Abstract
Cryptococcus neoformans is an opportunistic fungal pathogen with a propensity to infect the central nervous system of immune compromised individuals causing life-threatening meningoencephalitis. Cryptococcal biofilms have been described as a protective niche against microbial predators in nature and shown to enhance resistance against antifungal agents and specific mediators of host immune responses. Based on the potential importance of cryptococcal biofilms to its survival in the human host and in nature, these studies were designed to investigate those factors that mediate biofilm formation by C. neoformans. We observed that C. neoformans preferentially grew as planktonic cells when cultured under specific conditions designed to mimic growth within host tissues (37°C, neutral pH, and ~5% CO2) or phagocytes (37°C, acidic pH, and ~5% CO2) and as biofilms when cultured under conditions such as those encountered in the external environment (25–37°C, neutral pH, and ambient CO2). Altogether, our studies suggest that conditions similar to those observed in its natural habitat may be conducive to biofilm formation by C. neoformans.
Keywords: Biofilms, Cryptococcus neoformans, Cryptococcal biofilms, Fungal biofilms, Cryptococcus neoformans biofilms, Biofilm formation
Introduction
Cryptococcus neoformans, the etiological agent of cryptococcosis, is an opportunistic fungal pathogen with a predilection to invade the central nervous system (CNS) of immune compromised individuals where it causes life-threatening meningoencephalitis [1]. Human infection with C. neoformans is hypothesized to occur following the inhalation of aerosolized cryptococcal yeast or basidiospores that are commonly found in the environment [1, 2]. The wide distribution of C. neoformans in nature and among various hosts is likely a result of its ability to adapt to multiple unrelated extracellular and intra-cellular environments. Studies have postulated that adaptation of Cryptococcus species to survive and proliferate within multiple environmental predators results in enhanced virulence of the yeast within the human host [3–5]. Therefore, how Cryptococcus species have evolved to survive within several disparate environments has important implications as to its efficacy as a mammalian pathogen.
Biofilm formation is a common mechanism utilized by microorganisms to survive hostile environments, to colonize and seed new ecological niches, and to confer protection against predation [6, 7]. The pressures exerted on biofilms in the environment may also result in the derivation of planktonic cells that have a selective advantage for survival and proliferation in the environment and within a susceptible host. Indeed, experimental studies have shown that cryptococcal biofilms are less susceptible to anti-fungal agents [8] and to various other antimicrobial molecules produced by the immune system [9]. Clinically, cryptococcal biofilms have been observed on artificial medical implants [10–13].
Cryptococcal biofilms can readily be formed on glass surfaces and individual wells of polystyrene plates [8, 9, 14, 15] thus making it feasible to evaluate factors that affect the formation of cryptococcal biofilms. Based on the potential importance of cryptococcal biofilms to its survival in different niches, the studies presented herein were designed to investigate those factors that stimulate biofilm formation by C. neoformans. Our results demonstrate that C. neoformans biofilm formation predominantly occurs under conditions similar to those observed in nature suggesting that specific environmental stimuli regulate cryptococcal biofilm formation.
Materials and Methods
Strains
Cryptococcus neoformans strains H99 (serotype A, Mat α) and 145 (serotype A, Mat a) were recovered from 15% glycerol stocks stored at −80°C prior to use in the experiments described herein. The strains were maintained on yeast-extract-peptone-dextrose (YPD) medium (Bectin, Dickinson and Company, Sparks, MD).
Biofilm Formation
Cryptococcus neoformans strains H99 and 145 were grown for 18–20 h at 30°C with shaking in YPD broth (Becton, Dickinson and Company, Sparks, MD), harvested, and washed three times with sterile phosphate-buffered saline (PBS). Viable yeast were quantified using trypan blue dye exclusion in a hemacytometer and suspended at 1.0 × 107 cells/ml in Dubelcco's modified eagle media (DMEM) (GIBCO, Grand Island, NY) at pH 4.0 (pH adjusted using HCl) or pH 7.4. The DMEM used in our studies incorporates bicarbonate as the buffering agent. Biofilms were formed by pipetting 300 μl of each yeast suspension into individual wells of pre-sterilised, polystyrene, flat-bottomed, 24-well microtiter plates (Corning Incorporated, Corning, NY) and incubated at 25, 30, 35 or, 37°C for 48 h in ambient or 5% CO2. Alternatively, biofilms were formed in individual chambers of 8-well glass slides (Nalge Nunc International Corp., Naperville, IL) by dispensing standardized cell suspensions (250 μl of a suspension containing 1.0 × 107 cells/ml in DMEM, pH 4.0 or pH 7.4) into individual chambers and incubating at 25, 30, 35 or 37°C for 48 h in ambient or 5% CO2. After incubation, the wells were washed three times with sterile PBS to remove nonadhered cells. Preliminary studies showed no difference in using PBS to remove nonadhered cells compared to 0.05% Tween-20. Fungal cells that remained attached to the microtiter plate or glass slide surfaces were considered biofilms and were subsequently visualized using an inverted microscope fixed with a digital camera (Fisher Scientific Company LLC, Houston, TX) and documented using Micron Imaging software (Westover Scientific, Mill Creek, WA).
Measurement of Cryptococcal Biofilms
Biofilm formation was measured using crystal violet [16]. Briefly, biofilm-containing wells of 24-well microtiter plates were washed three times with 250 μl of sterile PBS and then air dried for 5 min. Each of the washed wells was thereafter stained with 100 μl of 0.3% aqueous crystal violet solution for 5 min. Next, each well was washed with sterile distilled water to remove excess stain and immediately destained with 200 μl of 100% ethanol for 5 min. Following destaining, 75 μl of destaining solution was transferred to a well of a new 96-well microtiter plate and the colorimetric readings of the crystal violet in the destaining solution were measured using a Benchmark Microplate Reader with Microplate Manager® 4.0 software (BioRad Laboratories, Hercules, CA) at 550 nm. Alternatively, biofilm-coated wells were washed three times with 250 μl of sterile PBS, and adhered cells removed by scraping individual wells with a sterile 1,000 μl pipette tip followed by resuspension in 500 μl of sterile PBS. The resulting yeast suspension was then quantified using Trypan blue dye exclusion in a hemacytometer. Wells containing no biofilms were used as negative controls for crystal violet staining (data not shown).
Scanning Electron Microscopy
For scanning electron microscopy (SEM), C. neofor-mans strain H99 biofilms were formed in 8-well glass slides (Nalge Nunc International Corp., Naperville, IL) by dispensing standardized cell suspensions (250 μl of a suspension containing 1.0 × 107 cells/ml in DMEM, pH 7.4 or pH 4.0) into individual chambers and incubating at 37°C for 48 h in ambient or 5% CO2. After incubation, the biofilms were washed with sterile PBS and placed in fixative (4% formaldehyde v/v, 1% glutaraldehyde v/v in PBS) overnight. The samples were rinsed in 0.1 M phosphate buffer (2 × 3 min) and then placed in 1% Zetterquist's osmium for 30 min. The samples were subsequently dehydrated in a series of ethanol washes (70% for 10 min, 95% for 10 min, and 100% for 20 min), then treated (2 × 5 min) with hexamethyldisilizane (HMDS: Polysciences Inc., Warrington, PA), and finally air dried in a desiccator. The specimens were coated with gold/palladium (40%/60%). After processing, samples were observed in a scanning electron microscope (Leo 435 VP) in high vacuum mode at 15 kV. The images were processed for display using Photoshop software (Adobe Systems Inc., Mountain View, Calif.).
Confocal Laser Scanning Microscopy (CLSM)
Cryptococcus neoformans strain H99 biofilms were formed as described above for SEM experiments. Biofilms were allowed to form for 48 h and were subsequently washed with sterile PBS and stained using the fluorescent stain FUN® 1 (Molecular Probes, Eugene, OR; 10 μM) following manufacturer's instructions. Both live and dead cells are labeled with this dye resulting in a diffusely distributed green fluorescence. However, metabolically active cells process this dye resulting in a shift from green to orange-red cylindrical intravascular structures. Stained biofilms were observed using a Zeis 510 Meta confocal scanning laser microscope system (Carl Zeis Advanced Imaging Microscopy; Jena, Germany). Image stacks of 210 × 210 μm2 in the X/Y were acquired using a 40× of 1.3 numerical aperture oil immersion lens with optimal Z intervals using excitation wavelengths of 488 (argon laser) and 543 (HeNe laser) and an emission band pass filter at 505–530 (for green) and >560 nm long pass filter (for red). All images were first deconvolved with Auto-quant (Media Cybernetics, Inc., Bethesda, MD) using an adaptive point spread function and then analyzed using the Imaris Suite (Bitplane Inc., Saint Paul, MN).
Statistical Analysis
The one-way analysis of variance (ANOVA) with the Tukey's post hoc test for multiple comparisons was used to detect statistically significant differences. Significant differences were defined as P < 0.05.
Results and Discussion
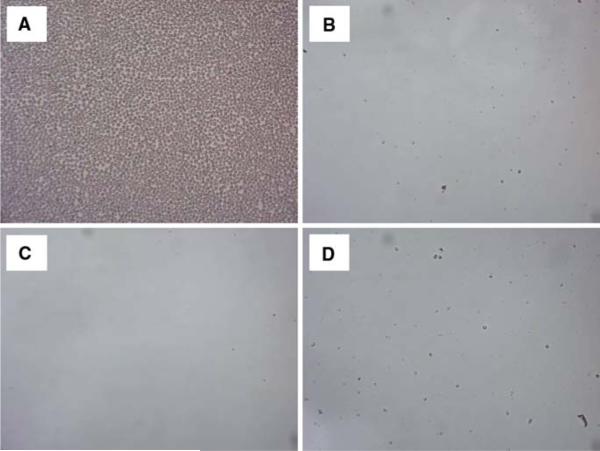
Survival of C. neoformans requires it to adapt to dramatically different environmental conditions ranging from that of its natural habitat to that encountered within mammals. Biofilm formation by Cryptococcus species has been demonstrated to provide a protective niche in inhospitable environments [7]. Consequently, our goal was to establish if biofilm production by C. neoformans strain H99 could occur under distinct conditions similar to those found in nature compared to the in vivo environments of the mammalian host. To this end, C. neoformans strain H99, one of the more virulent cryptococcal strains, was cultured in DMEM at physiological levels of pH and CO2 to mimic mammalian extracellular environments (pH 7.4 and 5% CO2) such as in lung alveolar spaces and the blood stream, as well as host intracellular environments (pH 4.0 and 5% CO2) such as within macrophage phagolysosomes. To mimic its natural habitat outside the human host, C. neoformans was also cultured in DMEM at pH 7.4 and ambient levels of CO2. In addition, we evaluated cryptococcal biofilm formation at pH 4.0 and ambient CO2 to allow us to differentiate the effect of pH and/or CO2 on biofilm development. Biofilm formation was observed using light microscopy (Fig. 1). Figure 1a shows that C. neoformans was able to form biofilms following incubation in DMEM at pH 7.4 for 48 h in ambient levels of CO2. In contrast, biofilms were not observed by light microscopy following incubation of C. neoformans strain H99 in DMEM at pH 7.4 or pH 4.0 for 48 h in the presence of 5% CO2, Fig. 1b, d, respectively. Likewise, no biofilm formation was observed following incubation of C. neoformans strain H99 in DMEM at pH 4.0 for 48 h in ambient levels of CO2 (<0.03% CO2) (Fig. 1c). We observed that C. neoformans within the wells displaying no biofilms appeared to grow as planktonic, single-celled populations (data not shown) indicating that lack of biofilms was not due to a non-specific growth defect. The pH of the media was observed not to change following incubation of the yeast under all conditions evaluated, suggesting that any interaction with the buffering agent, bicarbonate, and CO2 is negligible under our experimental conditions. Quantitative measurement of biofilms formed on polystyrene microtiter plates following incubation for 48 h in DMEM at pH 7.4 or pH 4.0 in the presence of 5% or ambient levels of CO2 was performed using crystal violet staining and also by hemacytometer counting of cells harvested from the biofilms. Similar to the results observed using light microscopy, biofilm formation by C. neoformans strain H99 following culture in DMEM at pH 7.4 for 48 h in ambient levels of CO2 were significantly higher compared to biofilms formed following incubation in DMEM at pH 7.4 or pH 4.0 for 48 h in the presence of 5% CO2 or in DMEM at pH 4.0 for 48 h in ambient levels of CO2 when measured using crystal violet assay (Fig. 2a) and hemacytometer counting (Fig. 2b). We conducted similar experiments using C. neoformans strain 145. Biofilm formation by C. neoformans strain 145 following culture in DMEM at pH 7.4 for 48 h in ambient levels of CO2 were significantly higher compared to biofilms formed following incubation in DMEM at pH 7.4 or pH 4.0 for 48 h in the presence of 5% CO2 (P < 0.001 under both conditions) or in DMEM at pH 4.0 for 48 h in ambient levels of CO2 (P < 0.001) when measured using crystal violet assay.
Fig. 1.
Light microscopy images of C. neoformans biofilm formation. C. neoformans strain H99 was cultured within individual chambers of 24-well microtiter plates in (a) DMEM, pH 7.4; ambient CO2, (b) DMEM, pH 7.4; 5% CO2, (c) DMEM, pH 4.0; ambient CO2, and (d) DMEM, pH 4.0; 5% CO2 for 48 h at 37°C. The glass chambers were subsequently washed with sterile PBS to remove nonadhered cells and viewed using an inverted microscope. Images were taken using a 40× power field. Images are representative of five separate experiments
Fig. 2.
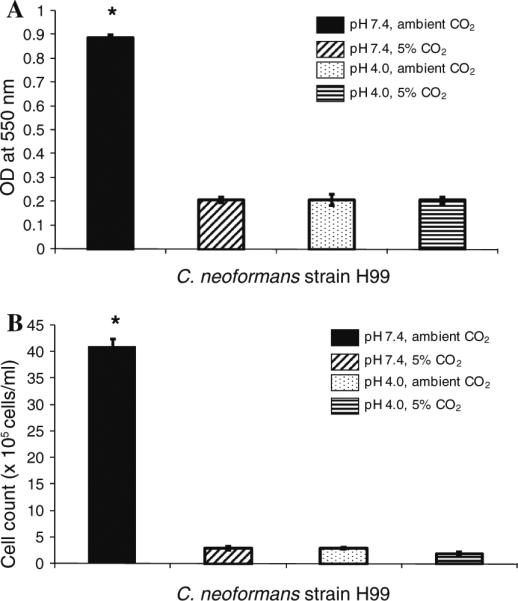
Quantification of C. neoformans biofilm formation. C. neoformans strain H99 was cultured within individual wells of 24-well microtiter plates at 37°C in DMEM, pH 4.0 or pH 7.4 for 48 h in ambient or 5% CO2. Following adhesion, the wells were washed to remove nonadhered cells and biofilm formation measured by crystal violet (CV) staining (a) and hemacytometer counting of adhered cells (b). Data shown is representative of three separate experiments involving three replicate wells for each. Results are presented as mean ± SEM. Asterisks indicate where significant increases (P < 0.001) in biofilms were observed compared to all other conditions measured
We visualized biofilms formed by C. neoformans strain H99 following incubation in DMEM at pH 7.4 at ambient levels of CO2 for 48 h within individual chambers on glass slides using a SEM. Figure 3 shows that the fixation and dehydration steps required to prepare the biofilms for SEM analysis seemed to have altered the architecture of the biofilm. Specifically, the polysaccharide capsule enclosing the individual yeast appeared to be detached from the surface of many of the cells; an observation previously made by other investigators studying cryptococcal biofilm formation [17]. Nevertheless, C. neoformans biofilms consisted of a dense network of yeast embedded in an exopolymeric matix. SEM images of the glass surfaces within the chambers used to incubate C. neoformans strain H99 under conditions observed to be unfavorable for biofilm formation (incubation in DMEM at pH 7.4 or pH 4.0 for 48 h in the presence of 5% CO2 or in DMEM at pH 4.0 for 48 h in ambient levels of CO2) were negative for biofilms (data not shown). Additionally, there was an absence of exopolymeric material coating the surface of chambers used to incubate C. neoformans strain H99 under conditions unfavorable for biofilm formation suggesting that establishment of this material on the surface is critical for biofilm formation.
Fig. 3.
Scanning electron microscopy image of C. neoformans strain H99 biofilms. C. neoformans strain H99 was induced to form biofilms by culture for 48 h at 37°C within individual chambers of 8-well glass slides in DMEM, pH 7.4 in ambient CO2. The biofilms were fixed prior to processing for SEM. Image is at a magnification of 15,000×
In addition, biofilm thickness and metabolic activity of the cells within the biofilms were visualized by confocal laser scanning microscopy (CLSM) following FUN-1 staining. Figure 4 shows a compilation of a series of horizontal (x–y) sections taken across the length of the biofilm, thus forming a three-dimensional reconstruction of the 48 h old biofilm. Measurements of the FUN-1 stained biofilm indicated that it was approximately 17.0 μm thick and contained predominantly metabolically active (red, FUN-1 stained) yeasts.
Fig. 4.
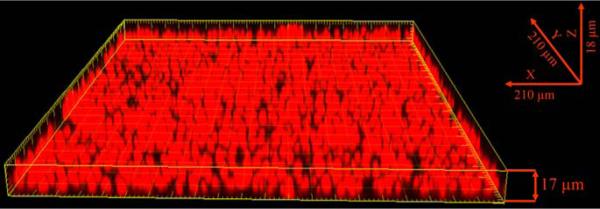
Confocal microscopy image of C. neoformans biofilm. Biofilms were allowed to form within individual chambers of 8-well glass slides for 48 h at 37°C in DMEM, pH 7.4 in ambient CO2. The glass slide was subsequently washed with sterile PBS and stained using the fluorescent stain FUN® 1. Metabolically active cells fluoresce red as shown in the image. Three-dimensional reconstruction of the biofilm was accomplished using software to compile images of sections in the xy plane taken along the z axis
Biofilm formation by C. neoformans at environmental levels of pH and CO2 suggests that cryptococcal biofilms can be formed in its natural habitat. To verify that C. neoformans biofilms could be formed at different environmental temperatures C. neoformans strain H99 was cultured in DMEM at pH 7.4 for 48 h in ambient levels of CO2 at 25, 30, and 35°C and visualized by light microscopy and confocal laser scanning microscopy (CLSM) following FUN-1 staining. Similar to previous studies with C. neoformans strain B3501 [15], biofilm formation by C. neoformans strain H99 was similar at 25, 30, and 35°C when observed by light microscopy (data not shown). Figure 5 shows the 3D stacks of the cells comprising the biofilms formed at 25, 30, and 35°C all with a similar thickness of approximately 16 μm, suggesting that temperature does not have a significant impact on biofilm development by C. neoformans strain H99.
Fig. 5.
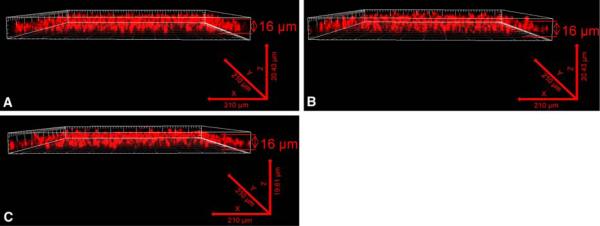
Effect of temperature on C. neoformans biofilm formation. C. neoformans strain H99 was cultured within individual chambers of 8-well glass slides in DMEM, pH 7.4 and incubated at 25 (a), 30 (b), or, 35°C (c) for 48 h in ambient CO2. Following adhesion, the glass slidess were washed with sterile PBS and stained using the fluorescent stain FUN® 1. Three-dimensional reconstruction of the biofilms was accomplished using software to compile images of sections in the xy plane taken along the z axis
Microbial biofilms have historically been considered to provide a protective niche against predation [7, 18–20]. We did not observe biofilm formation by C. neoformans strain H99 during culture in CO2 and pH conditions similar to those found within lung alveoli, the blood stream, or phagolysosomes of macrophages. However, we note that environments within the host involve a complex interplay of many factors in addition to the limited set of parameters investigated herein. Biofilm formation within the mammalian host may be influenced by various serum components and interactions between the yeast and mammalian cells, specifically those tasked with immune surveillance, of the host. Additionally, the yeast population may need to adapt in response to anti-fungal therapy. Nonetheless, cryptococcal biofilm formation may be described as an “environmental trait” along with mating as previously suggested [21]. Mating by C. neoformans has only been observed under environmental conditions similar to those found outside of the mammalian host and not in vivo. Charaterization of cryptococcal biofilm formation as a “virulence” trait together with C. neoformans capsule production may also be appropriate, if we consider in vitro studies showing that cryptococcal biofilms are less susceptible to anti-fungal agents [8] and various other antimicrobial molecules produced by the immune system [9]. Recently, Rittershaus et al. suggested glucosylceramide (GlyCer) synthase as essential for growth of C. neoformans in the blood and alveolar spaces of the lung [22]. Interestingly, GlyCer synthase was shown to be required for growth of C. neoformans cells in 5% CO2 at pH 7.4 suggesting a link between virulence and pH/CO2 sensing. Therefore, environmental sensing by C. neoformans appears to operate on multiple levels outside and inside the human host for survival of the organism in potentially hostile niches.
In conclusion, our studies suggest that biofilm formation by C. neoformans strain H99 preferentially occurs under environmental conditions similar to those observed in its natural habitat and outside of its human host. Biofilm formation may thus serve as a protected niche assuring survival of cryptococci in the environment. The capacity of C. neoformans to adapt to its environment and ensure its continued existence may help to explain its persistence in the environment and world-wide distribution.
Acknowledgments
We will like to thank Jose Lopez-Ribot Pharm.D., Ph.D. for critical reading of the manuscript. This work was supported by grants RO1 AI071752–03 and 5G12RR013646–09 from the National Institutes of Allergy and Infectious Diseases (NIAID) and the National Center for Research Resources (NCRR), respectively, of the National Institutes of Health (NIH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID, NCRR, or the National Institutes of Health.
Contributor Information
Sailatha Ravi, Department of Biology, University of Texas at San Antonio, One UTSA Circle, San Antonio, TX 78249-0062, USA.
Christopher Pierce, Department of Biology, University of Texas at San Antonio, One UTSA Circle, San Antonio, TX 78249-0062, USA; South Texas Center for Emerging Infectious Diseases, The University of Texas at San Antonio, San Antonio, TX, USA.
Colleen Witt, Department of Biology, University of Texas at San Antonio, One UTSA Circle, San Antonio, TX 78249-0062, USA; The Research Center for Minority Institutions Advanced Imaging Center, The University of Texas at San Antonio, San Antonio, TX, USA.
Floyd L. Wormley, Jr., Department of Biology, University of Texas at San Antonio, One UTSA Circle, San Antonio, TX 78249-0062, USA South Texas Center for Emerging Infectious Diseases, The University of Texas at San Antonio, San Antonio, TX, USA.
References
- 1.Mitchell TG, Perfect JR. Cryptococcosis in the era of AIDS–100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8(4):515–48. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kidd SE, Chow Y, Mak S, Bach PJ, Chen H, Hingston AO, et al. Characterization of environmental sources of the human and animal pathogen Cryptococcus gattii in British Columbia, Canada, and the Pacific Northwest of the United States. Appl Environ Microbiol. 2007;73(5):1433–43. doi: 10.1128/AEM.01330-06. doi: 10.1128/AEM.01330-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steenbergen JN, Nosanchuk JD, Malliaris SD, Casadevall A. Cryptococcus neoformans virulence is enhanced after growth in the genetically malleable host Dictyostelium discoideum. Infect Immun. 2003;71(9):4862–72. doi: 10.1128/IAI.71.9.4862-4872.2003. doi: 10.1128/IAI.71.9.4862-4872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steenbergen JN, Shuman HA, Casadevall A. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc Natl Acad Sci USA. 2001;98(26):15245–50. doi: 10.1073/pnas.261418798. doi:10.1073/pnas.261418798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadevall A, Steenbergen JN, Nosanchuk JD. ‘Ready made’ virulence and ‘dual use’ virulence factors in pathogenic environmental fungi—the Cryptococcus neoformans paradigm. Curr Opin Microbiol. 2003;6(4):332–7. doi: 10.1016/s1369-5274(03)00082-1. doi: 10.1016/S1369-5274(03)00082-1. [DOI] [PubMed] [Google Scholar]
- 6.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2(2):95–108. doi: 10.1038/nrmicro821. doi:10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 7.Joubert LM, Wolfaardt GM, Botha A. Microbial exo-polymers link predator and prey in a model yeast biofilm system. Microb Ecol. 2006;52(2):187–97. doi: 10.1007/s00248-006-9063-7. doi:10.1007/ s00248-006-9063-7. [DOI] [PubMed] [Google Scholar]
- 8.Martinez LR, Casadevall A. Susceptibility of Cryptococcus neoformans biofilms to antifungal agents in vitro. Antimicrob Agents Chemother. 2006;50(3):1021–33. doi: 10.1128/AAC.50.3.1021-1033.2006. doi: 10.1128/AAC.50.3.1021-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez LR, Casadevall A. Cryptococcus neoformans cells in biofilms are less susceptible than planktonic cells to antimicrobial molecules produced by the innate immune system. Infect Immun. 2006;74(11):6118–23. doi: 10.1128/IAI.00995-06. doi:10.1128/IAI.00995-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh TJ, Schlegel R, Moody MM, Costerton JW, Salcman M. Ventriculoatrial shunt infection due to Cryptococcus neoformans: an ultrastructural and quantitative microbiological study. Neurosurgery. 1986;18(3):373–5. doi: 10.1227/00006123-198603000-00025. doi:10.1097/00006123-198603000-00025. [DOI] [PubMed] [Google Scholar]
- 11.Banerjee U, Gupta K, Venugopal P. A case of prosthetic valve endocarditis caused by Cryptococcus neoformans var. neoformans. J Med Vet Mycol. 1997;35(2):139–41. doi:10.1080/02681219780001031. [PubMed] [Google Scholar]
- 12.Braun DK, Janssen DA, Marcus JR, Kauffman CA. Cryptococcal infection of a prosthetic dialysis fistula. Am J Kidney Dis. 1994;24(5):864–7. doi: 10.1016/s0272-6386(12)80683-4. [DOI] [PubMed] [Google Scholar]
- 13.Penk A, Pittrow L. Role of fluconazole in the long-term suppressive therapy of fungal infections in patients with artificial implants. Mycoses. 1999;42(Suppl 2):91–6. doi: 10.1111/j.1439-0507.1999.tb00021.x. [DOI] [PubMed] [Google Scholar]
- 14.Martinez LR, Christaki E, Casadevall A. Specific antibody to Cryptococcus neoformans glucurunoxylomannan antagonizes antifungal drug action against cryptococcal biofilms in vitro. J Infect Dis. 2006;194(2):261–6. doi: 10.1086/504722. doi: 10.1086/504722. [DOI] [PubMed] [Google Scholar]
- 15.Martinez LR, Casadevall A. Cryptococcus neoformans biofilm formation depends on surface support and carbon source and reduces fungal cell susceptibility to heat, cold, and UV light. Appl Environ Microbiol. 2007;73(14):4592–601. doi: 10.1128/AEM.02506-06. doi:10.1128/AEM.02506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peeters E, Nelis HJ, Coenye T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J Microbiol Methods. 2008;72(2):157–65. doi: 10.1016/j.mimet.2007.11.010. doi:10.1016/j.mimet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 17.Martinez LR, Casadevall A. Specific antibody can prevent fungal biofilm formation and this effect correlates with protective efficacy. Infect Immun. 2005;73(10):6350–62. doi: 10.1128/IAI.73.10.6350-6362.2005. doi:10.1128/IAI.73.10.6350-6362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matz C, McDougald D, Moreno AM, Yung PY, Yildiz FH, Kjelleberg S. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc Natl Acad Sci USA. 2005;102(46):16819–24. doi: 10.1073/pnas.0505350102. doi: 10.1073/pnas.0505350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matz C, Kjelleberg S. Off the hook—how bacteria survive protozoan grazing. Trends Microbiol. 2005;13(7):302–7. doi: 10.1016/j.tim.2005.05.009. doi:10.1016/j.tim.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Begun J, Gaiani JM, Rohde H, Mack D, Calderwood SB, Ausubel FM, et al. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog. 2007;3(4):e57. doi: 10.1371/journal.ppat.0030057. doi:10.1371/journal.ppat.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell AP. Fungal CO2 sensing: a breath of fresh air. Curr Biol. 2005;15(22):R934–6. doi: 10.1016/j.cub.2005.10.064. doi:10.1016/j.cub.2005.10.064. [DOI] [PubMed] [Google Scholar]
- 22.Rittershaus PC, Kechichian TB, Allegood JC, Merrill AH, Jr, Hennig M, Luberto C, et al. Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans. J Clin Invest. 2006;116(6):1651–9. doi: 10.1172/JCI27890. doi: 10.1172/JCI27890. [DOI] [PMC free article] [PubMed] [Google Scholar]