. Emerging integrase inhibitor resistance mutations in raltegravir-treated HIV-1-infected patients with low-level viremia. AIDS. 2011;25:665–9. doi: 10.1097/QAD.0b013e3283445834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mackie NE, Phillips AN, Kaye S, Booth C, Geretti A-M. Antiretroviral drug resistance in HIV-1-infected patients with low-level viremia. The Journal of infectious diseases. 2010;201:1303–7. doi: 10.1086/651618. [DOI] [PubMed] [Google Scholar]
- 16.Sungkanuparph S, Groger RK, Overton ET, Fraser VJ, Powderly WG. Persistent low-level viraemia and virological failure in HIV-1-infected patients treated with highly active antiretroviral therapy. HIV medicine. 2006;7:437–41. doi: 10.1111/j.1468-1293.2006.00403.x. [DOI] [PubMed] [Google Scholar]
- 17.Karlsson AC, Younger SR, Martin JN, Grossman Z, Sinclair E, Hunt PW, et al. Immunologic and virologic evolution during periods of intermittent and persistent low-level viremia. AIDS. 2004;18:981–9. doi: 10.1097/00002030-200404300-00005. [DOI] [PubMed] [Google Scholar]
- 18.Sklar PA, Ward DJ, Baker RK, Wood KC, Gafoor Z, Alzola CF, et al. Prevalence and clinical correlates of HIV viremia (‘blips’) in patients with previous suppression below the limits of quantification. AIDS. 2002;16:2035–41. doi: 10.1097/00002030-200210180-00008. [DOI] [PubMed] [Google Scholar]
- 19.Nettles RE, Kieffer TL, Kwon P, Monie D, Han Y, Parsons T, et al. Intermittent HIV-1 Viremia (Blips) and Drug Resistance in Patients Receiving HAART. JAMA: the journal of the American Medical Association. 2005;293:817. doi: 10.1001/jama.293.7.817. [DOI] [PubMed] [Google Scholar]
- 20.Lo Re V, Gasink L, Kostman JR, Leonard D, Gross R. Natural history of patients with low-level HIV viremia on antiretroviral therapy. AIDS patient care and STDs. 2004;18:436–42. doi: 10.1089/1087291041703692. [DOI] [PubMed] [Google Scholar]
- 21.Greub G, Cozzi-Lepri A, Ledergerber B, Staszewski S, Perrin L, Miller V, et al. Intermittent and sustained low-level HIV viral rebound in patients receiving potent antiretroviral therapy. AIDS. 2002;16:1967–9. doi: 10.1097/00002030-200209270-00017. [DOI] [PubMed] [Google Scholar]
- 22.Nettles RE, Kieffer TL, Simmons RP, Cofrancesco J, Moore RD, Gallant JE, et al. Genotypic Resistance in HIV-1–Infected Patients with Persistently Detectable Low-Level Viremia while Receiving Highly Active Antiretroviral Therapy. Clinical infectious diseases. 2004;39:1030–1037. doi: 10.1086/423388. [DOI] [PubMed] [Google Scholar]
- 23.Aleman S, Söderbärg K, Visco-Comandini U, Sitbon G, Sönnerborg A. Drug resistance at low viraemia in HIV-1-infected patients with antiretroviral combination therapy. AIDS. 2002;16:1039–44. doi: 10.1097/00002030-200205030-00010. [DOI] [PubMed] [Google Scholar]
- 24.Swenson LC, Gonzalez-Serna A, Min JE, Woods CK, Montaner JSG, Li JZ, et al. HIV Drug Resistance Occurring During Low-Level Viremia Is Associated with Subsequent Virologic Failure. Antiviral therapy. 2013;18:A1–A145. doi: 10.1097/QAD.0000000000000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jordan MR, Winsett J, Tiro A, Bau V, Berbara RS, Rowley C, et al. HIV Drug Resistance Profiles and Clinical Outcomes in Patients with Viremia Maintained at Very Low Levels. World Journal of AIDS. 2013;03:71–78. doi: 10.4236/wja.2013.32010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li JZ, Gallien S, Do TD, Martin JN, Deeks S, Kuritzkes DR, et al. Prevalence and significance of HIV-1 drug resistance mutations among patients on antiretroviral therapy with detectable low-level viremia. Antimicrobial agents and chemotherapy. 2012;56:5998–6000. doi: 10.1128/AAC.01217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woods CK, Brumme CJ, Liu TF, Chui CKS, Chu AL, Wynhoven B, et al. Automating HIV drug resistance genotyping with RECall, a freely accessible sequence analysis tool. Journal of clinical microbiology. 2012;50:1936–42. doi: 10.1128/JCM.06689-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agents in HIV-infected adults and adolescents. 2013;5:1–240. [Google Scholar]
- 29.Grennan JT, Loutfy MR, Su D, Harrigan PR, Cooper C, Klein M, et al. Magnitude of virologic blips is associated with a higher risk for virologic rebound in HIV-infected individuals: a recurrent events analysis. The Journal of infectious diseases. 2012;205:1230–8. doi: 10.1093/infdis/jis104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clinical infectious diseases. 2006;42:1608–18. doi: 10.1086/503914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winters B, Montaner J, Harrigan PR, Gazzard B, Pozniak A, Miller MD, et al. Determination of clinically relevant cutoffs for HIV-1 phenotypic resistance estimates through a combined analysis of clinical trial and cohort data. Journal of acquired immune deficiency syndromes. 2008;48:26–34. doi: 10.1097/QAI.0b013e31816d9bf4. [DOI] [PubMed] [Google Scholar]
- 32.Winters B, Van Craenenbroeck E, Van Der Borght K, Lecocq P, Villacian J, Bacheler L. Clinical cut-offs for HIV-1 phenotypic resistance estimates: update based on recent pivotal clinical trial data and a revised approach to viral mixtures. Journal of Virological Methods. 2009;162:101–108. doi: 10.1016/j.jviromet.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 33.Swenson LC, Pollock G, Wynhoven B, Mo T, Dong W, Hogg RS, et al. “Dynamic range” of inferred phenotypic HIV drug resistance values in clinical practice. PloS one. 2011;6:e17402. doi: 10.1371/journal.pone.0017402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laprise C, de Pokomandy A, Baril J-G, Dufresne S, Trottier H. Virologic Failure Following Persistent Low-level Viremia in a Cohort of HIV-Positive Patients: Results From 12 Years of Observation. Clinical infectious diseases. 2013;57:1489–96. doi: 10.1093/cid/cit529. [DOI] [PubMed] [Google Scholar]
- 35.Charpentier C, Landman R, Laouenan C, Joly V, Hamet G, Damond F, et al. Virological Outcome of Patients Displaying Persistent Low-level Viremia Comprised between 20 and 50 Copies/mL. CROI; Seattle, WA: 2012. Abstract 349. [DOI] [PubMed] [Google Scholar]
- 36.Do T, Duncan J, Butcher A, Liegler T. Comparative frequencies of HIV low-level viremia between real-time viral load assays at clinically relevant thresholds. Journal of clinical virology. 2011:8–11. doi: 10.1016/j.jcv.2011.09.022. [DOI] [PubMed] [Google Scholar]
- 37.Chao C, Tang B, Towner W, Silverberg MJ, Hurley L, Horberg M. Short-term clinical outcomes among treatment-experienced HIV-positive patients with early low level viremia. AIDS patient care and STDs. 2012;26:253–5. doi: 10.1089/apc.2012.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmer S, Maldarelli F, Wiegand A, Bernstein B, Hanna GJ, Brun SC, et al. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3879–84. doi: 10.1073/pnas.0800050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deeks SG. Durable HIV treatment benefit despite low-level viremia: reassessing definitions of success or failure. JAMA: the journal of the American Medical Association. 2001;286:224–6. doi: 10.1001/jama.286.2.224. [DOI] [PubMed] [Google Scholar]
- 40.Gandhi RT, Deeks SG. Plasma HIV-1 RNA levels during antiretroviral therapy: how low is low enough? Clinical infectious diseases. 2012;54:733–5. doi: 10.1093/cid/cir933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montaner JSG, Richman DD, Hammer SM. Poor Agreement between 2 Assays for Measuring Low Levels of HIV-1 Viral Load. Clinical Infectious Diseases. 2009;49:1283–1284. doi: 10.1086/605688. [DOI] [PubMed] [Google Scholar]
- 42.Manavi K. The significance of low-level plasma HIV viral load on COBAS TaqMan HIV-1 assays for patients with undetectable plasma viral load on COBAS Amplicor monitor version 1.5. HIV clinical trials. 2008;9:283–6. doi: 10.1310/hct0904-283. [DOI] [PubMed] [Google Scholar]
- 43.Lima VD, Gill VS, Yip B, Hogg RS, Montaner JSG, Harrigan PR. Increased resilience to the development of drug resistance with modern boosted protease inhibitor-based highly active antiretroviral therapy. The Journal of infectious diseases. 2008;198:51–8. doi: 10.1086/588675. [DOI] [PubMed] [Google Scholar]
- 44.Harrigan PR, Hogg RS, Dong WWY, Yip B, Wynhoven B, Woodward J, et al. Predictors of HIV drug-resistance mutations in a large antiretroviral-naive cohort initiating triple antiretroviral therapy. The Journal of infectious diseases. 2005;191:339–47. doi: 10.1086/427192. [DOI] [PubMed] [Google Scholar]
- 45.Lima V, Harrigan R, Montaner JSG. Increased reporting of detectable plasma HIV-1 RNA levels at the critical threshold of 50 copies per milliliter with the Taqman assay in comparison to the Amplicor assay. Journal of acquired immune deficiency syndromes. 2009;51:3–6. doi: 10.1097/QAI.0b013e31819e721b. [DOI] [PubMed] [Google Scholar]
- 46.Gill VS, Lima VD, Zhang W, Wynhoven B, Yip B, Hogg RS, et al. Improved virological outcomes in British Columbia concomitant with decreasing incidence of HIV type 1 drug resistance detection. Clinical infectious diseases. 2010;50:98–105. doi: 10.1086/648729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.