Abstract
Circadian clocks confer advantages by restricting biological processes to certain times of day through the control of specific phased outputs. Control of temperature signalling is an important function of the plant oscillator, but the architecture of the gene network controlling cold signalling by the clock is not well understood. Here we use a model ensemble fitted to time-series data and a corrected Akaike Information Criterion (AICc) analysis to extend a dynamic model to include the control of the key cold-regulated transcription factors C-REPEAT BINDING FACTORs 1–3 (CBF1, CBF2, CBF3). AICc was combined with in silico analysis of genetic perturbations in the model ensemble, and selected a model that predicted mutant phenotypes and connections between evening-phased circadian clock components and CBF3 transcriptional control, but these connections were not shared by CBF1 and CBF2. In addition, our model predicted the correct gating of CBF transcription by cold only when the cold signal originated from the clock mechanism itself, suggesting that the clock has an important role in temperature signal transduction. Our data shows that model selection could be a useful method for the expansion of gene network models.
Keywords: Arabidopsis, circadian clock, cold acclimation, temperature, gene network model
Introduction
Circadian clocks provide organisms with competitive advantages through the generation of rhythmic expression of key regulatory genes that time the peak of activity of essential functions. The endogenous circadian rhythm can be entrained to the external environment to enable organisms to predict daily changes and initiate responses. Central to the study of circadian clocks is not only understanding the architecture of the oscillators themselves, but also the way in which the clock affects output pathways that confer survival advantages, the maintenance of output phase and the integration of timing information into signalling pathways (Farré and Weise, 2012). A common feature of the initiation of circadian outputs is the co-option of DNA-binding transcription factors within the transcription–translation feedback loops that characterise eukaryotic circadian systems for the regulation of gene expression not associated with the oscillator itself (Suárez-López et al., 2001; Mikkelsen and Thomashow, 2009; Nusinow et al., 2011).
The Arabidopsis clock is characterised by a series of interlocking transcription/translation feedback loops (Figure 1) in which two morning-expressed and partially redundant transcription factors, LATE ELONGATED HYPOCOTYL (LHY) AND CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) repress the expression of a series of evening-expressed components, most importantly TIMING OF CAB EXPRESSION1 (TOC1) and the Evening Complex consisting of LUX ARRYTHMO (LUX), EARLY FLOWERING 3 and EARLY FLOWERING 4 (ELF3 and ELF4; Figure 1). The complexity of the plant oscillator means that our state-of-the-art understanding of clock architecture employs dynamic ordinary differential equation approaches to generate and test hypotheses (Locke et al., 2006; Pokhilko et al., 2010, 2012). Arabidopsis clock models have been successfully linked to output genes to generate and test hypotheses for photoperiodic signal generation (Salazar et al., 2009; Song et al., 2012).
Figure 1.
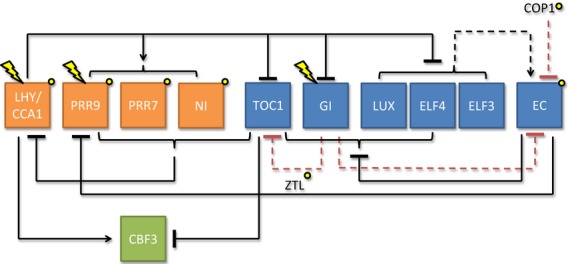
The architecture of the Arabidopsis circadian clock model used in this study (Pokhilko et al., 2012) and proposed new connections to CBF mRNA transcription.
Given the increasingly sophisticated understanding of gene networks underlying circadian function, a key bottleneck is understanding how to most effectively identify the mechanisms through which circadian outputs are produced. An important function of the Arabidopsis circadian clock is the regulation of temperature-responsive gene expression. The cold-induced expression of three redundant transcription factors known as C-REPEAT BINDING FACTORs (CBFs; also known as DREB1; Liu et al., 1998) is a key step in plant responses to cold. CBFs elicit tolerance to freezing temperatures by initiating a signal transduction cascade culminating in the transcription of COR genes that encode proteins that enhance freezing survival (Stockinger et al., 1997). The ability to survive freezing is altered in plants with compromised circadian clocks, with lhy cca1 double mutants showing increased sensitivity to cold, whereas prr5 prr7 prr9 triple mutants show a strong enhanced ability to withstand freezing temperatures (Nakamichi et al., 2009; Espinoza et al., 2010; Dong et al., 2011). CBF expression is also under circadian regulation with peak expression occurring 7–8 h after subjective dawn (Harmer et al., 2000), even though maximum cold-induced expression of CBFs occurs approximately 4 h earlier (Fowler et al., 2005). The reason for circadian regulation of the pathway is not clear, but a recent study suggests that temperature information mediated by the alternative splicing of CCA1 can be transduced to promote freezing tolerance (Seo et al., 2012). It is currently unclear how important this temperature signalling is compared with other known regulators of CBFs, such as INDUCER OF CBF EXPRESSION 1 (ICE1).
Several possible mechanisms for the circadian control of CBF expression have been proposed, including direct activation by LHY and CCA1 (Espinoza et al., 2010; Dong et al., 2011) and inhibition by PRR5, PRR7 and PRR9 (Nakamichi et al., 2009). Of the proposed modes of regulation, only CCA1 has been shown to bind the CBF promoters, and the data indicate that that this interaction is direct (Dong et al., 2011). In this study we use dynamic modelling to extend existing models of the Arabidopsis circadian clock (Pokhilko et al., 2012) to predict the nature of the molecular connections through which CBFs are regulated by the circadian clock. Using dynamic simulation and application of a modified corrected Akaike Information Criterion (AICc; Akaike, 1974; Hurvich and Tsai, 1989) model selection algorithm we compare various possible models of circadian control of CBF3 expression by optimised fitting of model parameters to time-series data. We show that AICc selects a model which not only recapitulates published data on CBF3 expression in Arabidopsis circadian clock mutants, but also predicts a previously unknown direct regulation by TOC1 and the Evening Complex. We show that this dual regulation of CBFs by CCA1 and evening-phased genes predicts the known circadian gating of the cold-regulation of CBFs if the cold signal comes from CCA1 activation (Seo et al., 2012) but not if the cold signal is external to the clock mechanism, suggesting that the circadian clock has an important role in temperature signal transduction.
Results
Construction of models describing the regulation of CBF3 expression by the Arabidopsis circadian oscillator
The most current Arabidopsis clock model contains representations of nine gene products that act in a multiple loop structure to control circadian period (Pokhilko et al., 2012; Figure 1, and hereafter referred to as the P2012 model). Under ambient conditions three CBF genes (1–3) are expressed daily with a phase of peak expression 8 h after dawn, a phase which is robust against variation of the photoperiod (Mockler et al., 2007; Figure 2). For simplicity we chose to model the control of CBF3 expression because CBF3 has the most robust expression under ambient temperatures. In the P2012 model the morning-expressed LHY and CCA1 transcription factors are represented by a single variable LHY. CCA1 has been shown previously to bind the CBF promoters, but LHY was also found to be necessary for normal CBF expression (Dong et al., 2011). Because this work showed that in the absence of LHY and CCA1 CBF expression was greatly reduced, we focussed on models that included a link promoting CBF expression by LHY. In addition to the simple model in which CBF3 transcription was activated by LHY/CCA1 alone, we constructed a range of models including activation and repression of expression by various combinations of clock components (Table 1; see Supplementary Information for methods, Figures S1–S3 and Tables S1–S3). We focussed on model containing direct links to PRR proteins, as prr5 prr7 prr9 triple mutants have been shown to have highly up-regulated CBF expression (Nakamichi et al., 2009). Although PRR5 is not explicitly represented as a variable in P2012, variable NI has an expression pattern and function consistent with PRR5 and was used as a proxy. While we could not construct every conceivable model, we analysed a total of 13 variants (Table 1). We fixed parameters in the P2012 model at their published values and represented the control of CBF3 expression with Hill functions for the simulation of transcription and exponential degradation kinetics (Pokhilko et al., 2010; see Supplementary Information). Simple models in which CBF3 transcription was regulated by one factor consisted of three novel unconstrained parameters. For those simulating CBF3 regulation with two clock components this number rose to 4. This increase in parameter number adds flexibility, but models of increased complexity are penalised by the AICc analysis. Models with more than three connections between the clock components and CBF3 were not considered because this radically increases the number of possible architectures. However, we make this assumption only for practical reasons and of course models with more than three connections cannot be ruled out. These models were optimised by fitting to existing time-series data describing CBF3 expression in 12 h light/12 h dark cycles (Mockler et al., 2007) using Systems Biology Software Infrastructure (SBSI, see Supplementary Information for methods). We did not consider models in which clock components modify mRNA decay kinetics because in Arabidopsis all the proteins considered in this study have been shown previously to affect transcription.
Figure 2.
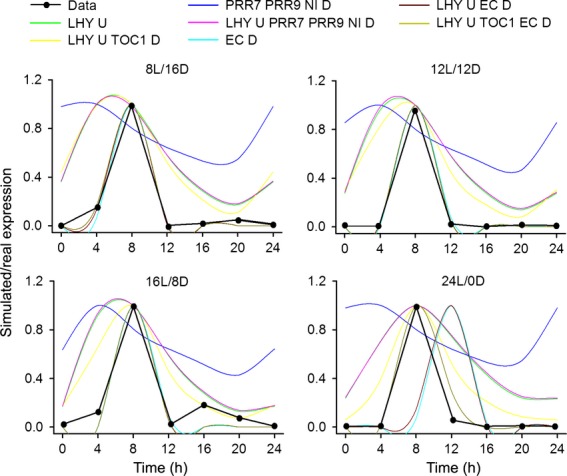
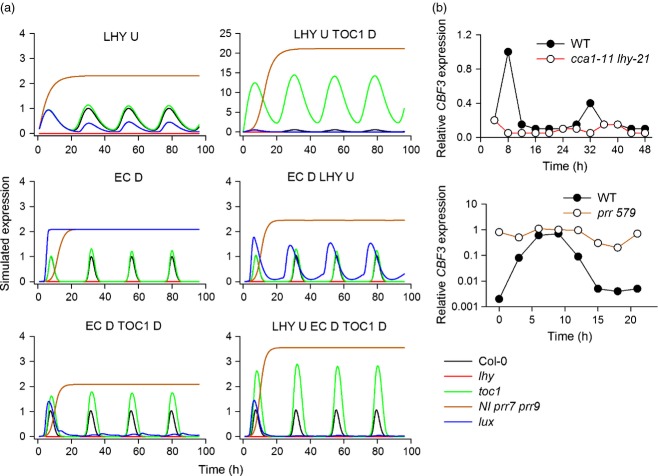
Simulation of the best model fit for each variant (coloured lines) against experimentally determined CBF3 expression (black line) in four indicated light regimes, either short days (8L/16D), 12 h light, 12 h dark (12L/12D), long days (16L/8D) or constant light 24L/0D. U indicated CBF3 transcription up-regulated by variable, D indicates CBF3 transcription down-regulated by variable.
Table 1.
Model selection by AICc, with low scores indicating the most favoured model, prefers architectures which confer regulation of CBFs by LHY, TOC1 and the Evening Complex (EC)
| Rank | Model | AICc score | AICc weight | Features |
|---|---|---|---|---|
| 1 | EC↓:TOC1↓:LHY/CCA1↑ | −807.29 | 1 | Good fit to waveform and phase |
| 2 | EC↓: TOC1↓ | −746.64 | 0 | Good fit to waveform but not phase in 24L |
| 3 | EC↓:LHY/CCA1↑ | −732.40 | 0 | Good fit to waveform but not phase in 24L |
| 4 | EC↓ | −700.14 | 0 | Good fit to waveform but not phase in 24L |
| 5 | LHY/CCA1↑:TOC1↓ | −316.76 | 0 | Good fit to phase in all light dark cycles and 24L, poor fit to waveform |
| 6 | TOC1↓ | −316.47 | 0 | Good fit to phase in LD and 24L only, poor fit to waveform |
| 7 | LHY/CCA1↑ | −257.29 | 0 | Good fit to phase in LD and 24L only, poor fit to waveform |
| 8 | LHY/CCA1↑:NI↓:PRR7↓:PRR9↓ | −220.30 | 0 | Good fit to phase in LD and 24L only, poor fit to waveform |
| 9 | EC↑ | −105.04 | 0 | Good fit to neither phase or waveform |
| 10 | NI↓:PRR7↓:PRR9↓ | −76.48 | 0 | Good fit to neither phase or waveform |
| 11 | NI↓ | 2.80 | 0 | Good fit to neither phase or waveform |
| 12 | PRR7↓ | 4.16 | 0 | Good fit to neither phase or waveform |
| 13 | PRR9↓ | 10.27 | 0 | Good fit to neither phase or waveform |
AICc analysis considered in parallel matches to data in the four photoperiodic regimes outlined in Figure 2. Upward pointing arrows indicate transcriptional up-regulation of CBF3 by preceding variable, downward pointing arrows indicate inhibition. 24L, constant light conditions; LD, long days.
Model selection using the corrected Akaike Information Criterion
AICc scores were calculated for all model variants and ranked, comparing model fit to CBF expression in four light regimes (see Supplementary Information; Table S1). The four highest ranking models were parameterised to closely match experimental data for CBF3 expression in light dark cycles (Figure 2). By extending the analysis to introduce parameter uncertainty, the same four models continued to be the most probable (see Supplementary Information). These all included inhibition of expression by the Evening Complex (EC), which appeared necessary to generate the sharp peak in CBF3 expression observed 8 h after dawn. Models without the EC inhibition could in general match phase, but not the sharpness of the waveform of expression. This included a model in which CBF expression was controlled by LHY alone, but although this could match phase, the broad peak of CBF3 activation did not resemble that observed in experimental data. Models that were very unsuccessful included control by direct inhibition by PRRs, suggesting that up-regulation of CBF3 expression in prr5 prr7 prr9 triple mutants (Nakamichi et al., 2009) is not necessarily caused by loss of direct inhibition of transcription by these genes. Importantly, models which included inhibition by the EC alone or in combination with LHY activation predicted a 4 h phase delay in CBF3 expression in constant light that was not observed in the experimental data (Figure 2). However, the model with the highest AICc weight, inhibition by the EC and TOC1, combined with activation by LHY, was notable in its unique ability to match not only the sharp peak in CBF3 expression but also was capable of reproducing the phase stability in light/dark cycles and constant light. This model also retains the only directly validated link from a circadian clock component to CBF3 expression, that of CCA1 (represented in variable LHY). Thus our analysis predicted that the simple waveform of CBF expression requires the complex interaction of various components to reproduce oscillations seen in experimental data. In this model CBF3 transcriptional activation by LHY is rapidly inhibited by the action of TOC1 at dusk, whereas the primary function of the EC is to repress transcription over dawn during the early period of LHY activity (Figure 3).
Figure 3.
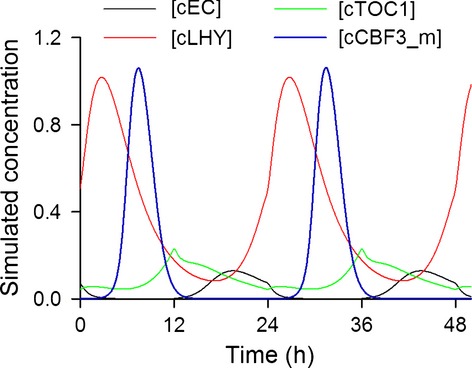
Simulated protein abundances of clock species proposed to control CBF3 expression. Activation by LHY in the morning is first prevented by the repressive action of EC. During the afternoon loss of EC allows CBF3 expression but this declines in a feed-forward manner because the loss of LHY reduces transcription both directly and indirectly via de-repression of the repressor, TOC1. As TOC1 levels decline through the night, the role of repressor is resumed by the EC. Simulation uses 12 h light/12 h dark cycles.
Simulation of CBF3 expression in single and multiple mutants of oscillator components
Successful models for the circadian control of CBF3 expression must reproduce the known effects of clock perturbation on CBF3 gene expression (Nakamichi et al., 2009; Dong et al., 2011). Therefore we tested whether model architectures qualitatively predicted experimentally determined CBF3 expression patterns in clock mutants. The most robust predictions in the literature are those of Dong et al. (2011), who showed that CBF mRNA oscillates with only trace levels in the lhy cca1 double mutant, and that of Nakamichi et al. (2009) who showed constitutive high expression of CBFs in prr5 prr7 prr9 triple mutants. Our parameterisation of multiple models allowed us to understand which variants could effectively simulate the effects of known circadian clock perturbations on CBF3 expression (Figure 4).
Figure 4.
Simulation of CBF3 mRNA expression in constant light in simulated circadian clock gene mutant backgrounds in the five most favoured models, compared with the model of the only known direct connection of the clock mechanism with CBFs, that of LHY/CCA1. Models shown are simulated in constant light. (a) Simulation of lhy cca1 double mutants, ni prr7 prr9 triple mutants, toc1, and lux mutants are shown in each model. D indicates transcriptional down-regulation by variable, U-, up-regulation. (b) Real data comparing CBF3 expression in lhy cca1 double mutants in constant light (re-drawn from Dong et al., 2011), and prr5 prr7 prr9 triple mutants in 12 h white light 12 h darkness (re-drawn from Nakamichi et al., 2009) for comparison.
Interestingly, a simple model in which CBF3 expression was controlled by LHY up-regulation was sufficient to fulfil both criteria, with expression abolished in lhy cca1 double mutants and elevated in ni prr7 prr9 triple mutants. In our optimised parameter set for a model including LHY up-regulation and TOC1 inhibition, the effect of PRR loss was dramatically magnified. All models including the EC inhibition of CBF3 expression also reproduced the down-regulation of CBF3 expression in lhy mutants, and the up-regulation in prr mutants, suggesting that these two observations alone could in principle be explained by multiple architectures involving either expression promotion by a morning-phased component or inhibition by an evening-phased component (or both). As these predictions did not prove discriminatory between different models we continued to simulate the effects of genetic perturbations with unknown effects on CBF3 expression, specifically loss of TOC1 or LUX (essential EC component) in each of the most favoured models (Figure 4). For simulations of toc1 mutants the behaviour of our models divided broadly into two classes. In the first class, which included LHY up-regulation alone, EC inhibition alone, and EC inhibition combined with LHY up-regulation, CBF expression was predicted to be similar to wild type in toc1 mutants. The second class included models in which TOC1 directly inhibited CBF3 expression, in combination with EC inhibition, LHY up-regulation or both. These models all predicted an increase in CBF3 expression in toc1 mutants of differing magnitude: LHY up-regulation in combination with TOC1 inhibition models showed a 12-fold increase in CBF3 expression, whereas the model including EC inhibition, TOC1 inhibition and LHY activation predicts a 2–3-fold increase. Most models predicted that LUX loss-of-function should lead to lower CBF3 expression, with the exception of models that relied exclusively on the EC for down-regulation, which predicted an increase. Together these results led to a hypothesis that could be tested experimentally: A modest rise in CBF3 expression in toc1 mutants, and a decrease in expression in lux mutants would imply that models including both TOC1 and EC inhibition were most accurate in simulating control of CBF3 expression. In contrast, if CBF3 expression is inhibited by the EC alone, we should see no change in CBF3 expression in toc1 mutants, and an increase in lux mutants. Models relying exclusively on TOC1 for inhibition of CBF3 expression should lead to extreme overexpression of CBF3 in toc1 mutants.
TOC1 and Evening Complex components are direct inhibitors of CBF3 expression
In order to probe the role of TOC1 and the EC in the control of CBF transcription real-time qPCR was used to examine CBF expression in the toc1-101 and lux-2 mutant backgrounds in light/dark cycles under ambient temperatures (Figure 5(a)). In our hands published CBF1 primers also possibly mis-primed from CBF3 (Figures S4 and S5) so expression data from these primers should be treated with caution. This analysis showed that CBF1, CBF2 and CBF3 expression were elevated approximately 2–3-fold in the toc1-101 mutant compared with wild type. This observation supports a model architecture requiring LHY activation, with TOC1 and EC inhibition because only this model predicts an increase in CBF3 levels in the toc1-101 mutant consistent with the experimental data. This very same model was also favoured by AICc analysis (Table 1). The increase in CBF expression in toc1-101 also effectively rules out models that do not include TOC1 as an inhibitory factor, as these models predict wild type expression levels of CBFs in toc1 mutants. This prediction was general to all models without TOC1 inhibiting CBF3 expression, rather than specific to particular parameter sets (Figure 4(a)).
Figure 5.
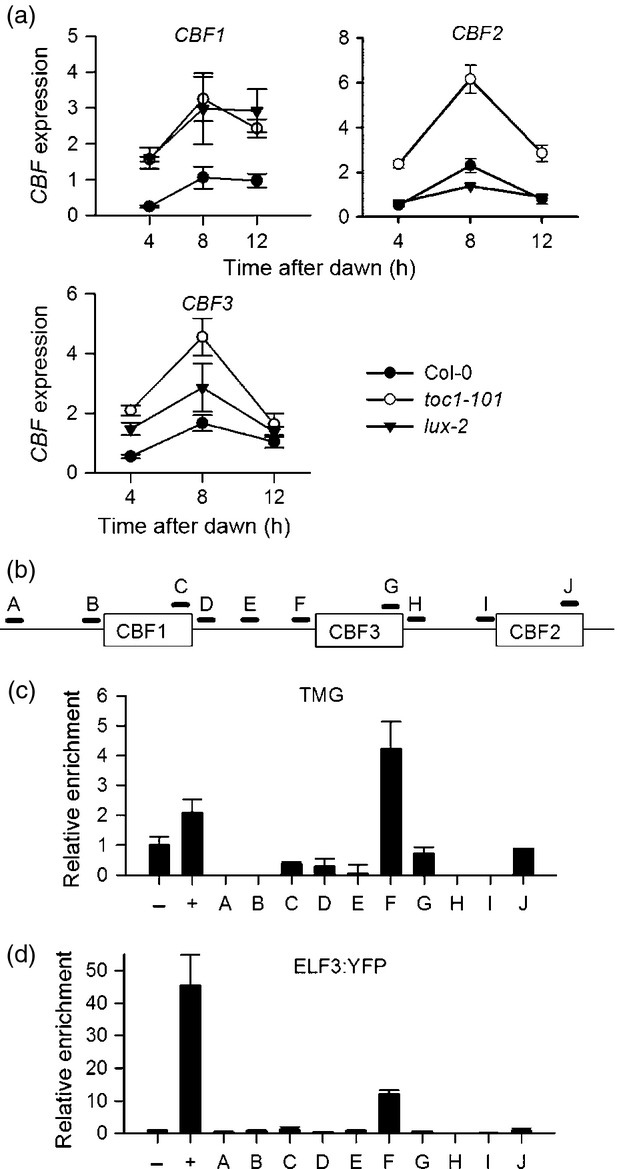
TOC1 and Evening Complex components directly regulate CBF transcription in Arabidopsis. (a) Real-time RT-PCR to show CBF expression in toc1-101 and lux-2 mutants compared with wild type control. Data points represent the mean and standard error of three biological replicates per genotype. (b) Cartoon to show the CBF locus and the location of primer pairs used for ChIP analysis. Open rectangle indicates the CBF transcribed regions horizontal lines represent regions amplified in RT-PCR. (c) ChIP using TOC1-minigene (TMG) shows that TOC1 binds a CBF3 promoter region containing a putative t1me element (Gendron et al., 2012) but not elsewhere in the CBF locus. For all ChIP experiments data represents the mean and standard error of three replicates per locus. −ve control ACTIN2, +ve control TOC1 binding site in the LHY promoter. (d) ChIP to show binding of the EC protein ELF3:YFP to a region close to the TOC1 binding site in CBF3, but not elsewhere in the CBF locus. −ve control ACTIN2, +ve control EC binding site in the PRR9 promoter.
We also tested the role of the EC in the control of CBF expression by analysis of CBF mRNA levels in lux-2 mutants (Figure 5(a)). These results showed variation between the three CBF isoforms tested. CBF2 showed similar expression to wild type, but CBF1 and CBF3 showed an increase. These results are not suggested by any of the leading models, all of which predict low CBF3 expression in lux mutants, because lux mutants have low LHY levels (Hazen et al., 2005; Pokhilko et al., 2012). However, interpretation of the effect of LUX mutations on EC function may complicated by potential redundancy among transcription factors that can fulfil the role of LUX or cross-regulation of CBFs by other CBFs. Taken together, this shows that the EC has a role in the inhibition of CBF1 and CBF3, expression but not CBF2. Our data support the predictions from our AICc analysis of our model ensemble that in addition to activation by LHY/CCA1, inhibition of CBF expression by both TOC1 and the EC is necessary to explain known features of the regulation of CBF expression by the circadian clock, but that the EC has a role in the inhibition of CBF3, expression but not CBF2. It also suggests that the P2012 model may overemphasise the role of LUX in activating LHY and CCA1 expression (see Discussion).
Previously it had been shown that TOC1 can form a complex with the PIF7 protein, and TOC1 can modify the action of PIF7, which binds the G-box element of the CBF1 promoter (Kidokoro et al., 2009). However it is unclear whether TOC1 acts directly on CBFs, and the CBF3 promoter lacks the PIF7 G-box; this is not therefore a potential mechanism for the circadian control of CBF3 expression. It has since been shown that TOC1 can function as a sequence-specific DNA-binding transcription factor in vitro and in vivo (Gendron et al., 2012). Our analysis of CBF3 promoter sequences revealed one putative copy of the recently identified TOC1 DNA-binding (t1me) element close to the transcription start site of CBF3 which raised the possibility that TOC1 may directly bind to CBF promoters. Therefore chromatin immunoprecipitation (ChIP) was used to determine whether TOC1–YFP (Más et al., 2003) could bind at various points in the CBF locus, including regions across all three promoters (Figure 5(b,c)). We confirmed as a positive control that TOC1 binds to the LHY promoter t1me element promoter (Gendron et al., 2012), and also that TOC1–YFP immunoprecipitates are enriched for the putative TOC1-binding site in the CBF3 promoter (Figure 5(b,c)), but not in sequences downstream of CBF3, or in the negative control gene ACTIN 2 (Figure 5(c)). However, we could find no further sites at which we could confirm TOC1 binding at the CBF locus. Therefore our statistical analysis correctly predicted that TOC1 interacts directly with CBF3 promoter in a manner consistent with TOC1 acting as a transcriptional inhibitor, but no corresponding binding to the CBF1 or CBF2 loci could be detected.
We also searched the CBF locus for evidence of LUX binding sites (Nusinow et al., 2011). No putative LUX binding sites were observed close to the transcription start site of CBF3, although sequences resembling published consensus LUX binding sites were observed 1.8Kb and 5Kb upstream of the CBF3 start codon, and 1.6Kb upstream of the CBF1 start codon. These were examined for EC binding using ELF3:YFP transgenic lines (Figure 5(b,d)). We could find no evidence of EC binding to the putative LUX consensus binding regions, showing that the EC does not bind this area of the CBF1 or CBF3 promoters. However, we could observe clear enrichment of DNA close to the TOC1 binding site in the CBF3 promoter in ELF3:YFP immunoprecipitates (Figure 5d). These results are surprising, but raise the prospect that a modified ELF3-containing EC binds the CBF3 promoter close to the binding site of TOC1 to impart transcriptional repression. No further sites capable of binding ELF3 were detected throughout the CBF locus. Taken together, these results demonstrate that multiple evening-expressed clock proteins control CBF3 expression and that AICc analysis can be used to correctly predict connections to deterministic circadian clock models. However, they also suggest that there are other processes that provide the inhibition of CBF1 and CBF2 transcription at dawn as we could not find any evidence for direct binding of TOC1 or ELF3 to these promoters.
TOC1 negatively regulates non-acclimated freezing tolerance
Given that TOC1 is shown to be a negative regulator of CBF2 and CBF3 expression (Figure 5(a)) and that CBF expression induces cold acclimation and tolerance to freezing in Arabidopsis, our analysis predicts that toc1 mutants should have an increased tolerance to freezing when grown under ambient temperatures without cold acclimation. In contrast, the potential contribution of EC components to cold tolerance is less clear. To test the freezing tolerance of evening phase clock mutants, wild type, toc1-101, lux-2 and elf3-1 mutants grown at three physiologically relevant temperatures were subjected to freezing stress at either −3°C or −5°C in the absence of any prior acclimation period at cool temperatures (see Experimental Procedures). At −3°C survival of all lines was high, but −5°C treatments revealed different phenotypes among the mutants. Wild type survival increased as the plants were grown at lower temperatures (Figure 6), whereas the prr5 prr7 prr9 triple mutant showed high tolerance to freezing at all growth temperatures, as described previously (Nakamichi et al., 2009). toc1-101 mutants also showed an elevated survival rate at lower growth temperatures compared with higher temperatures, but at each growth temperature tested, toc1-101 showed significantly increased survival compared with wild type. Thus TOC1 is required for non-acclimated freezing tolerance in wild type plants. In contrast, lux-2 and elf3-1 showed freezing survival similar to wild type (Figure 6). This finding is surprising given that lux mutants exhibit very low LHY and CCA1 expression (Hazen et al., 2005), but this situation may be balanced by reduced direct inhibition of cold signalling by the EC.
Figure 6.
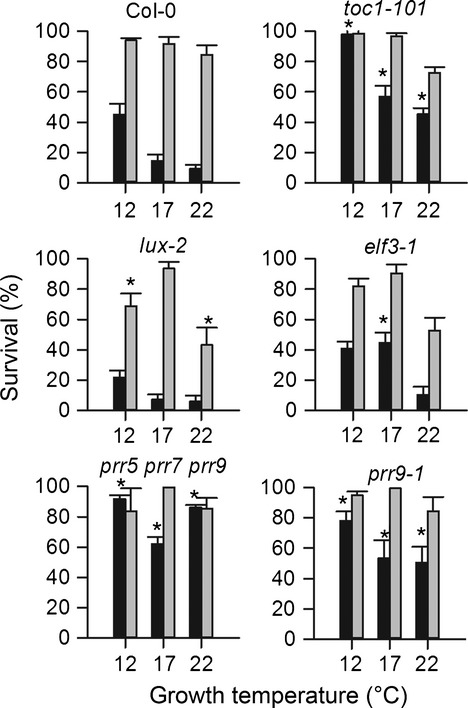
The role of TOC1 and the EC complex in freezing survival in Arabidopsis. Plants grown at either 12°C, 17°C or 22°C were frozen for 2 h at −5°C (black bars) and −3°C (grey bars) without prior cold acclimation. Data represent the mean and standard error of three independent experiments. Asterisks indicate a significant difference from wild type using Student's t-test (P < 0.01). By two-way analysis of variance (anova) both genotype (P = 0.0002) and growth temperature (P = 0.004) have highly significant effects on freezing survival.
Control of CBF3 expression by LHY, TOC1 and the Evening Complex allow observed gating of temperature signals
Cold induction of CBF transcription has been proposed to be mediated by several processes. Firstly, a clock-independent signal transduction cascade involving ICE1 and/or CAMTA transcription factors has been proposed to mediate transcriptional promotion in response to cold, which is then gated by independent circadian control of CBF expression (Chinnusamy et al., 2003; Fowler et al., 2005; Doherty et al., 2009). Alternatively, it has been suggested that CBFs receive a cold signal from the circadian clock itself, at least in part through CCA1, the abundance of which is affected by temperature-controlled splicing (Dong et al., 2011; Seo et al., 2012). To determine which of these hypotheses is supported by our model, we tested them individually. In our model in which CBF3 expression is controlled by LHY, TOC1, and EC, this cold-regulation of CCA1 can be simulated by a pulse of LHY protein (CL). The effects of a five-fold increase in CL on CBF mRNA levels were simulated every 4 h for 24 h (see Experimental Procedures). This was compared with a direct clock-independent cold induction of CBF transcription: as this signal is clock-independent it cannot affect the phase of CBF expression and therefore in this scenario cold-induced CBF expression follows a similar dynamic to CBF expression under ambient temperatures and was modelled by increasing CBF3 levels five-fold directly. We hypothesised that the correct model for the cold induction of CBF expression would reproduce the observed gating of peak responsiveness of CBF expression to cold, a time approximating to ZT4 (Fowler et al., 2005; Dong et al., 2011). Fowler et al. (2005) used northern blot analysis to analyse CBF induction by cold at 4, 10, 16 or 22 h after subjective dawn; we digitised these published data to provide a quantitative analysis of the magnitude of CBF gene expression gating across a subjective day (Figure 7). These data were compared with the phase of cold-induced CBF3 expression (data reproduced from Dong et al., 2011), and to our model predictions. Notably, our analysis not only predicted a peak gating of CBF3 expression in response to cold to ZT0-4 where cold induction of CBF occurred via LHY, but also closely replicated the magnitude of observed cold responses outside of this period (Figure 7). In contrast, circadian gating of a direct cold signal to CBF necessarily matches the peak phase of CBF expression, at ZT8, and predicts that CBF responsiveness to cold falls too sharply outside of the period of maximum induction. This analysis clearly shows that our simulations of temperature pulses through LHY accurately reproduce the pattern of observed cold induction whereas the pattern resulting from an external temperature signal acting additively to the circadian regulation does not. Because our model correctly predicts the phase of the gating of the transcriptional induction CBF expression by cold only when the temperature signal is assumed to originate within the circadian clock mechanism, we suggests that the circadian clock plays an important direct role in low-temperature signal transduction.
Figure 7.
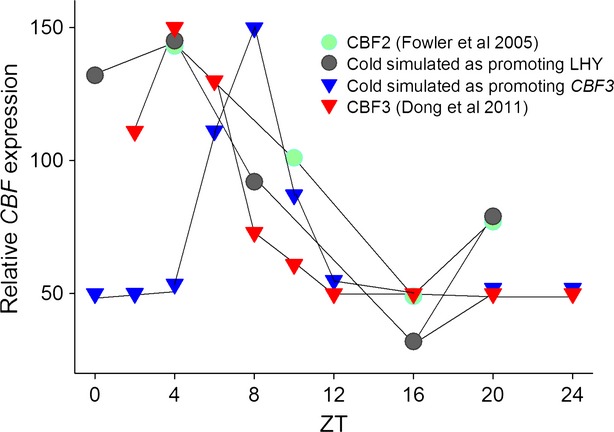
Prediction of the low-temperature gating of CBF expression closely matches experimental data if cold is assumed to increase the level of the LHY protein. Simulations for the model in which CBF expression is controlled by LHY, TOC1 and EC were scaled and plotted against data for the low-temperature induction of CBF2 expression, gathered from Fowler et al. (2005; closed circles) and the cold induction of CBF3 (Dong et al., 2011). Low temperature was simulated by increasing CLHY (open circles) or CCBF3 m (closed triangles, to simulate a cold signal arriving directly at the CBF locus) five-fold at the indicated times after subjective dawn.
Discussion
Circadian clocks play a central role in the coordination of environmental signalling pathways in plants, but given the vast numbers of clock-regulated genes simple methods are required to understand how various phased outputs are generated, and the functional significance of regulatory mechanisms. Here we extend a deterministic Arabidopsis circadian clock model and show that function connections that control CBF3 phase and waveform can be deduced in silico based on the statistical analysis of alternative model architectures by AICc. Because this work uses only simple time-series datasets based on transcriptomic analyses it represents a paradigm that can be applied to biological systems in general where large public datasets of gene expression are available. Our AICc analysis correctly predicted the existence of previously unknown connections between the clock and CBF3 expression, connections that could also be validated and shown to have physiological consequences for the plant's ability to resist cold. Our model also reproduces observed gating of CBF mRNA induction by cold (Fowler et al., 2005), but only if the low temperature is assumed to require the clock itself for signal transduction to CBF expression. This finding suggests that the plant circadian clock also has a role in temperature signal transduction, as in timing biological events.
Here we have shown that in addition to the activation of CBF transcription by LHY and CCA1, CBF3 transcription is also directly inhibited by at least two evening-phased components, TOC1 and ELF3, as part of the EC (Figure 5). Only models that include these three connections are able to maintain correct phase in variable photoperiods, and to qualitatively simulate the effects of mutation of clock components on CBF3 expression. CBF1 and CBF2 transcription is also subject to a similar inhibition by TOC1 (Figure 5), but we could find no evidence that TOC1 binds DNA close to CBF1 or CBF2. One possibility is that TOC1 binding to the neighbouring CBF3 promoter (immediately 3′ to CBF2) is sufficient to confer repression, while another is that TOC1 represses CBF1 and CBF2 indirectly through a second factor. This factor may be via PIF7 (Kidokoro et al., 2009), and it is possible that ChIP fails to detect binding via PIF7, either because the association is weak or affected by the presence of the YFP tag on the transgenic line used here. Analysis of CBF1 expression was complicated by the very close sequence similarity between CBF1 and CBF3 and the possible mis-priming of published CBF1 primers from CBF3 cDNA (Figure S4). Taken together it is clear from our data that repression by TOC1 is essential for normal CBF dynamics.
The EC, or some variant, also clearly binds the CBF3 locus and inhibits CBF3 expression (Figures5 and 6). Although this connection was predicted by our statistical analysis, it was not accurately simulated by P2012, possibly because further work is required to improve P2012, particularly to constrain the affect of EC loss on LHY and CCA1 protein levels. In our model EC loss can cause CBF up-regulation by loss of direct inhibition, and also down-regulation indirectly via down-regulation of LHY (Figure 1). The fact that CBF1 and CBF3 are up-regulated in EC mutants and CBF2 is, if anything, slightly down-regulated shows that there is probably more than one route through which EC components can influence CBF expression. It is likely that, for CBF3, loss of direct repression dominates whereas for CBF2 loss of LHY activation is more important, but is overestimated by the model. This highlights the differences in the regulation of the three CBF isoforms (Novillo et al., 2004) and also suggests that there exists a yet-unknown process for providing transcriptional inhibition of CBF2 during the night and over dawn. That ELF3 binds to a region of the CBF3 promoter with no obvious consensus LUX binding sites suggests that the ELF3 may associate with other DNA-binding components. For instance, ELF3 has been shown to complex with other proteins, notably phytochrome, in yeast and in vitro (Liu et al., 2001), and phytochrome has a role in the control of CBF expression (Franklin and Whitelam, 2007).
The ice1 mutation also preferentially inhibits the cold induction of CBF3, confirming that differences in the regulation of CBF informs exist (Chinnusamy et al., 2003). In addition, CBF2 has been shown to inhibit CBF1 and CBF3 expression after cold exposure (Novillo et al., 2004) and may also contribute to inhibition of expression towards dusk, along with TOC1 activity. Our data suggest that CBF3 expression is also the most tightly coupled to the circadian oscillator. We speculate that after duplication, pseudorandom subfunctionalisation caused by genetic drift is the most likely cause of the current specialisations of the CBF isoforms, in preference to invoking a particular selective advantage to the current state of regulation in Arabidopsis Col-0. In this view it is likely that the original single isoform contained most of the connections to the oscillator we observe at the CBF3 locus, some of which have been lost at CBF1 and CBF2.
Our quality control for the specificity of published CBF QPCR primers included expression analysis of CBF RNAi plants (Novillo et al., 2007). From this work (Figure S4), we found that published primers detected highly overexpressed CBF transcripts in the RNAi lines, and that primers for CBF1 reported elevated expression also in CBF3 RNAi plants that had a similar expression level to that detected by the CBF3 primers. We believe this finding indicates that the primers are most likely detecting cDNA synthesised from the overexpressed constructs, raising the suspicion that the CBF1 primers can also prime from CBF3 under some conditions and may not be specific. However, CBF3 RNAi may also profoundly elevate CBF1 expression.
Over one-third of Arabidopsis genes show a 24 h rhythm in their gene expression, suggesting control by the circadian clock (Harmer et al., 2000). Our successful use of model selection and parameterisation of time-series data from public microarray data suggests that this approach may have wider utility in the extension of gene network models in other systems where large transcriptomic time-series datasets exist.
Experimental Procedures
Plant materials
The lux-2 (Hazen et al., 2005) and TOC1–YFP (Más et al., 2003) have been described previously and were gifts from Steve Kay. The pELF3:ELF3:YFP line has been described previously (Dixon et al., 2011). CBF1 and CBF3 RNAi lines were a gift from Julio Salinas, as was the cbf2 mutant (Novillo et al., 2004, 2007). toc1-101 was a gift from Peter Quail (Kikis et al., 2005). All lines are in the wild type Columbia-0 background.
Model construction
See the Supplementary Information for details on model construction, parameterisation and analysis.
Gene expression and real-time PCR
Seedlings were grown for 10 days in 12 h white light/12 h dark cycles before harvest at the indicated time relative to dawn. Biological replicates were collected in triplicate and RNA extracted with an RNeasy Plant Mini Kit (Qiagen, Germantown, MD, USA) according to the manufacturer's instructions. cDNA was synthesised by Superscript Reverse Transcriptase from 3 μg total RNA using oligo-dT primers at 42°C for 1 h, and diluted 1/10 before use. Primers for the amplification of CBFs were CBF1: 5′-GGAGACAATGTTTGGGATGC-3′ and 5′-CGACTATCGAATATTAGTAACTCC-3′ (Dong et al., 2011); CBF2: 5′-CGACGGATGCTCATGGTCTT-3′ and 5′-TCTTCATCCATATAAAAC GCATCTTG-3′; CBF3: 5′-AATATGGCAGAAGGGATGCT-3′ and 5′-ACTCCATAACGATACGTCGT-3′. Primers for amplification of the control gene ACTIN2 were 5′- CGTTTCGCTTTCCTTAGTGTTA-3′ and 5′-AGCGAACGGATCTAGAGACTC-3′. PCR was performed on an ABI prism 3700 thermocycler using manufacturer's standard conditions.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed following the protocol in Gendrel et al. (2002) with modifications. In summary the following steps were altered. Seedlings were grown on MS agar plates at 22°C for 14 days with 12 hr white light/dark cycles and harvested at CT14. The chromatin was sheared to between 100 and 1000 bp in a Bioruptor UCD 200 (Diagenode) at high intensity for 10 min (cycles of 30 sec on/30 sec off) at 4°C after Lau et al. (2011). An aliquot of the chromatin was reserved at this point as the Input chromatin. Immunoprecipitation used equilibrated Dynabeads® Protein A (Invitrogen cat# 100-01D). The pre-cleared chromatin was transferred away from the beads and incubated with rotation over night at 4°C with a 1:1000 dilution of anti-GFP (Abcam ab290). The immunocomplexes were recovered from the beads by boiling for 10 min in the presence of 10% Chelex resin (BioRad cat# 142-1253) and the proteins removed using Proteinase K Solution (Invitrogen cat# AM2546) at 50°C. The reserved Input chromatin was also processed in parallel with Chelex and Proteinase K and then purified using QIAquick PCR purification Kit (Qiagen cat# 28104). qPCR on the ChIP and Input DNA was performed in triplicate using Brilliant III Ultra-Fast SYBR® Green QPCR Master Mix (Agilent cat# 600883) on a Mx3005P machine. Primer sequences for positive controls, the LUX binding site in the PRR9 promoter and the TOC1 binding site in the LHY promoter, have been described previously (Helfer et al., 2011; Gendron et al., 2012). Primer sequences can be found in Table S4.
Analysis of freezing sensitivity
Wild type and mutant plants were grown on MS medium in 16 h light 8 h dark cycles at 22°C, 17°C or 12°C in a Sanyo MLR 350 incubator before the assay until they had two true leaves. Plants were then frozen at −5°C or −3°C for 24 h in a Sanyo MIR 154 incubator, transferred to 4°C to recover for a further 24 h, and then returned to the growth temperature for 7 days. Survival was scored as the presence of green seedlings after 7 days of growth in three biological replicates of 50 seedlings per genotype.
Acknowledgments
The authors would like to acknowledge funding from BBSRC grant BB/F005296/2 and the Royal Society to Penfield and BBSRC grant BB/F005237/1 to Halliday.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Optimised new parameter values for each of the thirteen models.
Table S2. AICcU analysis results.
Table S3. Sensitivity heatmap for the expression of CBF3 at the indicated time after dawn.
Table S4. Primer sequences for chromatin Immunoprecipitation.
Generalised method used for comparing models.
Figure S2. Effect of τ2
Figure S3. Effect of d.
Figure S4. Published CBF1 Real-time PCR primers appear to prime from CBF1 and CBF3.
Figure S5. Alignment of published CBF1-specific primers with CBF1 and CBF3 cDNA sequences.
References
- Akaike H. A new look at the statistical model identification. IEEE Trans. Auto. Con. 1974;19:716–723. [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon LE, Knox K, Kozma-Bognar L, Southern MM, Pokhilko A, Millar AJ. Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr. Biol. 2011;21:120–125. doi: 10.1016/j.cub.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell. 2009;21:972–984. doi: 10.1105/tpc.108.063958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong MA, Farré EM, Thomashow MF. Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl Acad. Sci. USA. 2011;108:7241–7246. doi: 10.1073/pnas.1103741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza C, Degenkolbe T, Caldana C, Zuther E, Leisse A, Willmitzer L, Hincha DK, Hannah MA. Interaction with diurnal and circadian regulation results in dynamic metabolic and transcriptional changes during cold acclimation in Arabidopsis. PLoS ONE. 2010;5:e14101. doi: 10.1371/journal.pone.0014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré EM, Weise SE. The interactions between the circadian clock and primary metabolism. Curr. Opin. Plant Biol. 2012;15:293–300. doi: 10.1016/j.pbi.2012.01.013. [DOI] [PubMed] [Google Scholar]
- Fowler SG, Cook D, Thomashow MF. Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 2005;137:961–968. doi: 10.1104/pp.104.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Whitelam GC. Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat. Genet. 2007;39:1410–1413. doi: 10.1038/ng.2007.3. [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA. Dependence of Heterochromatic Histone H3 Methylation Patterns on the Arabidopsis Gene DDM1. Science. 2002;297:1871–1873. doi: 10.1126/science.1074950. [DOI] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl Acad. Sci. USA. 2012;109:3167–3172. doi: 10.1073/pnas.1200355109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl Acad. Sci. USA. 2005;102:10387–10392. doi: 10.1073/pnas.0503029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 2011;21:126–133. doi: 10.1016/j.cub.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurvich CM, Tsai CL. Regression and time series model selection in small samples. Biometrika. 1989;76:297–307. [Google Scholar]
- Kidokoro S, Maruyama K, Nakashima K, et al. The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol. 2009;151:2046–2057. doi: 10.1104/pp.109.147033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikis EA, Khanna R, Quail PH. ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 2005;44:300–313. doi: 10.1111/j.1365-313X.2005.02531.x. [DOI] [PubMed] [Google Scholar]
- Lau OS, Huang X, Charron JB, Lee JH, Li G, Deng XW. Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Mol. Cell. 2011;43:703–712. doi: 10.1016/j.molcel.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XL, Covington MF, Fankhauser C, Chory J, Wagner DR. ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell. 2001;13:1293–1304. doi: 10.1105/tpc.13.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JC, Kozma-Bognár L, Gould PD, Fehér B, Kevei E, Nagy F, Turner MS, Hall A, Millar AJ. Experimental validation of a predicted feedback loop in the multi-oscillator clock of Arabidopsis thaliana. Mol. Syst. Biol. 2006;2:59. doi: 10.1038/msb4100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P, Alabadí D, Yanovsky MJ, Oyama T, Kay SA. Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell. 2003;15:223–236. doi: 10.1105/tpc.006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Thomashow MF. A role for circadian evening elements in cold-regulated gene expression in Arabidopsis. Plant J. 2009;60:328–339. doi: 10.1111/j.1365-313X.2009.03957.x. [DOI] [PubMed] [Google Scholar]
- Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM. The DIURNAL project: Diurnal and circadian expression profiling, model-based pattern matching and promoter analysis. Cold Spring Harb. Symp. Quant. Biol. 2007;72:353–363. doi: 10.1101/sqb.2007.72.006. [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, Yamashino T, Saito K, Sakakibara H, Mizuno T. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 2009;50:447–462. doi: 10.1093/pcp/pcp004. [DOI] [PubMed] [Google Scholar]
- Novillo F, Alonso JM, Ecker JR, Salinas J. CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc. Natl Acad. Sci. USA. 2004;101:3985–3990. doi: 10.1073/pnas.0303029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novillo F, Medina J, Salinas J. Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl Acad. Sci. USA. 2007;104:21002–21007. doi: 10.1073/pnas.0705639105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A, Hodge SK, Stratford K, Knox K, Edwards KD, Thomson AW, Mizuno T, Millar AJ. Data assimilation constrains new connections and components in a complex, eukaryotic circadian clock model. Mol. Syst. Biol. 2010;6:416. doi: 10.1038/msb.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokhilko A, Fernández AP, Edwards KD, Southern MM, Halliday KJ, Millar AJ. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 2012;8:574. doi: 10.1038/msb.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar JD, Saithong T, Brown PE, Foreman J, Locke JC, Halliday KJ, Carré IA, Rand DA, Millar AJ. Prediction of photoperiodic regulators from quantitative gene circuit models. Cell. 2009;139:1170–1179. doi: 10.1016/j.cell.2009.11.029. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Park MJ, Lim MH, Kim SG, Lee M, Baldwin IT, Park CM. A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell. 2012;24:2427–2442. doi: 10.1105/tpc.112.098723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science. 2012;336:1045–1049. doi: 10.1126/science.1219644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl Acad. Sci. USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Optimised new parameter values for each of the thirteen models.
Table S2. AICcU analysis results.
Table S3. Sensitivity heatmap for the expression of CBF3 at the indicated time after dawn.
Table S4. Primer sequences for chromatin Immunoprecipitation.
Generalised method used for comparing models.
Figure S2. Effect of τ2
Figure S3. Effect of d.
Figure S4. Published CBF1 Real-time PCR primers appear to prime from CBF1 and CBF3.
Figure S5. Alignment of published CBF1-specific primers with CBF1 and CBF3 cDNA sequences.