Abstract
We compared the performance characteristics of a real-time PCR method, the LightCycler vanA/vanB detection assay (Roche Diagnostics Corporation, Indianapolis, Ind.) to that of Enterococcosel agar (BBL, Sparks, Md.) for direct detection of vancomycin-resistant enterococci (VRE) from 894 perianal stool swabs. For 421 of 894 swabs, the result for LightCycler PCR was compared to an Enterococcosel plate containing vancomycin at 6 μg/ml; for the remaining 473 swabs, the result for LightCycler PCR was compared to an Enterococcosel plate containing 8 μg/ml vancomycin. The LightCycler method produced considerably more positive results than either the Enterococcosel plate containing vancomycin at 6 μg/ml (n = 25 versus n = 11; sensitivity, 100%; specificity, 97%; positive predictive value [PPV], 42%; negative predictive value [NPV], 100%) or the Enterococcosel plate containing vancomycin at 8 μg/ml (n = 31 versus n = 10; sensitivity, 100%; specificity, 95%; PPV, 32%; NPV, 100%). When possible, additional testing, including culture, LightCycler PCR, and/or a conventional PCR method (PCR-restriction fragment length polymorphism assay), were performed on either the original specimens or original cultures or subsequent specimens for cases in which the original specimen was positive by LightCycler PCR but the Enterococcosel plate was negative. This additional testing demonstrated positive results for 7 of 14 (50%) evaluable discordant specimens which initially tested as LightCycler PCR positive but culture negative using the Enterococcosel plate containing vancomycin at 6 μg/ml and 12 of 17 (71%) evaluable discordant specimens which initially tested as LightCycler positive but culture negative using the Enterococcosel plate containing vancomycin at (8 μg/ml). These results demonstrate that the LightCycler VRE detection assay is considerably more sensitive than the standard culture method for detecting VRE directly from perianal swab specimens. The LightCycler assay also provides results much faster than culture (∼3.5 versus ≥72 h). The use of this test could have important implications for the effective control and prevention of nosocomial outbreaks of VRE.
Over the past decade vancomycin-resistant enterococci (VRE) have emerged as major nosocomial pathogens (5, 9, 17, 19). At least three gene cassettes and their associated phenotypes, VanA, VanB, and VanC, are responsible for vancomycin resistance in enterococci (14). In general, the MICs of vancomycin for strains with VanA phenotypes are relatively high, whereas those for strains with VanC phenotypes are relatively low. Because VanC resistance occurs rarely and MICs are usually ≤8 μl/ml, the clinical significance of VanC resistance is unknown (14). VanA and VanB phenotypes can occur in either Enterococcus faecalis or Enterococcus faecium strains. We and others have previously determined that based on DNA sequence information at least two additional vanB alleles exist, vanB2 and vanB3 (7, 15).
The U.S. Centers for Disease Control and Prevention and the Society of Healthcare Epidemiology of America have provided recommendations to prevent the spread of VRE in institutional settings (2b,10). These recommendations include screening of patients by perianal or rectal swab or fecal surveillance cultures to identify carriers of VRE and subsequent isolation or cohorting of VRE carriers. As well, patients experiencing active VRE infection are isolated. Such a policy, when activated at 32 health care facilities in a region of the Midwest United States, significantly reduced or eliminated the transmission of VRE in these facilities (12).
Screening feces or perianal or rectal swab specimens for VRE by culture may be challenging. Various selective culture media, both agar and broth, have been developed that contain various amounts of vancomycin (8 to 64 μl/ml) (2, 8). Limitations of these methods include the following: (i) other vancomycin-resistant organisms, including Leuconostoc spp., Pediococcus spp., and Lactobacillus spp., may grow with these media; (ii) some Enterococcus spp. with vanB phenotypes may not grow due to MICs in the 6 to 8 μl/ml range; (iii) the time requirements for VRE confirmation frequently exceed 48 h; and (iv) Enterococcus spp. may exist in a viable but nonculturable state (3b). Relevant to points ii and iv, it is not surprising that a high percentage of false-negative results (42%) has been reported when culture-based detection methods are used to detect VRE colonization in stool specimens (3).
Recently, a real-time PCR assay, the Roche LightCycler vanA/vanB detection assay (Roche Molecular Diagnostics, Indianapolis, Ind.) became commercially available. This test method uses fluorescent energy transfer (FRET) probes and the LightCycler instrument (Roche Diagnostics Corporation) and can detect VRE (vanA and vanB genes) directly from fecal or perianal swab samples. Extraction of specimens for this assay can also be conveniently performed using a specialized buffer (Stool Transport and Recovery [S.T.A.R.] buffer; Roche Diagnostics Corporation, Indianapolis, Ind.) and the automated MagNa Pure instrument (Roche Applied Science) as well as commercially available manual extraction methods. The objective of the present study was to compare the Roche LightCycler vanA/vanB detection assay to culture for detection of VRE from perianal swabs. Performance characteristics, as well as turn around time for results, were compared for each test method.
MATERIALS AND METHODS
Study design.
The study was approved by the Institutional Review Board of the Mayo Foundation. Before the clinical evaluation was conducted, one hundred archived clinical isolates of well-characterized enterococci were evaluated for the presence of vanA, vanB and vanB-2/3 genes using the LightCycler vanA/vanB detection assay. The LightCycler assay was performed on bacterial colonies growing on blood agar plates. Results generated were compared to those obtained using a conventional multiplex PCR-restriction fragment length polymorphism (RFLP) assay developed in our laboratory (14) and agar dilution antimicrobial susceptibility testing. For the clinical evaluation perianal swabs were collected from March 2002 to March 2003 from 948 adult and pediatric patients at high risk for VRE colonization. These patients included solid organ transplant recipients, patients with malignancy and/or patients requiring intensive care. Fifty-four of 948 (6%) patients were excluded from the evaluation because these patients or their guardians declined to provide permission to use their specimens and medical histories for evaluation (Minnesota statute 144.335). Two comparison studies which used the same LightCycler PCR assay but different culture methods were performed for 421 swabs and 473 swabs, respectively. For the first culture method a VRE culture plate was prepared using vancomycin at 6 μg/ml and Enterococcosel agar (BBL, Sparks, Md.). For the second culture method, vancomycin at 8 μg/ml and Enterococcosel agar were used. We evaluated both concentrations of vancomycin as either of these concentrations have been recommended by previous investigators (2, 8) and the higher concentration (8 μg/ml) vancomycin may inhibit some VanB phenotypes.
Archived bacterial isolates.
The one hundred archived clinical isolates of Enterococcus spp. were previously identified by the Mayo Clinic Microbiology Laboratory over the time period 1992 to 1995 and stored at −70°C. These enterococcal isolates were classified by demonstrating 6.5% NaCl tolerance and growth on bile-esculin agar with esculin hydrolysis. Identification to the species levels of Enterococci was based on fermentation of sugars, arginine hydrolysis, motility, pigmentation and growth on tellurite agar. Susceptibility testing was performed by an agar dilution method following guidelines of the National Committee for Clinical Laboratory Standards (11). Mueller-Hinton agar with vancomycin concentrations of 2 through 256 μg/ml and teicoplanin concentrations of 8 and 16 μg/ml were used, and the MICs were determined after 24 h of incubation at 35°C.
(i) Culture and DNA extraction.
Each Enterococcus sp. isolate was inoculated to a Trypticase soy agar plate (Becton Dickinson Microbiology Systems, Sparks, Md.) containing 5% sheep blood and incubated at 35°C for 24 h. Three colonies were transferred into 100 μl of sterile water and boiled in a dry heat block at 100°C for 10 min. The suspension was centrifuged at 20,800 × g for 1 min.
(ii) Multiplex PCR-RFLP.
A single multiplex PCR was performed using colonies growing on blood agar plates as previously described (14). This assay detects the presence of vanA, vanB, van-C1, or vanC-2 genes. The PCR products generated were digested with the enzyme MspI at 37°C overnight. The digested PCR products were electrophoresed on a 3% Nu Sieve agarose gel containing ethidium bromide to discriminate among the van genes.
Clinical study. (i) Collection of specimens.
All specimens were collected using a swab collection and transport system (Culture Swab; Becton Dickinson Microbiology Systems, Cockeysville, Md.). The perianal specimen was obtained by swabbing the anal verge area without rectal insertion.
(ii) Perianal swab culture.
All perianal swabs were inoculated to an Enterococcosel culture plate (containing vancomycin at either 6 or 8 μg/ml) prior to processing for DNA testing. The plates were examined after incubation for 24 and 48 h for the presence of black colonies (bile esculin positive) which were Gram stained. Gram-positive cocci were plated to a blood agar plate for isolation and after 24 h were Gram stained and checked for catalase and pyrrolinodyl peptidase activity. The catalase-negative and pyrrolinodyl peptidase-positive isolates were reported as Enterococcus spp., and susceptibility testing was performed. Those isolates for which the MIC of vancomycin was ≥32 μg/ml were reported as VRE. For quality control purposes, each new lot of Enterococcosel agar was tested with vancomycin-resistant and -susceptible Enterococcus faecalis strains as recommended by the National Committee for Clinical Laboratory Standards (11).
(iii) Swab DNA extraction.
After culture was performed the swab was swirled in a 1.5-ml screw-cap tube containing approximately 250 μl of 0.1-mm-diameter zirconia-silica beads (Biospec Products, Inc., Bartlesville, Okla.) and 300 μl of sterile distilled water. The capped tube was processed on a FastPrep Instrument (Qbiogene, Inc., Carlsbad, Calif.) for 30 s at 6.5. The tube was centrifuged at maximum speed for 1 min, and 100 μl of the supernatant was added to 100 μl of commercially available S.T.A.R. buffer (Roche Diagnostics Corporation) in a MagNA Pure sample cartridge. The DNA was extracted with the automated MagNA Pure LC instrument using the Total Nucleic Acid isolation kit (Roche Applied Science). Quality control for DNA extraction was assessed as follows. A positive control plasmid at a concentration of 100 copies/μl in S.T.A.R. buffer was stored at room temperature. This control, along with a negative control of 200 μl of S.T.A.R. buffer, was included in all MagNA Pure extraction runs.
LightCycler vanA/vanB PCR for archived bacterial isolates and clinical study.
For each reaction mixture, 5 μl of the boiled lysate (from colonies) or extracted DNA (from swabs) was added to 15 μl of the PCR reagent mix. The PCR reagent mix contains the following: 2 μl of LightCycler FastStart DNA Master Hybridization Probe mix with enzyme, 7 μl of sterile water, and 2 μl of MgCl2 supplied with the FastStart reagents (Roche Diagnostics Corporation catalog no. 3 003 248), 2 μl of vanA/vanB Primer/Hybridization probe (Roche catalog no. 3 334 961), and 2 μl of the LightCycler vanA/vanB Recovery Template (Roche catalog no. 3 334 970). The LightCycler instrument in which color compensation was installed was programmed as described in Table 1. LightCycler software versions 3.1 and 3.5 were used for all the experiments. Sterile water was used as a negative control. A positive control, LightCycler vanA/vanB, Template DNA (Roche catalog no. 3 334 988) and a negative control were included in each run.
TABLE 1.
Programming of the LightCycler instrument
| Program name | Analysis mode | No. of cycles | Temp (°C) | Time (s) | Temp transition rate (°C/s) | Signal acquisition |
|---|---|---|---|---|---|---|
| Initial | None | 1 | 95 | 600 | 20 | None |
| PCR | Quantification | 45 | 95 | 10 | 20 | None |
| 55 | 15 | 20 | Single | |||
| 72 | 15 | 20 | None | |||
| Melt | Melt | 1 | 95 | 0 | 20 | None |
| Analysis | 59 | 20 | 20 | None | ||
| 45 | 20 | 0.2 | None | |||
| 85 | 0 | 0.2 | Continuous | |||
| Cool | None | 1 | 40 | 10 | 20 | None |
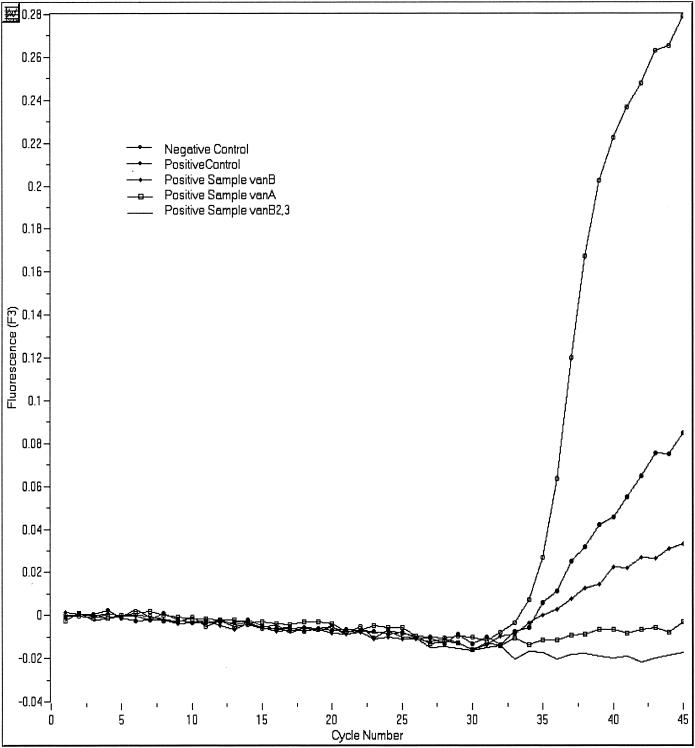
The LightCycler vanA/vanB recovery template has the same sequence as the PCR product, except the probe region has been replaced with a synthetic sequence complementary to the recovery template probes and labeled with a LC-Red705 dye. The recovery template is added in low concentrations to the reagent mix and is amplified along with the target DNA using the same primers. Detection of the target DNA labeled with a LC-Red640 dye is performed in channel 2 and detection of the recovery template is in channel 3. In most cases, the recovery template is amplified in all samples including the negative control. If neither the target DNA nor the recovery template is positive, it is assumed that inhibition of the amplification has occurred and the test for that sample is not valid. However, if the target DNA is amplified but the recovery template is not, it is assumed the target DNA is present in greater amounts and the positive result is valid. Amplification of the recovery template is not necessary if the target DNA is in high concentrations. In prior spiking experiments performed in our laboratory, we have determined that the addition of the recovery template to the reaction mix does not decrease sensitivity of this assay (compete with target DNA).
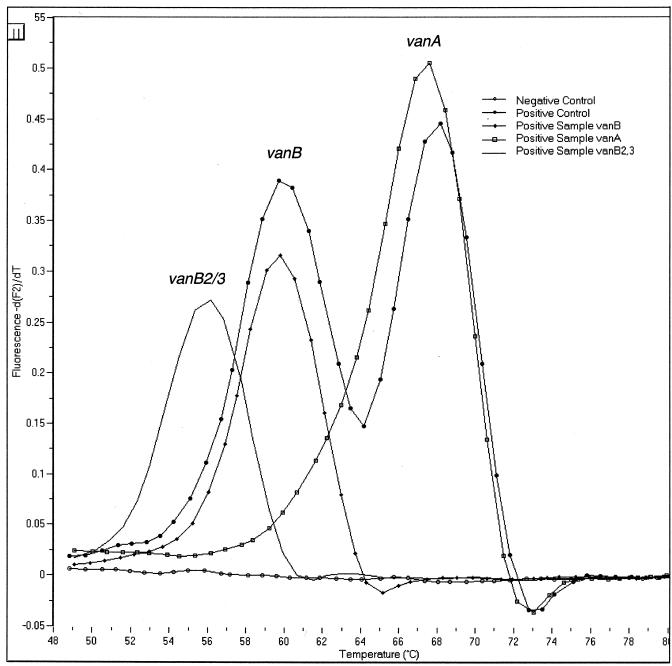
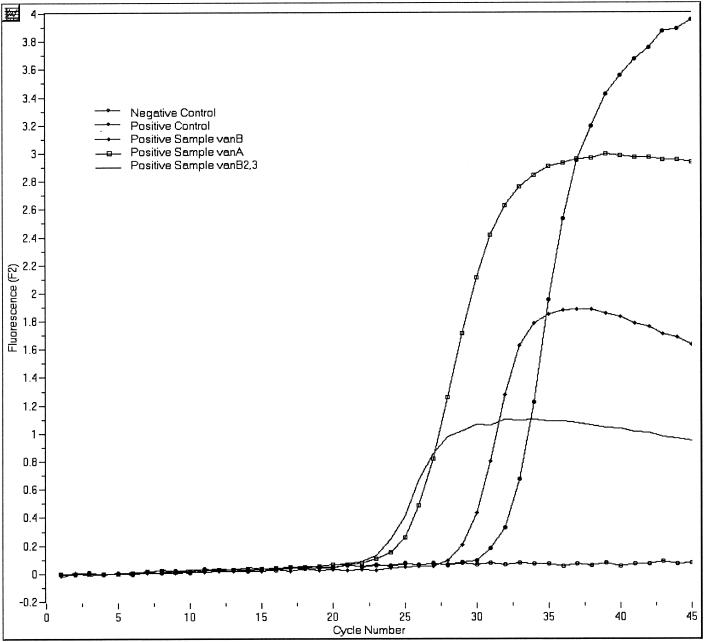
The LightCycler instrument amplifies and monitors the fluorescent development of the target nucleic acid after each cycle. A melting curve analysis was used to differentiate the vanA, vanB, and vanB-2/3 gene targets. Representative results for analyses of real-time data are shown in Fig. 1 and 2. A representative melting curve analysis, which confirms the identification of the amplicon as belonging to one of the VRE genotypes, is shown in Fig. 3. The analytical sensitivity of the assay is less than 50 organisms per reaction. The LightCycler vanA/vanB detection assay is designed to detect and differentiate vanA, vanB, and vanB-2/3 in a single tube using two primer and probe sets.
FIG. 1.
Melting curve analysis for LightCycler vanA/vanB detection assay. The melting temperatures (means ± standard deviations) corresponding to the vancomycin-resistant genotypes were determined by melting curve analysis and are as follows: vanA, 67 ± 2.5°C; vanB, 60 ± 2.0°C; and vanB-2,3, 56 ± 2.0°C.
FIG. 2.
Detection of VRE DNA with the LightCycler instrument and the LightCycler vanA/vanB Primer/Hybridization probes. Positive results are indicated by an upward-deflecting curve as seen here in both the positive control and the positive sample. F2 refers to the fluorescence emission for the LC-Red640.
FIG. 3.
Quantitative representation or cycling curve analysis of recovery template (internal control) for the LightCycler PCR assay. The recovery template FRET probe has a reporter dye (LC-Red640) different from that of the FRET probe used to detect target DNA in the sample (LC-Red640) and is detected in channel F3 of the LightCycler instrument. This quality control step indicates whether inhibition of the PCR occurred in any of the patient samples or the positive and negative controls. Amplification of the recovery template should occur with each of these analyses except when the amount of target DNA in the patient's sample significantly exceeds that of the recovery template DNA.
Additional testing for discordant results: cultures, conventional PCR (multiplex PCR-RFLP), and LightCycler PCR.
For perianal swab specimens that tested positive by LightCycler PCR but were negative by the Enterococcosel culture method, additional testing was performed providing the original swab was available, the original Enterococcosel plate was retrievable, and/or there was enough extracted DNA available from the original specimen. All additional testing, except multiplex PCR-RFLP analysis, was conducted within 5 to 7 days of the initial testing of specimen; PCR-RFLP analysis was performed on some DNA extracts up to 18 months after initial testing of the specimen. Original perianal swabs and original Enterococcosel culture plates were stored at 4°C; DNA extracts were stored at −20°C.
Additional testing included (i) culture of the original perianal swab into heart infusion (HI) broth (Becton Dickinson Microbiology Systems) with subculture of HI broth to both an Entercoccosel plate and CNA plate (Becton Dickinson Microbiology Systems) (the same concentration of vancomycin [either 6 or 8 μg/ml] was used for this Enterococcosel subculture plate as that of the original Enterococcosel agar plate used for the specimen); (ii) PCR using the LightCycler vanA/vanB detection assay of a swab of undifferentiated bacterial growth, if present, from the original Enterococcosel plate; (iii) PCR using the LightCycler vanA/vanB detection assay of a swab of undifferentiated bacterial growth, if present, from the Enterococcosel and/or CNA subculture plates inoculated from the HI broth; (iv) Multiplex PCR-RFLP testing of DNA extracted from the original perianal swab (see method for multiplex PCR-RFLP above); and/or (v) standard Enterococcosel plate culture of a new (subsequent) perianal swab specimen from the same patient.
For the discordant patient specimens, all the results for microbiology tests that were ordered 120 days prior to the date the VRE study culture was ordered were reviewed. Other cultures from which VRE were isolated were noted.
Specificity panel evaluation for LightCycler PCR assay.
The specificity of the LightCycler vanA/vanB detection assay was determined by evaluation of DNA extracted from pure cultures of a variety of gram-positive and gram-negative bacteria (see list of organisms below in Results). These bacteria included many gram-positive enterococci as well as other bacteria and parasites that are considered normal flora, colonizers, or cause of infection in the gastrointestinal tract.
Analytical sensitivity for LightCycler PCR assay.
To determine analytical sensitivity, dilutions of the plasmid control were tested in triplicate.
Analysis of data for clinical evaluation.
For each of the evaluations using the two different concentrations of vancomycin in the Enterococcosel plate, the results for the LightCycler PCR were compared to the results for Enterococcosel plate culture to determine sensitivities, specificities, and positive and negative predictive values. Therefore, the Enterococcosel plate culture was considered the gold standard.
Statistical analysis.
Confidence intervals for sensitivity, specificity, and positive and negative predictive values were based on exact binomial probabilities.
Assessment of assay time requirements.
The approximate time required to complete each test, including specimen processing and assay time, was recorded.
RESULTS
Specificity panel evaluation for Lightcycler PCR assay.
Archived clinical isolates of the following nonenterococcal bacteria were tested and were negative by the LightCycler PCR assay: Actinomyces pyogenes, Aeromonas hydrophila, Bacteroides distasonis, Bacteroides fragilis, Bacteroides thetaiotaomicrons, Bacteroides vulgatus, Citrobacter freundii, Clostridium perfringens, Escherichia vulneris, Enterobacter cloacae, Escherichia coli, Escherichia coli O142:K86(B):H6, Escherichia coli O157:H7, Escherichia coli O7:K1(L):NM, Escherichia coli O70:K:H42, Escherichia hermanii, Eubacterium lentum, Fusobacterium gonidiformii, Fusobacterium nucleatum, group B streptococcus, Klebsiella pneumoniae, Mycobacterium chelonae, Mycobacterium gordonae, Mycobacterium intracellulare, Mycobacterium tuberculosis, Plesiomonas shigelloides, Proteus mirabilis, Pseudomonas aeruginosa, Salmonella group B, Salmonella species, Shigella dysenteriae, Shigella flexneri, Shigella sonnei, Staphylococcus aureus, Staphylococcus epidermidis, Streptococcus bovis, Streptococcus viridans, and Yersinia enterocolitica. The following vancomycin-resistant isolates were tested and found negative by the LightCycler assay: Lactobacillus species (two isolates), Leuconostoc species (two isolates), Pediococcus species. The following intestinal parasites were tested for and were negative by the LightCycler assay: Cryptosporidium parvum, Cryptosporidium species, Dientamoeba fragilis, Encephalitozoon cuniculi, Encephalitozoon hellum, Entamoeba histolytica, Entamoeba moshkovskii, and Septata intestinalis.
The following Enterococcus spp. were evaluated for the presence of either the vanA or vanB genes and found to be negative by the LightCycler PCR assay: five isolates of Enterococcus gallinarum containing the vanC-1 gene, four isolates of Enterococcus casseliflavus containing the vanC-2,3 gene, one isolate of Enterococcus faecium, and one isolate of Enterococcus faecalis with the vanC-1 gene. Enterococcal isolates tested which did not contain the van genes were as follows: Enterococcus faecium (2 isolates), Enterococcus faecalis (11 isolates), Enterococcus raffinosus, Enterococcus avium, and E. casseliflavus. As expected strains of enterococci containing the vanC gene did not produce a signal by melt curve analysis with the VRE detection assay.
Analytical sensitivity for LightCycler PCR assay.
The analytical sensitivity was determined to be less than 10 targets/μl (50 copies/reaction tube).
Clinical evaluation. (i) LightCycler assay versus Enterococcosel plate containing vancomycin at 6 μg/ml.
Among 421 perianal swabs, 11 (3%) were identified as positive by culture and 25 (6%) were identified as positive by LightCycler PCR (vanA [n = 9], vanB [n = 2], and vanB-2/3 [n = 14]). PCR inhibition was determined by lack of recovery template (internal control) amplification for 5 of 421 (1.2%) specimens evaluated using the LightCycler method. The sensitivity, specificity, and positive and negative predictive values for the LightCycler versus culture comparison are shown in Table 2. The mean and median for LightCycler crossing points (the cycle number at which the assay was positive) for concordant results (n = 11) were 28.7 and 26, respectively, and for discordant results (n = 13) were 30 and 30, respectively.
TABLE 2.
Sensitivities, specificities, and predictive values for LightCycler PCR assay compared to Enterococcosel screening plates for detection of VRE from perianal swabsa
| Vancomycin concn (μg/ml) | Sensitivity | Specificity | Positive predictive value | Negative predictive value |
|---|---|---|---|---|
| 6b | 11/11 (100) (59-100) | 396/410 (97) (94-98) | 10/24 (42) (22-63) | 396/397 (100) (99-100) |
| 8c | 10/10 (100) (69-100) | 442/463 (95) (93-97) | 10/31 (32) (17-51) | 442/442 (100) (99-100) |
First set of values in parentheses are percentages. Second set of values in parentheses are 95% confidence intervals.
The prevalence rate of positive cultures was 2.6%.
The prevalence rate of positive cultures was 2.0%.
(ii) LightCycler assay versus Enterococcosel plate containing vancomycin at 8 μg/ml.
Among 473 perianal swabs, 10 (2.1%) were identified as positive by culture and 31 (6.6%) were identified as positive by LightCycler (vanA [n = 13], vanB [n = 5], and vanB-2/3 [n = 13]). PCR inhibition was detected for 1 of 295 (0.3%) specimens evaluated by the LightCycler method. The sensitivity, specificity, and positive and negative predictive value for the LightCycler method compared to culture are shown in Table 2.
The mean and median for LightCycler crossing points for concordant results (n = 9) were 25.3 and 24, respectively, and those for discordant results (n = 16) were 28.7 and 29, respectively.
(iii) Additional testing for discordant results: cultures, conventional PCR (multiplex PCR-RFLP), and LightCycler PCR.
Fourteen discordant results (LightCycler positive, culture negative) were noted for specimens screened using the Enterococcosel plate containing vancomycin at 6 μg/ml. Original specimen and/or original Enterococcosel plates were available for all of these cases. Additional testing demonstrated positive results for 7 of 14 (50%) of these cases.
Twenty-one discordant results (LightCycler positive, culture negative) were observed for specimens screened using the Enterococcosel plate containing vancomycin at 8 μg/ml. Original specimens or original Enterococcosel culture plates were available for additional testing for 16 of these 21 cases; additional testing demonstrated positive results for 11 of 16 of these cases. For one additional case for which the original specimen and original Enterococcosel plate were not available, a new specimen was obtained and was found to be positive by Enterococcosel plate culture. In summary, positive results were noted for 12 of 17 (71%) of available discordant cases for which an Enterococcosel plate containing vancomycin at 8 μg/ml was used.
Assessment of assay time requirements.
Approximately 2.5 h was required to complete specimen processing (extraction of nucleic acid from the sample), and 1 h was required for analysis and reporting of results (total time requirement = 3.5 h). Due to the automation of extraction (MagNA Pure) and analysis (LightCycler), the actual hands-on time was approximately 20 to 25 min for a full run of 32 samples.
DISCUSSION
The results of the present study suggest that the Roche vanA/vanB LightCycler detection assay is considerably more sensitive than culture for detecting VRE in surveillance specimens obtained using perianal swabs. Regardless of whether Enterococcosel agar containing either vancomycin at either 6 or 8 μg/ml was used as the culture standard, the LightCycler PCR method produced more than twice the number of positive results produced by culture.
Our protocol for evaluating discordant positive results for the LightCycler compared with culture showed that many of the samples that were LightCycler PCR positive but culture negative represented true-positive samples. Several possibilities exist which could explain cases where the LightCycler PCR method was positive but the culture negative. Nonviable enterococci may have been present in specimens which were shed from the small intestine or upper region of the colon. Alternatively, viable enterococci may have been present but in a nonculturable state as recently described by del Mar Lleò and colleagues (3b). Another possibility is that some vanB-containing enterococci may have been inhibited by vancomycin at 8 or 6 μg/ml. Discordant results occurred more frequently with vanB gene detection (with vancomycin at 8 μg/ml, 17 of 20 [58%]; with vancomycin at 6 μg/ml, 12 of 14 [86%]). Finally, recovery of VRE may relate to the amount or type of specimen cultured. Relevant to this last point, D'Agata and colleagues showed that a single screening culture for VRE from rectal swabs, was only 58% sensitive compared to serial quantitative stool cultures and skin cultures (3). Therefore, sampling of enough specimen or different specimen types for our patients may have enhanced the recovery of VRE and further reconciled the number of discordant results.
For some of our discordant samples, additional testing of the original DNA extract using PCR-RFLP was negative. This is expected, as PCR-RFLP, which requires visual inspection of bands on gels, is less sensitive than real-time PCR, which automatically measures fluorescence of hybridized probes. Also, most of the samples tested were from DNA extracts of the original specimen stored for up to 18 months. Degradation of target DNA likely occurred in these samples.
Unfortunately there is no clinical gold standard for assessing the accuracy of tests for detecting VRE in perianal specimen. Because the presence of VRE in perianal swabs indicates colonization and not disease, arbitration of discordant PCR positive versus culture-negative results by medical history review is not possible.
The LightCycler VRE assay can be performed in considerably less time than culture. In our practice, VRE culture results for perianal swabs require a minimum of 72 h. In contrast, LightCycler PCR, including extraction of nucleic acid from the specimen requires 3.5 h. Considering both the enhanced sensitivity and turnaround time for results for LightCycler PCR, use of this assay in place of culture should have significant implications for infection control. Because of the speed and ease of performance of the LightCycler PCR test, it is conceivable that all patients could be screened for VRE before admission to healthcare institutions, including hospitals and nursing homes. This strategy would be similar to the “search and destroy” strategy recently reported as an effective and cost-saving method for preventing nosocomial outbreaks of methicillin-resistant Staphylococcus aureus (MRSA) infections in a Dutch hospital (20). At that healthcare facility, patients who are considered at high risk for MRSA are isolated until nasal cultures are demonstrated to be negative for MRSA. If the same strategy were used for VRE, it is possible that similar results for VRE control and cost savings would be realized. However, if one screens patients before admission with a real-time PCR assay, one should also eliminate the additional costs incurred for quaranting patients (non-VRE carriers) until culture-based susceptibility results are available. Indeed, in a “search and destroy” approach some patients may require isolation for 3 days or longer before results could be confirmed for culture-based methods.
Other studies have shown that the more rapid provision of bacteria identification and antibiotic susceptibility results can be cost saving. Doern and colleagues demonstrated that same-day versus overnight provision of results for bacterial identification and antimicrobial susceptibility to physicians at their institution resulted in statistically significantly fewer laboratory studies ordered per patient and a statistically significant savings per patient hospitalization of ∼$4,000. Over a year's time this represented a total cost savings of $2,403,162 (4). Barenfanger and colleagues showed in a similarly designed study that provision of more rapid results for bacterial identification and antimicrobial susceptibility decreased length of hospital stay for patients an average of 2.0 days, decreased the mortality rate from 9.6 to 7.9% and resulted in an annual cost savings of $4,189,500 (1).
Other investigators have developed PCR assays for the direct detection of van genes from rectal or perirectal swabs. Satake and colleagues (18) developed a multiplex PCR assay to detect vanA, vanB, vanC-1, and vanC-2 genes. Samples were extracted using two commercially available column technologies in a sequential fashion (QIAamp tissue kit column; QIAGEN Inc., Chatsworth, Calif.; Centerisep gel filtration column, Princeton Separation, Inc., Adelphia, N.J.). PCR and detection were accomplished using a conventional thermocycler and gel electrophoresis, respectively. No vanB-containing enterococci were isolated by culture. Compared to culture the sensitivity and specificity of the vanA assay were 88.5 and 99.6%, respectively. The vanA gene was detected in one sample from which no enterococci were isolated. No internal control (recovery template) was used to assess PCR inhibition. Paule and colleagues (16) showed more impressive results using a multiplex PCR vanA and vanB assay. DNA was extracted from rectal or perianal swabs using a MasterPure DNA purification kit (Epicentre Technologies, Madison, Wis.), amplified using conventional thermocycling and amplified product was detected by direct visualization of 1.5% agarose gels stained with SYBR green I (Molecular Probes, Eugene, Oreg.). This manual PCR assay detected statistically significantly more VRE from either rectal or perianal swabs than the standard Enterococcosel plate containing vancomycin at 6 μg/ml.
Recently, Pallidino and colleagues (13) used a real-time LightCycler PCR assay to detect vanA and vanB genes directly from rectal swabs. DNA extraction was performed using the QIAmp DNA Stool Mini kit (QIAGEN, Inc.) and the assay was developed using hybridization probes. This real-time PCR assay was positive for 45 of 100 specimens; in contrast a positive result was obtained for 43 of 100 specimens using a standard agar plate culture method. This assay was also used to test enrichment broth cultures for these same specimens. For this determination, 88 of 100 specimens were positive by PCR, representing a 95% increase in sensitivity. The PCR inhibition rate for DNA extracts from negative rectal swabs spiked with a vanB containing Enterococcus faecium strain was 55%. It is likely that this amount of inhibition contributed to the relative lack of sensitivity for the direct specimen PCR method versus the enrichment broth PCR method. If one assumes for this study that due to inhibition of PCR ∼50% of true positives were not detected, then if the inhibition rate is decreased to ∼1%, twice as many specimens (90 versus 45) specimen would test PCR-positive. This improvement represents a 100% increase in sensitivity which is similar to the results we found for our real-time PCR assay versus the standard culture method using a vancomycin concentration of 6 μg/ml in the Entercoccosel agar plate. We detected slightly over twice as many positives by PCR versus culture using either the culture plate containing vancomycin at 6 or 8 μl/ml. This represented an increase of 109 and 141%, respectively. The inhibitory effects of stool on PCR are well known. In our experience the use of S.T.A.R. buffer and the MagNA Pure instrument significantly decreases inhibition. In fact, in the present study, the inhibition rate was ∼1%. The use of the MagNA Pure also automates the extraction process. The time required for specimen extraction of 32 samples is approximately 2.5 h, and the assay run time is ∼1 h. Because both of those processes are automated, actual hands-on time is ∼20 to 25 min.
The LightCycler VRE detection assay has the potential to be used to identify VRE cultivated on conventional agar plate media or in blood culture bottles. With this approach the identification process for VRE could be shortened by 24 h or more. Elsayed and colleagues (6) recently showed that by using a similar real-time PCR method that MRSA could be detected 24 to 36 h sooner directly from blood culture bottles that were smear-positive for gram-positive cocci; appropriate therapies could therefore be offered 24 to 36 h sooner. We are currently evaluating the ability of this assay for identifying VRE in blood culture bottles that are positive for gram-positive cocci by Gram staining.
In summary, the results of the present study suggest that the Lightcycler VRE detection assay is considerably more sensitive than the standard Enterococcosel culture method for detecting VRE from perianal swabs. The LightCycler method also provides results much faster than culture (∼3.5 versus ≥72 h). The use of this test method could have important implications for the effective control and prevention of nosocomial outbreaks of VRE.
Acknowledgments
We thank Laura Onken for the efforts in preparing the manuscript.
REFERENCES
- 1.Barenfanger J., C. Drake, and G. Kacich. 1994. Clinical and financial benefits of rapid bacterial identification and antimicrobial susceptibility testing. J. Clin. Microbiol. 37:1415-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barton A. L., and G. V. Doern. 1995. Selective media for detecting gastrointestinal carriage of vancomycin-resistant enterococci. Diagn. Microbiol. Infect. Dis. 23:119-122. [DOI] [PubMed] [Google Scholar]
- 2b.Centers for Disease Control and Prevention. 1995. Recommendations for preventing the spread of vancomycin resistance. Recommendations of the Hospital Infection Control Practices Advisory Committee (HICPAC). Morb. Mortal. Wkly. Rep. Recomm. Rep. 44:1-13. [PubMed] [Google Scholar]
- 3.D'Agata, E. M. C., S. Gautam, W. K. Green, and Y. W. Tang. 2002. High rate of false-negative results of the rectal swab culture method in detection of gastrointestinal colonization with vancomycin-resistant enterococci. Clin. Infect. Dis. 34:167-172. [DOI] [PubMed] [Google Scholar]
- 3b.del Mar Lleò, M., B. Bonato, C. Signoretto, and P. Canepari. 2003. Vancomycin resistance is maintained in enterococci in the viable but nonculturable state and after division is resumed. Antimicrob Agents Chemother. 47:1154-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doern, G. V., R. Vautour, M. Gaudet, and B. Levy. 1994. Clinical impact of rapid in vitro susceptibility testing and bacterial identification. J. Clin. Microbiol. 32:1757-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edmond, M. B., J. F. Ober, D. L. Weinbaum, M. A. Pfaller, T. Hwang, M. D. Sanford, and R. P. Wenzel. 1995. Vancomycin-resistant Enterococcus faecium bacteremia: risk factors for infection. Clin. Infect. Dis. 20:1126-1133. [DOI] [PubMed] [Google Scholar]
- 6.Elsayed, S., B. L. Chow, N. L. Hamilton, D. B. Gregson, J. D. D. Pitout, and D. L. Church. 2003. Development and validation of a molecular beacon probe-based real-time polymerase chain reaction assay for rapid detection of methicillin resistance in Staphylococcus aureus. Arch. Pathol. Lab. Med. 127:845-849. [DOI] [PubMed] [Google Scholar]
- 7.Gold, H. S., S. Unal, E. Cercenado, C. Thauvin-Eliopoulos, G. M. Eliopoulos, C. B. Wennersten, and R. C. Moellering. 1993. A gene conferring resistance to vancomycin but not to teicoplanin in isolates of Enterococcus faecalis and Enterococcus faecium demonstrates homology with vanB, vanA, and vanC genes in enterococci. Antimicrob. Agents Chemother. 37:1604-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landman, D., J. M. Quale, E. Oydna, B. Willey, V. Ditore, M. Zaman, K. Patel, G. Saurina, and W. Huang. 1996. Comparison of five selective media for identifying fecal carriage of vancomycin-resistant enterococci. J. Clin. Microbiol. 34:751-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linden, P. K., A. W. Pasculle, R. Manez, D. J. Kramer, J. J. Fung, A. D. Pinna, and S. Kusne. 1996. Differences in outcomes for patients with bacteremia due to vancomycin-resistant Enterococcus faecium or vancomycin-susceptible E. faecium. Clin. Infect. Dis. 22:663-670. [DOI] [PubMed] [Google Scholar]
- 10.Muto, C. A., J. A. Jernigan, B. E. Ostrowsky, H. M. Richet, W. R. Jarvis, J. M. Boyce, and B. M. Farr. 2003. SHEA guideline for preventing nosocomial transmission of multidrug-resistant strains of Staphylococcus aureus and Enterococcus. Infect. Control Hosp. Epidemiol. 24:362-386. [DOI] [PubMed] [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial susceptibility testing; thirteenth informational supplement. Approved standard M100-S13. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 12.Ostrowsky, B. E., W. E. Trick, A. H. Sohn, S. B. Quirk, S. Holt, L. A. Carson, B. C. Hill, M. J. Arduino, M. J. Kuehnert, and W. R. Jarvis 2001. Control of vancomycin-resistant enterococcus in health care facilities in a region. N. Engl. J. Med. 344:1427-1433. [DOI] [PubMed] [Google Scholar]
- 13.Palladino, S., I. D. Kay, J. P. Flexman, I. Boehm, A. M. G. Costa, E. J. Lambert, and K. J. Christiansen. 2003. Rapid detection of vanA and vanB genes directly from clinical specimens and enchrichment broths by real-time multiplex PCR assay. J. Clin. Microbiol. 41:2483-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel, R., J. R. Uhl, P. Kohner, M. K. Hopkins, and F. R. Cockerill III. 1997. Multiplex PCR detection of vanA, vanB, vanC-1, and vanC-2/3 genes in enterococci. J. Clin. Microbiol. 35:703-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel, R., J. R. Uhl, P. Kohner, M. K. Hopkins, J. M. Steckelberg, B. Kline, and F. R. Cockerill III. 1998. DNA sequence variation with vanA, vanB, vanC-1, and vanC-2/3 genes of clinical Enterococcus isolates. Antimicrob. Agents Chemother. 42:202-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paule, S. M., W. E. Trick, F. C. Tenover, M. Lankford, S. Cunningham, V. Stosor, R. L. Cordell, and L. R. Peterson. 2003. Comparison of PCR assay to culture for surveillance detection of vancomycin-resistant enterococci. J. Clin. Microbiol. 41:4805-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rice, L. B. 2001. Emergence of vancomycin-resistant enterococci. Emerg. Infect. Dis. 7:183-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Satake, S., N. Clark, D. Rimland, F. S. Nolte, and F. C. Tenover. 1997. Detection of vancomycin-resistant enterococci in fecal samples by PCR. J. Clin. Microbiol. 35:2325-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vergis, E. N., M. K. Hayden, J. W. Chow, D. R. Snydman, M. J. Zervos, P. K. Linden, M. M. Wagener, B. Schmitt, and R. R. Muder. 2001. Determinants of vancomycin resistance and mortality rates in enterococcal bacteremia. a prospective multicenter study. Ann. Intern. Med. 135:484-492. [DOI] [PubMed] [Google Scholar]
- 20.Vriens, M., H. Blok, A. Fluit, A. Troelstra, C. van der Werken, and J. Verhoef. 2002. Costs associated with a strict policy to eradicate methicillin-resistant Staphylococcus aureus in a Dutch university medical center: a 10-year survey. Eur. J. Clin. Microbiol. Infect. Dis. 21:782-786. [DOI] [PubMed] [Google Scholar]