Abstract
Background
Early embryo patterning is orchestrated by tightly regulated morphogen gradients. The Nodal morphogen patterns the mesendoderm, giving rise to all endoderm and head and trunk mesoderm. High Nodal concentrations favor endoderm differentiation while lower promote mesoderm differentiation. Nodal signaling is controlled by both positive and negative feedback regulation to ensure robust developmental patterning.
Results
Here we identify odd skipped related1 (osr1), a zinc finger transcription factor, as a new element in Nodal feedback regulation affecting endoderm development. We show that osr1 expression in zebrafish germ ring mesendoderm requires Nodal signaling; osr1 expression was lost in embryos lacking Nodal signaling. Conversely, osr1 expression was ectopically induced by the activation of Nodal signaling. Furthermore we demonstrate that osr1 responds directly to Nodal signaling. Additionally, osr1 knockdown generated excess endoderm cells marked by sox32 expression while expression of osr1 mRNA was not affected in sox32-deficient embryos.
Conclusions
Our findings identify osr1 as a Nodal-induced, negative feedback regulator of Nodal signaling that acts at the earliest stages of endoderm differentiation to limit the number of endoderm progenitors. As such, we propose that osr1 represents a novel network motif controlling the output of Nodal signaling to regulate mesendoderm patterning. (195)
Keywords: odd skipped related1, osr1, nodal, endoderm development
Introduction
During zebrafish gastrulation, secreted TGFβ proteins of the Nodal family induce mesoderm and endoderm in the germ ring mesendoderm, a region equivalent to the node in amniotes (Warga and Nüsslein-Volhard, 1999; Dougan et al., 2003; Schier, 2003). The zebrafish Nodal-related genes cyclops (cyc) and squint (sqt) are expressed in the first two rows of marginal cells and act as potent inducers of endoderm and mesoderm (Erter et al., 1998; Feldman et al., 1998). Loss of Nodal signaling either in sqt;cyc double mutants or mutants lacking the maternal and zygotic contribution of one-eyed pinhead (oep), a Nodal co-receptor (MZoep), results in complete loss of endoderm and head and trunk mesoderm (Feldman et al., 1998; Gritsman et al., 1999).
Nodal agonists, together with the diffusible Lefty antagonists, establish an embryonic morphogen gradient on the vegetal-animal pole axis during gastrulation (Schier and Talbot, 2005). Zebrafish endoderm progenitors are specified closest to the source of Nodal signaling where high concentrations of Nodal are required to induce expression of sox32 (casanova), a key factor in endoderm determination (Alexander et al., 1999a; Poulain et al., 2006). Mesoderm progenitors are intermingled with endoderm progenitors at the blastoderm margin and extend further toward the animal pole where the expression of genes such as no tail (ntl) requires lower concentrations of Nodal (Piccolo et al., 1999; Rodaway et al., 1999; Warga and Nüsslein-Volhard, 1999). Nodal morphogen gradients have been modeled on the concept of a reaction-diffusion gradient (Muller et al., 2012) where a auto-inducing, short range Nodal stimulus is antagonized by the long range diffusible antagonist Lefty (Chen and Schier, 2001). Nodal signaling via Alk receptor activation and Smad2 phosphorylation can also be antagonized at the level of transcription. Inhibition of the Smad co-factor Foxh1 by the broadly expressed co-repressor DRAP1 interrupts auto-induction of nodal and limits the range of Nodal signaling in mouse embryos (Iratni et al., 2002). Ectodermally expressed factors such as Ectodermin and Serum Response Factor also inhibit transcriptional responses to Nodal, restricting Nodal activity to the mesendoderm (Iratni et al., 2002; Yun et al., 2007; Morsut et al., 2010). Despite this progress in unraveling molecular mechanisms underlying Nodal signaling dynamics, it is likely that additional factors act in mesendoderm to modulate outcomes of Nodal tissue patterning.
Odd skipped related 1 (osr1), a zinc finger transcription factor, homolog of the Drosophila pair-rule gene odd, initially gained attention in vertebrates for its distinctive expression in the intermediate mesoderm and the essential role it plays in kidney formation in mice and zebrafish (Wang et al., 2005; James et al., 2006; Mudumana et al., 2008). Loss of function studies in zebrafish revealed an additional role for osr1 in endoderm patterning, as loss of osr1 caused an increase in the number of cells expressing endoderm markers sox17 and foxa2 (Mudumana et al., 2008). Consistent with its function in regulating endoderm differentiation, zebrafish osr1 is expressed in the mesendoderm prior to gastrulation, and in endoderm during gastrulation (Mudumana et al., 2008), suggesting it may be induced by Nodal signaling.
Here we report that osr1 is induced in mesendoderm by Nodal signaling, most likely as a direct Nodal target gene. In addition to the previously reported negative regulation of sox17- and foxa2-positive endoderm differentiation (Mudumana et al., 2008), we show that osr1 activity is required to limit the number of sox32 expressing endoderm cells. Taken together, our results define a Nodal-induced, negative regulator of endoderm-specific Nodal signaling that shapes embryonic responses to the Nodal morphogen gradient.
Results
Nodal signaling is required for osr1 expression in the mesendoderm
osr1 gene expression is transient and dynamic throughout early zebrafish development. At four hours post fertilization (hpf) until tailbud stage, osr1 is expressed first in mesendoderm progenitors, and subsequently in the forming endoderm. After gastrulation, osr1 is expressed in lateral plate mesoderm (Tena et al., 2007; Mudumana et al., 2008). To test whether early mesendoderm expression of osr1 is controlled by Nodal signaling, we manipulated Nodal signaling and assayed osr1 gene expression by in situ hybridization.
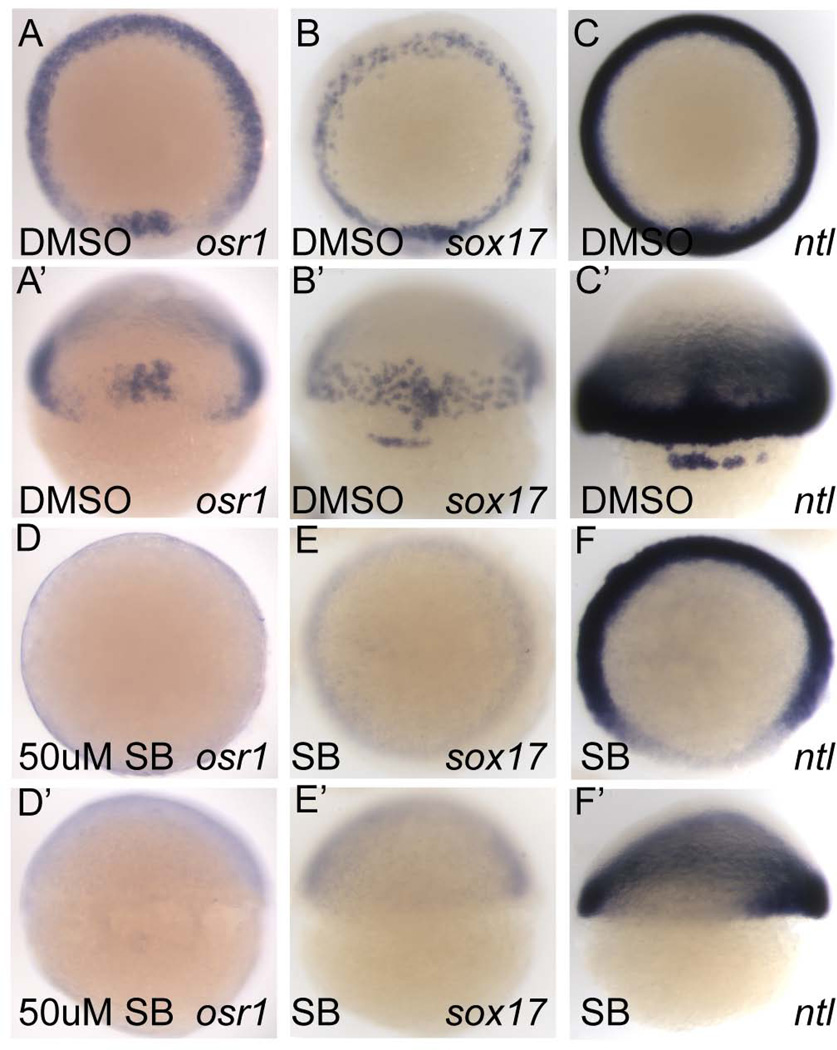
Pharmacological inhibition of Nodal signaling using the Activin type I receptor inhibitor SB505124 (DaCosta Byfield et al., 2004) starting at the zebrafish mid-blastula transition (MBT) is known to phenocopy the cyc;sqt double mutant phenotype and block all endoderm and mesoderm development, while progressively later treatment results in a progressively less severe phenotype (Hagos and Dougan, 2007). We treated embryos with SB505124 starting at the MBT (2.75 hpf) and assayed osr1 expression at shield stage. Compared to wildtype embryos (Fig. 1A, A'), SB505124 treated embryos lacked all osr1 expression (Fig. 1D, D'). sox17 expression and the dorsal expression of ntl, both dependent on Nodal signaling (Feldman et al., 1998; Gritsman et al., 1999), was also blocked by SB505124 treatment, confirming a complete inhibition of Nodal signaling (Fig. 1B, B' and E, E'; C, C' and F, F', respectively).
Figure 1. Pharmacological inhibition of Nodal signaling by SB505124 prevents osr1 expression.
(A–F) Animal pole views, (A'–F') dorsal views. osr1 expression in the germ ring (A) and shield (A') is absent in embryos treated with 50µM SB505124 from 2.5 hpf until the embryo was fixed at shield stage (6 hpf, D, D'). sox17 expression (B, B') in the endoderm and forerunner cells (B') is also absent in SB505124 treated embryos (E, E') demonstrating that a complete block of Nodal signaling is achieved by SB505124. ntl expression (C, C') requires Nodal signaling only in the most dorsal region (F, F').
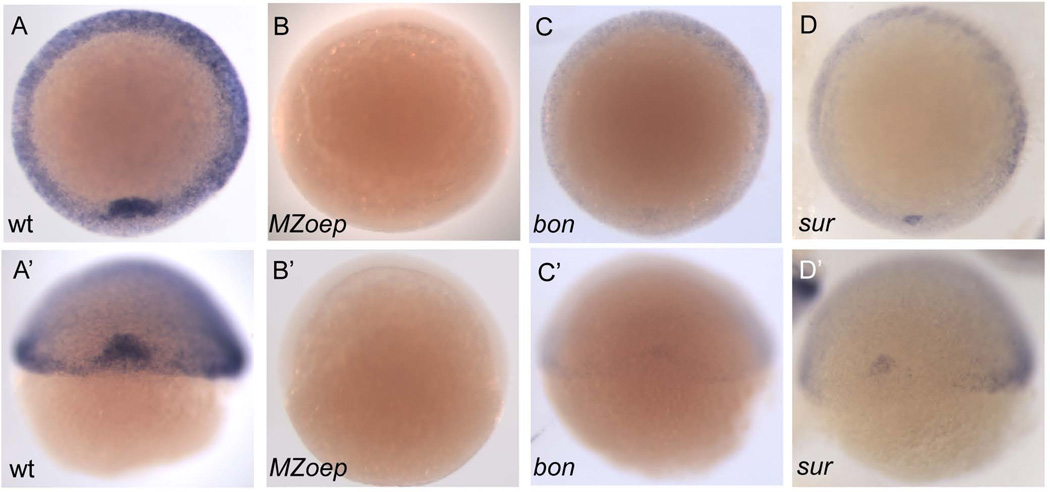
osr1 expression was also strongly affected by mutations in genes encoding zebrafish Nodal signaling components. MZoep mutants lack both maternal and zygotic function of the one-eyed pinhead (oep) gene, an essential co-receptor of Nodal signaling, and are therefore insensitive to Nodal signaling (Gritsman et al., 1999). At shield stage MZoep mutant embryos lacked all osr1 expression (Fig. 2A,A' and B, B', Table 1). Mixer and FoxH1 are transcription factors that mediate Nodal signaling; mutations in either of these genes result in a partial reduction of Nodal signaling (Alexander and Stainier, 1999; Pogoda et al., 2000). osr1 expression in both bonnie and clyde (bon; mixer) and schmalspur (sur; foxh1) mutants was significantly decreased in germ ring mesendoderm and shield tissue but not completely ablated (Fig. 2C, C' and D, D', Table 1). The results indicate that Nodal signaling is required for osr1 expression in the mesendoderm, and that both Mixer and FoxH1 are required to establish the full expression domain of osr1.
Figure 2. osr1 expression is disrupted in Nodal signaling mutants.
(A–D) dorsal view (A’–D’) view of the animal pole. At shield stage osr1 is expressed in the shield and germ ring of the embryo (A, A’). Maternal zygotic one eyed pinhead (MZoep) mutants, which lack all Nodal signaling, show a complete loss of osr1 expression (B, B’). Mutants bonnie and clyde(bon) and schmalspur(sur), which have reduced Nodal signaling, show a reduction of osr1 expression (C, C’ and D, D’, respectively).
Table 1.
Nodal signals are required for normal osr1 expression in the germ ring or at shield stage
| Mutant background | n | Embryos showing normal expression of osr1 |
Embryos showing reduced expression of osr1 |
|---|---|---|---|
| MZ one eyed pinhead | 10 | 0% | 100% |
| bonnie and clyde | 184 | 80% | 20% |
| schmalspur | 39 | 56% | 44% |
Embryos from heterozygous or homozygous intercrosses of the genotype indicated were analyzed at shield stage for the expression of osr1.
osr1 is a direct target of Nodal signaling
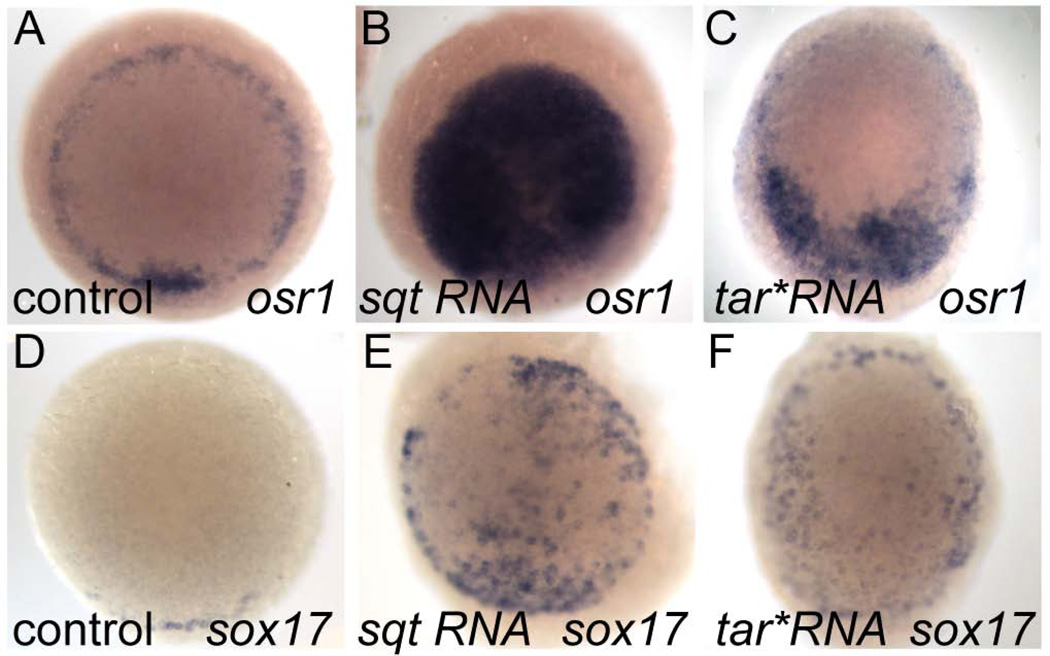
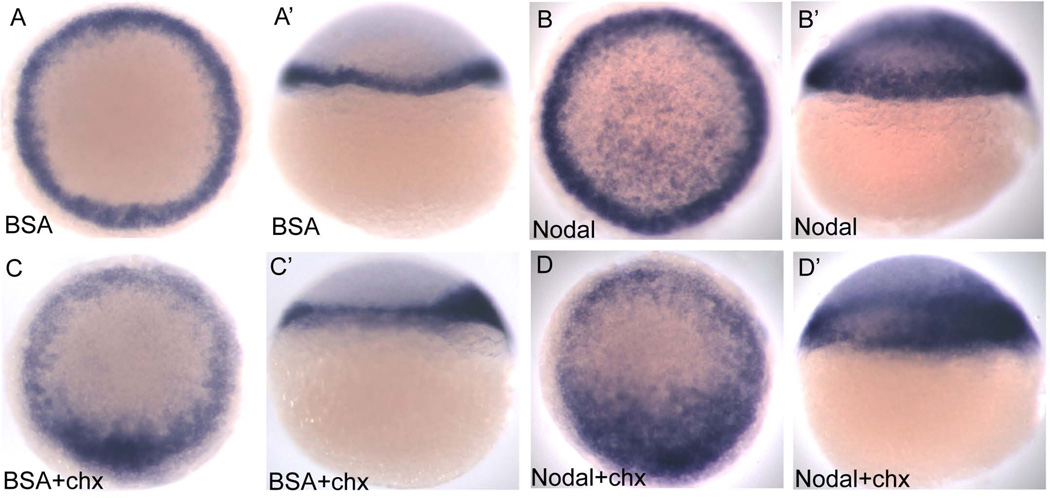
To determine whether osr1 is a target of Nodal signaling we first tested whether ectopic activation of Nodal signaling by injection of mRNA encoding the Nodal ligand squint or a constitutively active type I activin receptor, taram-a (tar*), could induce osr1 gene expression. (Dickmeis et al., 2001). Both squint and tar* RNA induced ectopic osr1 expression in more than 80% of injected embryos while control injections had no effect (Table 2, Fig. 3 A–C). Ectopic Nodal activation in injected embryos was confirmed by sox17 expression (Fig 3 D–E). To test whether osr1 is an "immediate early" target of Nodal signaling we injected recombinant mouse Nodal protein into the animal pole region of zebrafish embryos at 4 hpf and assessed embryos for ectopic expression of osr1 one hour later. Nodal induced ectopic, animal pole expression of osr1, even in embryos pre-incubated with the protein synthesis inhibitor cycloheximide (Ragheb et al., 1999) (Fig. 4A,A'; B, B' and D, D', Table 2). We conclude that osr1 induction by Nodal signaling does not require protein synthesis, indicating that osr1 belongs to the class of direct transcriptional targets of Nodal signaling that also includes cas, bon, gata5 and mezzo (Poulain and Lepage, 2002).
Table 2.
Nodal signals induce ectopic expression of osr1.
| Nodal signal | n | Embryos showing normal expression of osr1 |
Embryos showing ectopic expression of osr1 |
|---|---|---|---|
| squint RNA | 12 | 17% | 83% |
| TARAM-A RNA | 54 | 13% | 87% |
| mouse Nodal protein | 11 | 27% | 73% |
RNA was injected at 1–2 cell stage. Nodal protein was injected in the animal pole at 4hpf.
Figure 3. osr1 expression is induced by ectopic Nodal signaling.
Embryos were injected with either squint or tar*/Acvr1b RNA at 1-cell stage fixed at 4.5 hpf. (A–F) animal pole views. (A) osr1 is normally expressed in the margin of the embryo. Both squint and tar* can induce ectopic expression of osr1 in animal pole cells. Ectopic expression of sox17 (D–F) in these embryos confirms ectopic activation of Nodal signaling.
Figure 4. osr1 is a direct target of Nodal signaling.
(A, B, C, D) animal pole view; (A’, B’, C’, D’) lateral view, dorsal to the right. 4 hpf embryos were injected with 2 ng recombinant mouse Nodal protein (B, B’) or 400 ng BSA (A, A’). Half of the injected embryos were treated with 50µg/ml cycloheximide (C, C’, D, D’), and all were fixed after 1 hr and 20 minutes. Nodal protein strongly induces ectopic osr1 expression in the animal pole of the embryo (B, B’). Ectopic expression is induced in the presence of cycloheximide indicating that osr1 is a direct target of Nodal signaling (D, D’).
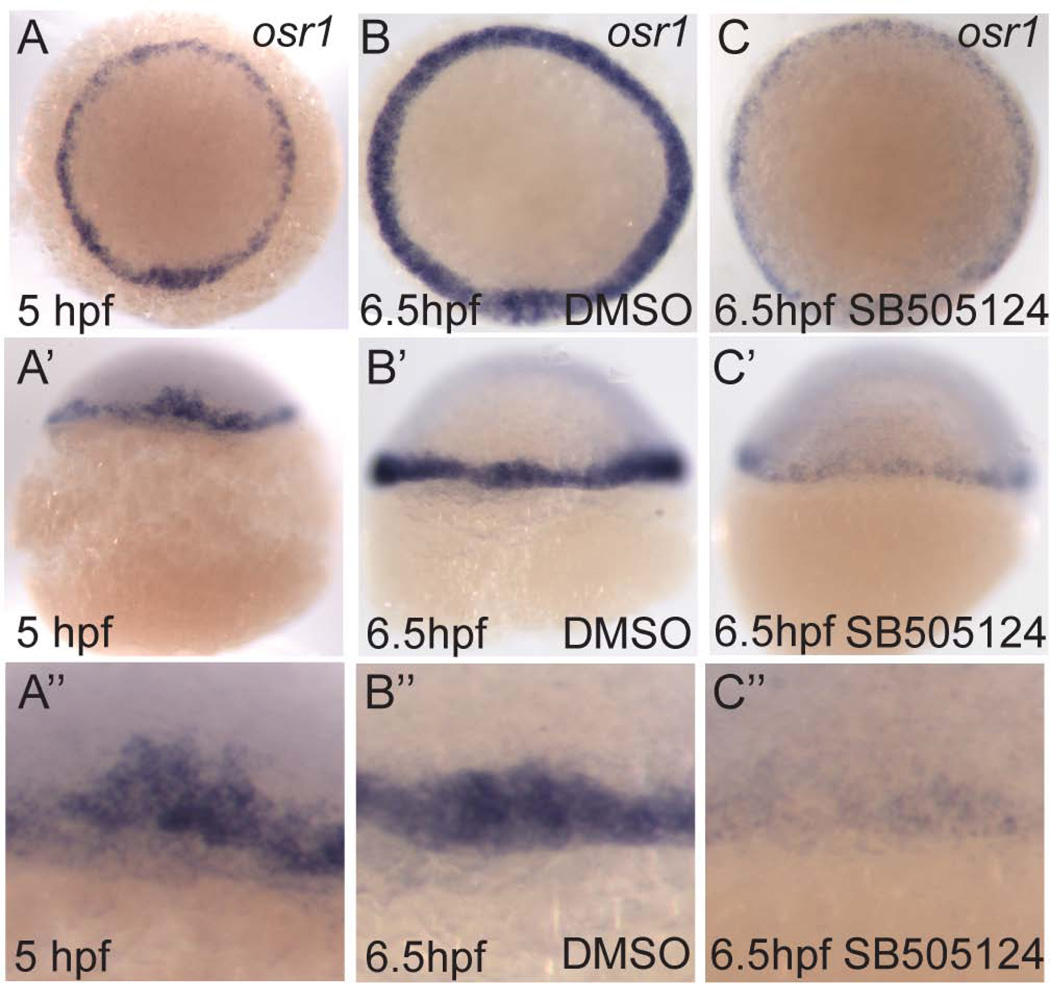
While the data show that osr1 can be ectopically induced by Nodal, the loss of osr1 expression in Nodal signaling mutants could be due to a simple absence of mesendoderm cells, as opposed to a requirement for Nodal in osr1 induction; i.e. osr1 could just be a marker of mesendoderm. To confirm a requirement for Nodal signaling in osr1 gene regulation independent of Nodal's role in maintaining mesendoderm and to further explore the dynamics of osr1 gene expression, we tested the effects of Nodal inhibition after osr1 transcription had been initiated in mesendoderm. Inhibition of Nodal signaling using SB505124 treatment starting at 5 hpf, when the majority of mesendoderm precursors have been specified (Hagos and Dougan, 2007), dramatically diminished the expression of osr1 (Fig. 5 A-A", B-B", and C-C"). These results indicate that continued Nodal signaling is required to maintain osr1 mesendoderm expression and, in addition, suggest that osr1 mRNA is unstable and is degraded after inhibition of Nodal signaling. Taken together our results indicate that osr1 is a direct target of Nodal signaling and that maintenance of its expression in the mesendoderm requires Nodal signaling activity.
Figure 5. Nodal signaling is required to maintain osr1 expression.
(A–C) Animal pole views. At 5 hpf (A) osr1 is expressed in the margin of the embryo. Embryos treated with 50µM SB505124 from 5 hpf to 6.5 hpf show a dramatic reduction in levels of osr1 mRNA (C, C’, C’’) as compared to DMSO treated controls (B, B’, B’’) indicating that osr1 mRNA is degraded in the absence of Nodal signaling.
Osr1 functions in parallel with sox32 in the Nodal signaling hierarchy
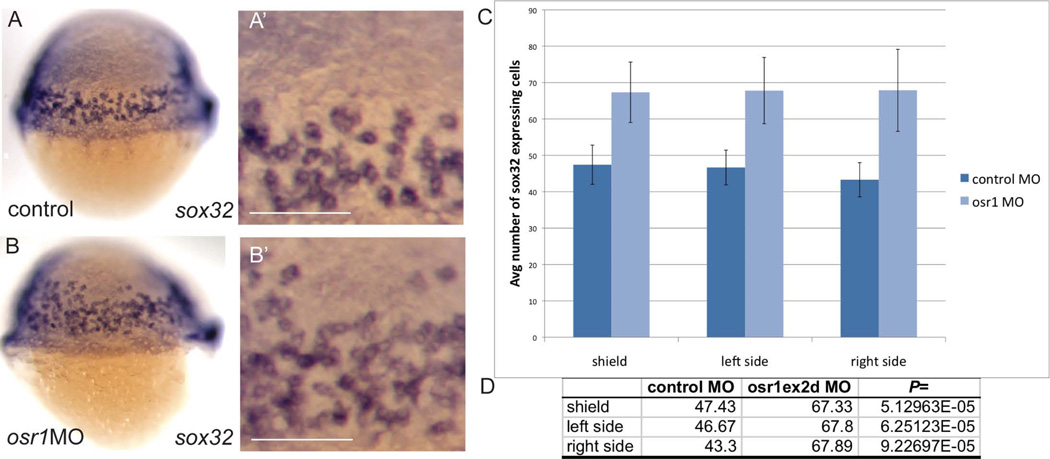
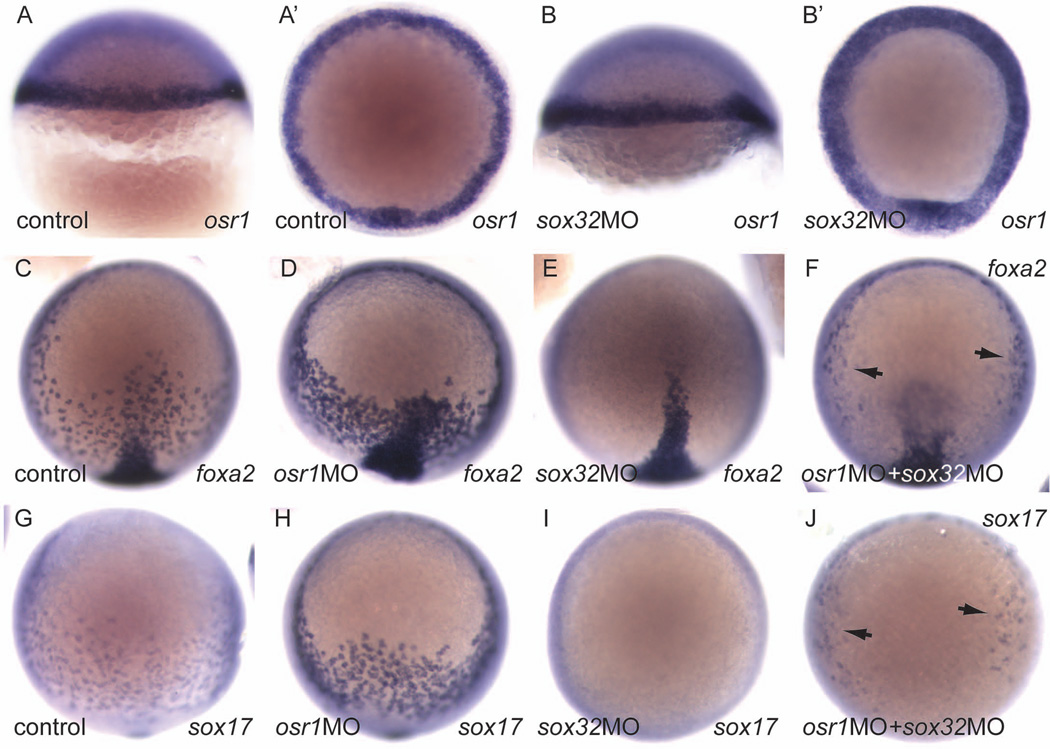
We have shown previously that osr1-deficiency results in an expansion of endoderm expressing sox17 and foxa2 (Mudumana et al., 2008). Both of these genes are downstream of sox32 in the endoderm induction cascade, as sox32/cas mutants lack all endoderm expression of sox17 and foxa2 (Alexander et al., 1999b). To test whether osr1 acts at a primary step in Nodal-induced endoderm differentiation or whether it functions downstream of sox32 to regulate a secondary wave of endoderm gene expression, we assayed the effect of osr1 knockdown on sox32 expression. osr1 knockdown significantly increased the number of sox32-expressing endoderm cells as judged by cell counts in whole mount sox32 in situ hybridization embryos (Fig. 6 A–D). The expression of two additional Nodal immediate early genes, bon and gata5, as well as two Nodal ligand genes, cyc and sqt, was also examined in osr1 morphants. However the expression of these genes was not significantly affected by loss of osr1 (data not shown), suggesting that osr1 exerts its effect on mesendoderm patterning primarily by regulating sox32 expression. The results suggest that osr1 normally acts at a primary step to limit the output of Nodal signaling on endoderm cell differentiation and control the number of sox32-expressing endoderm progenitors. If osr1 is an immediate-early Nodal induced gene and acts at the initial steps of endoderm specification, then elimination of other immediate-early endoderm genes such as sox32 should not affect osr1 expression. We therefore generated sox32-deficient embryos by morpholino knockdown and assayed osr1 expression by in situ hybridization. Our results show that mesendoderm osr1 expression is not affected by sox32 knockdown (Fig. 7A,A', B,B'), indicating that osr1 functions upstream or in parallel with sox32 to limit endoderm differentiation. As we have previously reported (Mudumana et al., 2008), osr1 knockdown increases the number of foxa2 (Fig. 7 C,D) and sox17 (Fig. 7 G,H) expressing endoderm cells. Interestingly, while sox32 knockdown was sufficient to eliminate foxa2 and sox17 endoderm expression (Fig. 7 E, I), simultaneous knockdown of osr1 in sox32-deficient embryos partially restored foxa2 (Fig. 7F) and sox17 (Fig. 7J) expression, suggesting that residual, low level Sox32 expression in sox32 morphants may be sufficient to drive endoderm cell differentiation after removal of osr1 inhibition.
Figure 6. Loss of osr1 increases the number of sox32 expressing endoderm cells.
sox32 expressing cells were counted in osr1 morphants and embryos injected with a control morpholino. (A) and (B) show lateral views of control and osr1 morphant embryos, respectively (dorsal is on the right). Cells were counted in identical sized windows (area = 0.05mm2) (A', B') on both lateral sides and the dorsal side (shield). Average number of cells per 0.05mm2 on each side is shown in (C). Welch's t-test analysis indicates that osr1 morphant embryos show a significant increase in the number of sox32 expressing cells (D). n=7 (control); n=10 (osr1 MO). Scale bar in (A', B') = 0.1mm.
Figure 7. osr1 expression is independent of sox32 and acts to limit endodermal cell differentiation.
(A, B) lateral views, dorsal on the right. (A', B', animal pole views, dorsal bottom). osr1 expression in the germ ring (A, A') is unaffected in sox32 morphant embryos, which lack all endoderm derivatives. (C–J, animal pole views, dorsal bottom). foxa2 expression in control (C), osr1 knockdown (D), sox32 knockdown (E), and combined osr1, sox32 knockdown (F) reveals increased foxa2 expressing cells in osr1- and combined osr1-, sox32-deficient embryos. sox17 expression in control (G), osr1 knockdown (H), sox32 knockdown (I), and combined osr1, sox32 knockdown (J) reveals increased sox17 expressing cells in osr1- and combined osr1-, sox32-deficient embryos.
Discussion
osr1 is a direct target of Nodal signaling
We report that the zinc finger transcription factor osr1 is expressed in the mesendoderm of zebrafish embryos and is a direct target of Nodal signaling. Ectopic activation of Nodal signaling with either squint or taram-a* mRNA or the recombinant mouse Nodal protein induces ectopic activation of osr1 in animal pole cells. This induction is direct because ectopic activation was observed in the absence of protein synthesis. Maintenance of osr1 gene expression requires continued Nodal signaling, and the level of osr1 mRNA is decreased in late blastula stage embryos treated with the Nodal inhibitor SB505124 (Fig. 2.5). Our results add osr1 to the short list of direct Nodal targets that includes cas, bon, gata5 and mezzo (Poulain and Lepage, 2002). However, unlike previously described Nodal targets, which are inducers of mesendoderm fates, osr1 acts as a repressor of the endoderm fate. Thus, osr1 represents a novel class of Nodal target genes and introduces an additional layer of regulation of the cellular response to Nodal.
Prior to our work little was known about the regulation of osr1 expression. One study conducted in mice found that runx2, an osteogenic transcription factor, and ikzf1, a transcriptional regulator of hematopoietic cell differentiation, can bind the osr1 promoter repressing gene expression (Yamauchi et al., 2008). In chicken, intermediate concentrations of BMP can induce osr1 expression in primitive streak explants, but this induction is not direct, and does not occur in the presence of cycloheximide (James and Schultheiss, 2005). Both types of regulatory mechanisms might play a role in regulating osr1 expression in the lateral plate mesoderm in zebrafish. This area of the embryo is known to require BMP signaling for cell fate specification, and gives rise to blood progenitors. However, osr1 is more likely to be a marker of differentiation in this tissue as opposed to a direct target of BMP signaling.
Osr1 limits expression of sox32, a key inducer of endoderm
Endoderm formation in zebrafish requires the function of the sox32 gene; embryos lacking sox32 lack all endoderm derivatives (Alexander et al., 1999b). sox32 expression is induced by the combined action of the transcription factors bon and gata5 (Aoki et al., 2002) and is a direct target of Nodal signaling (Poulain and Lepage, 2002). Previously, loss of osr1 function has been shown to increase the number of cells expressing endoderm markers sox17 and foxa2 (Mudumana et al., 2008). These genes are expressed in cells committed to the endoderm lineage and both genes require sox32 for their induction in endoderm (Reiter et al., 2001). In order to define the position of Osr1 activity in the Nodal signaling hierarchy we examined expression of bon, gata5, sqt, cyc, and sox32 in osr1 morphants and found that while the expression of bon, gata5, sqt, and cyc was not affected by the loss of osr1, the number of endoderm cells expressing sox32 was increased. Based on this observation we propose that osr1 functions in parallel with sox32 specifically in endoderm development since both osr1 and sox32 are direct Nodal targets and osr1 deficiency de-represses sox32 expression. This conclusion is strengthened by our observation that osr1 expression is not affected by sox32 deficiency. Based on this, one would predict that ectopic expression of osr1 would reduce the number of sox32 expressing cells and inhibit endoderm formation. However, in multiple experiments we were unable to observe an effect of osr1 over-expression on sox32 expression (data not shown), suggesting that osr1 by itself is not sufficient to repress sox32. It is likely that additional co-factors or repressors are required for osr1 to limit endoderm development.
osr1 is a novel negative feedback Nodal signaling component
The gradient of Nodal activity that patterns the mesendoderm is formed by both positive and negative regulation of Nodal signaling (Schier, 2009). Nodal patterning is shaped by positive auto-induction of Nodal ligands, expression of the diffusible Nodal antagonist Lefty, as well as repression of Nodal signaling by factors expressed in neighboring tissues (Yun et al., 2007; Schier, 2009; Zhao et al., 2013). Nodal auto-induction and short range signaling coupled with expression of the diffusible long range inhibitor Lefty, results in a steep Nodal activity gradient and confers field-size independent regulation on the nodal morphogen gradient as predicted by reaction-diffusion models (Muller et al., 2012). Transcriptional inhibition of Nodal signaling by factors expressed in overlapping or neighboring tissues sharpens the boundaries different tissue layers to ensure correct embryonic patterning (Yun et al., 2007; Zhao et al., 2013). Our finding that osr1 is a nodal-induced, negative feedback regulator of endoderm cell differentiation introduces a new component to Nodal signaling dynamics. The negative feedback activity of osr1 on endoderm development is conceptually similar to the more broadly acting SnoN in TGFβ signaling, where a ligand (TFGβ), induces expression of a repressor (SnoN) of its own downstream signaling activity (Smad2/4; (Stroschein, 1999)). As such, osr1 may be considered an element of an endoderm-specific Nodal signaling incoherent feed forward loop (Milo et al., 2002). An incoherent feed forward loop is composed of two parallel but antagonistic regulatory pathways (Milo et al., 2002). The outcome of activation of gene targets coupled directly with delayed expression of a repressor (protein) can be to change a monotonic increase in target output to a transient pulse of target gene expression (Mangan and Alon, 2003). Incoherent feed forward loops have also been associated with oscillatory network motifs and to confer "fold-change", as opposed to simple amplitude, driven signaling (Goentoro et al., 2009). Whether Nodal signaling in zebrafish endoderm cell differentiation functions in a pulsatile or frequency encoded fashion as has been reported for Smad4 nuclear localization and Activin signaling in Xenopus embryos (Warmflash et al., 2012) and BMP2 induced gene expression in cultured myoblasts (Schul et al., 2013) may be a fruitful avenue for future investigation.
Experimental Procedures
Zebrafish maintenance, Mutant strains
Wild-type zebrafish were maintained according to established protocols (Westerfield, 1995). Embryos were collected from crosses of wild-type Tü/AB adults, grown at 28°C in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2·2H2O, 0.33 mM MgSO4) and fixed at the indicated developmental stages. Embryos were staged according to Kimmel et al. (Kimmel et al., 1995). Homozygous mutant embryos were obtained from an incross of mutant heterozygotes. The following mutant strains were used: bonnie and clydem435 (Kikuchi et al., 2000); MZoeptz57 (Hammerschmidt et al., 1996; Gritsman et al., 1999), sur/foxh1m768 (Pogoda et al., 2000).
Whole mount in situ hybridization
Whole-mount in situ hybridization using DIG-labeled riboprobes was carried out as previously described (Mudumana et al., 2004). Photographs were acquired using a Leica MZ12 microscope equipped with a Spot Image digital camera. Image contrast, brightness, and sharpness were adjusted with Adobe Photoshop CS3 in accordance with ethical guidelines for image manipulation established by the Journal of Cell Biology.
Plasmid constructs and morpholino injection
The following probes were used: odd skipped related 1/osr1(Mudumana et al., 2008), sox32/casanova (Alexander et al., 1999b), sox17 (Alexander and Stainier, 1999), ntl (Schulte-Merker et al., 1992), foxa2/axial (Strahle et al., 1993), oep (Zhang et al., 1998), cyc (Rebagliati et al., 1998), bonnie and clyde/bon (Kikuchi et al., 2000), gata5/faust (Reiter et al., 2001).
Morpholinos (Gene Tools) were resuspended in sterile water, stored at −20°C as a 1 or 2mM stock solutions and diluted before use to the appropriate concentration. For injection, morpholino oligonucleotides were diluted in 0.1 M KCl, 0.01M HEPES and 0.2% Phenol Red and injections were performed using a nanoliter2000 microinjector (World Precision Instruments). Injection concentrations ranged from 0.2 mM to 0.75 mM and injection volume was set at 4.6 nl/embryo (1.4–7.4 ng/embryo). Embryos were injected at the 1–2 cell stage. Efficiency of the morpholino’s effect on RNA splicing was confirmed by RT-PCR.
osr1ex2d (osr1MO), ATCTCATCCTTACCTGTGGTCTCTC;
osr1ATG: GGAGCGTCTTACTACCCATGACTAA;
osr1CoMO: AATCAGTACCCATCATTCTGCGAGG (Mudumana et al., 2008);
sox32ATG, CAGGGAGCATCCGGTCGAGATACAT (Dickmeis et al., 2001);
RNA Synthesis
Capped mRNA was synthesized using the mMESSAGE mMACHINE system (Ambion) from the following previously described templates: squint (Feldman et al., 1998; Gritsman et al., 1999)) and taram-a (Peyrieras et al., 1998). Embryos were injected at the 1–4 cell stage with sqt (650pg), tar* (650pg) synthetic mRNAs.
Protein injection and chemical inhibitor treatment
Recombinant mouse Nodal protein (R&D Systems, Minneapolis, MN; cat no. 3115-ND) was reconstituted to a concentration of 100 µg/ml in sterile 1mM HCL containing 0.1% BSA. Embryos were grown until 4 hpf and injected into the animal pole using a nanoliter2000 microinjector (World Precision Instruments). For pre-incubation with cycloheximide, chorions were perforated with a pulled glass capillary needle 0.5 hr prior to protein injection and cycloheximide was added to E3 medium to a final concentration of 50µg/ml. After injection, embryos were returned to cycloheximide-containing medium. 10 mg/ml BSA was injected in the same manner as a control. To confirm efficacy of cycloheximide treatment, a subset of treated embryos were maintained for an extended period in medium containing cycloheximide or DMSO. Cycloheximide treated embryos arrested prior to entering gastrulation, consistent with a requirement for protein synthesis for gastrulation, and eventually died whereas control embryos developed normally.
For Nodal inhibitor studies, chorions were perforated prior to start of treatment for better penetration. For pharmacological inhibition of Nodal signaling SB505124 (2-(5-benzo[1,3]dioxol-5-yl-2-tert-butyl-3Himidazol-4-yl)-6-methylpyridine hydrochloride) was obtained from Sigma-Aldrich and was stored as a 10 mM stock solution, dissolved in DMSO at −20°C. For drug treatments, embryos were kept at 28.5 °C until start of treatment, chorions were perforated prior to start of drug treatment and embryos were split into treatment groups in a 24-well plate. Egg medium (E3) was replaced with 0.5ml E3 + 50µM SB505124 or DMSO starting at MBT (2.75 hpf) or 5 hpf, depending on the experiment, and incubated in the dark at 28.5°C until fixation. In all experiments, some embryos in each experiment were allowed to develop until 24 hpf and examined morphologically to verify the efficacy of the treatment. All experiments were performed at least three times. For SB505124, the lowest dose that reproduced the sqt;cyc phenotype has been reported to range from 30–50 µM, depending on the age of the drug (Hagos and Dougan, 2007). We found that 50 µM consistently and reliably inhibited Nodal signaling.
Bullet points.
odd-skipped related 1 is a zinc finger transcription factor expressed in zebrafish mesendoderm.
osr1 is a direct target of Nodal signal transduction.
osr1 negatively regulates endoderm development.
osr1 is a novel component of an endoderm nodal signaling incoherent feedforward loop.
Acknowledgements
We thank Dr. Alex Schier for the gift of MZoep embryos. This work was supported by NIH Grant R01 DK071041 to I.A.D.
Grant Sponsor: NIH R01 DK071041
References
- Alexander J, Rothenberg M, Henry GL, Stainier DY. casanova plays an early and essential role in endoderm formation in zebrafish. Developmental Biology. 1999a;215:343–357. doi: 10.1006/dbio.1999.9441. [DOI] [PubMed] [Google Scholar]
- Alexander J, Rothenberg M, Henry GL, Stainier DY. casanova plays an early and essential role in endoderm formation in zebrafish. Dev Biol. 1999b;215:343–357. doi: 10.1006/dbio.1999.9441. [DOI] [PubMed] [Google Scholar]
- Alexander J, Stainier DY. A molecular pathway leading to endoderm formation in zebrafish. Curr Biol. 1999;9:1147–1157. doi: 10.1016/S0960-9822(00)80016-0. [DOI] [PubMed] [Google Scholar]
- Aoki TO, David NB, Minchiotti G, Saint-Etienne L, Dickmeis T, Persico GM, Strahle U, Mourrain P, Rosa FM. Molecular integration of casanova in the Nodal signalling pathway controlling endoderm formation. Development. 2002;129:275–286. doi: 10.1242/dev.129.2.275. [DOI] [PubMed] [Google Scholar]
- Chen Y, Schier AF. The zebrafish Nodal signal Squint functions as a morphogen. Nature. 2001;411:607–610. doi: 10.1038/35079121. [DOI] [PubMed] [Google Scholar]
- DaCosta Byfield S, Major C, Laping NJ, Roberts AB. SB-505124 is a selective inhibitor of transforming growth factor-beta type I receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2004;65:744–752. doi: 10.1124/mol.65.3.744. [DOI] [PubMed] [Google Scholar]
- Dickmeis T, Mourrain P, Saint-Etienne L, Fischer N, Aanstad P, Clark M, Strahle U, Rosa F. A crucial component of the endoderm formation pathway, CASANOVA, is encoded by a novel sox-related gene. Genes Dev. 2001;15:1487–1492. doi: 10.1101/gad.196901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan ST, Warga RM, Kane DA, Schier AF, Talbot WS. The role of the zebrafish nodal-related genes squint and cyclops in patterning of mesendoderm. Development. 2003;130:1837–1851. doi: 10.1242/dev.00400. [DOI] [PubMed] [Google Scholar]
- Erter CE, Solnica-Krezel L, Wright CV. Zebrafish nodal-related 2 encodes an early mesendodermal inducer signaling from the extraembryonic yolk syncytial layer. Dev Biol. 1998;204:361–372. doi: 10.1006/dbio.1998.9097. [DOI] [PubMed] [Google Scholar]
- Feldman B, Gates MA, Egan ES, Dougan ST, Rennebeck G, Sirotkin HI, Schier AF, Talbot WS. Zebrafish organizer development and germ-layer formation require nodal-related signals. Nature. 1998;395:181–185. doi: 10.1038/26013. [DOI] [PubMed] [Google Scholar]
- Goentoro L, Shoval O, Kirschner MW, Alon U. The incoherent feedforward loop can provide fold-change detection in gene regulation. Mol Cell. 2009;36:894–899. doi: 10.1016/j.molcel.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritsman K, Zhang J, Cheng S, Heckscher E, Talbot WS, Schier AF. The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell. 1999;97:121–132. doi: 10.1016/s0092-8674(00)80720-5. [DOI] [PubMed] [Google Scholar]
- Hagos EG, Dougan ST. Time-dependent patterning of the mesoderm and endoderm by Nodal signals in zebrafish. BMC Dev Biol. 2007;7:22. doi: 10.1186/1471-213X-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt M, Pelegri F, Mullins MC, Kane DA, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Haffter P, Heisenberg CP, Jiang YJ, Kelsh RN, Odenthal J, Warga RM, Nusslein-Volhard C. Mutations affecting morphogenesis during gastrulation and tail formation in the zebrafish, Danio rerio. Development. 1996;123:143–151. doi: 10.1242/dev.123.1.143. [DOI] [PubMed] [Google Scholar]
- Iratni R, Yan YT, Chen C, Ding J, Zhang Y, Price SM, Reinberg D, Shen MM. Inhibition of excess nodal signaling during mouse gastrulation by the transcriptional corepressor DRAP1. Science. 2002;298:1996–1999. doi: 10.1126/science.1073405. [DOI] [PubMed] [Google Scholar]
- James RG, Kamei CN, Wang Q, Jiang R, Schultheiss TM. Odd-skipped related 1 is required for development of the metanephric kidney and regulates formation and differentiation of kidney precursor cells. Development. 2006;133:2995–3004. doi: 10.1242/dev.02442. [DOI] [PubMed] [Google Scholar]
- James RG, Schultheiss TM. Bmp signaling promotes intermediate mesoderm gene expression in a dose-dependent, cell-autonomous and translation-dependent manner. Developmental Biology. 2005;288:113–125. doi: 10.1016/j.ydbio.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Trinh LA, Reiter JF, Alexander J, Yelon D, Stainier DY. The zebrafish bonnie and clyde gene encodes a Mix family homeodomain protein that regulates the generation of endodermal precursors. Genes Dev. 2000;14:1279–1289. [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci U S A. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- Morsut L, Yan KP, Enzo E, Aragona M, Soligo SM, Wendling O, Mark M, Khetchoumian K, Bressan G, Chambon P, Dupont S, Losson R, Piccolo S. Negative control of Smad activity by ectodermin/Tif1gamma patterns the mammalian embryo. Development. 2010;137:2571–2578. doi: 10.1242/dev.053801. [DOI] [PubMed] [Google Scholar]
- Mudumana SP, Hentschel D, Liu Y, Vasilyev A, Drummond IA. odd skipped related1 reveals a novel role for endoderm in regulating kidney versus vascular cell fate. Development. 2008;135:3355–3367. doi: 10.1242/dev.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudumana SP, Wan H, Singh M, Korzh V, Gong Z. Expression analyses of zebrafish transferrin, ifabp, and elastaseB mRNAs as differentiation markers for the three major endodermal organs: liver, intestine, and exocrine pancreas. Dev Dyn. 2004;230:165–173. doi: 10.1002/dvdy.20032. [DOI] [PubMed] [Google Scholar]
- Muller P, Rogers KW, Jordan BM, Lee JS, Robson D, Ramanathan S, Schier AF. Differential diffusivity of Nodal and Lefty underlies a reaction-diffusion patterning system. Science. 2012;336:721–724. doi: 10.1126/science.1221920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrieras N, Strahle U, Rosa F. Conversion of zebrafish blastomeres to an endodermal fate by TGF-beta-related signaling. Curr Biol. 1998;8:783–786. doi: 10.1016/s0960-9822(98)70303-3. [DOI] [PubMed] [Google Scholar]
- Piccolo S, Agius E, Leyns L, Bhattacharyya S, Grunz H, Bouwmeester T, De Robertis EM. The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature. 1999;397:707–710. doi: 10.1038/17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogoda HM, Solnica-Krezel L, Driever W, Meyer D. The zebrafish forkhead transcription factor FoxH1/Fast1 is a modulator of nodal signaling required for organizer formation. Curr Biol. 2000;10:1041–1049. doi: 10.1016/s0960-9822(00)00669-2. [DOI] [PubMed] [Google Scholar]
- Poulain M, Fürthauer M, Thisse B, Thisse C, Lepage T. Zebrafish endoderm formation is regulated by combinatorial Nodal, FGF and BMP signalling. Development. 2006;133:2189–2200. doi: 10.1242/dev.02387. [DOI] [PubMed] [Google Scholar]
- Poulain M, Lepage T. Mezzo, a paired-like homeobox protein is an immediate target of Nodal signalling and regulates endoderm specification in zebrafish. Development. 2002;129:4901–4914. doi: 10.1242/dev.129.21.4901. [DOI] [PubMed] [Google Scholar]
- Ragheb JA, Deen M, Schwartz RH. The destabilization of IL-2 mRNA by a premature stop codon and its differential stabilization by trans-acting inhibitors of protein synthesis do not support a role for active translation in mRNA stability. J Immunol. 1999;163:3321–3330. [PubMed] [Google Scholar]
- Rebagliati MR, Toyama R, Fricke C, Haffter P, Dawid IB. Zebrafish nodal-related genes are implicated in axial patterning and establishing left-right asymmetry. Developmental Biology. 1998;199:261–272. doi: 10.1006/dbio.1998.8935. [DOI] [PubMed] [Google Scholar]
- Reiter JF, Kikuchi Y, Stainier DY. Multiple roles for Gata5 in zebrafish endoderm formation. Development. 2001;128:125–135. doi: 10.1242/dev.128.1.125. [DOI] [PubMed] [Google Scholar]
- Rodaway A, Takeda H, Koshida S, Broadbent J, Price B, Smith JC, Patient R, Holder N. Induction of the mesendoderm in the zebrafish germ ring by yolk cell-derived TGF-beta family signals and discrimination of mesoderm and endoderm by FGF. Development. 1999;126:3067–3078. doi: 10.1242/dev.126.14.3067. [DOI] [PubMed] [Google Scholar]
- Schier A, Talbot W. Molecular genetics of axis formation in zebrafish. Annu. Rev. Genet. 2005;39:561–613. doi: 10.1146/annurev.genet.37.110801.143752. [DOI] [PubMed] [Google Scholar]
- Schier AF. Nodal signaling in vertebrate development. Annu Rev Cell Dev Biol. 2003;19:589–621. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
- Schier AF. Nodal morphogens. Cold Spring Harb Perspect Biol. 2009;1:a003459. doi: 10.1101/cshperspect.a003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schul D, Schmitt A, Regneri J, Schartl M, Wagner TU. Burst BMP triggered receptor kinase activity drives Smad1 mediated long-term target gene oscillation in C2C12 cells. PLoS One. 2013;8:e59442. doi: 10.1371/journal.pone.0059442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Merker S, Ho RK, Herrmann BG, Nusslein-Volhard C. The protein product of the zebrafish homologue of the mouse T gene is expressed in nuclei of the germ ring and the notochord of the early embryo. Development. 1992;116:1021–1032. doi: 10.1242/dev.116.4.1021. [DOI] [PubMed] [Google Scholar]
- Strahle U, Blader P, Henrique D, Ingham PW. Axial, a zebrafish gene expressed along the developing body axis, shows altered expression in cyclops mutant embryos. Genes Dev. 1993;7:1436–1446. doi: 10.1101/gad.7.7b.1436. [DOI] [PubMed] [Google Scholar]
- Stroschein SL. Negative Feedback Regulation of TGF- Signaling by the SnoN Oncoprotein. Science. 1999;286:771–774. doi: 10.1126/science.286.5440.771. [DOI] [PubMed] [Google Scholar]
- Tena JJ, Neto A, de la Calle-Mustienes E, Bras-Pereira C, Casares F, Gómez-Skarmeta JL. Odd-skipped genes encode repressors that control kidney development. Developmental Biology. 2007;301:518–531. doi: 10.1016/j.ydbio.2006.08.063. [DOI] [PubMed] [Google Scholar]
- Wang Q, Lan Y, Cho ES, Maltby KM, Jiang R. Odd-skipped related 1 (Odd 1) is an essential regulator of heart and urogenital development. Developmental Biology. 2005;288:582–594. doi: 10.1016/j.ydbio.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warga RM, Nüsslein-Volhard C. Origin and development of the zebrafish endoderm. Development. 1999;126:827–838. doi: 10.1242/dev.126.4.827. [DOI] [PubMed] [Google Scholar]
- Warmflash A, Zhang Q, Sorre B, Vonica A, Siggia ED, Brivanlou AH. Dynamics of TGF-beta signaling reveal adaptive and pulsatile behaviors reflected in the nuclear localization of transcription factor Smad4. Proc Natl Acad Sci U S A. 2012;109:E1947–E1956. doi: 10.1073/pnas.1207607109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) 3rd Edition. Eugene, OR: University of Oregon Press; 1995. p. 385. [Google Scholar]
- Yamauchi M, Kawai S, Kato T, Ooshima T, Amano A. Odd-skipped related 1 gene expression is regulated by Runx2 and Ikzf1 transcription factors. Gene. 2008;426:81–90. doi: 10.1016/j.gene.2008.08.015. [DOI] [PubMed] [Google Scholar]
- Yun CH, Choi SC, Park E, Kim SJ, Chung AS, Lee HK, Lee HJ, Han JK. Negative regulation of Activin/Nodal signaling by SRF during Xenopus gastrulation. Development. 2007;134:769–777. doi: 10.1242/dev.02778. [DOI] [PubMed] [Google Scholar]
- Zhang J, Talbot WS, Schier AF. Positional cloning identifies zebrafish one-eyed pinhead as a permissive EGF-related ligand required during gastrulation. Cell. 1998;92:241–251. doi: 10.1016/s0092-8674(00)80918-6. [DOI] [PubMed] [Google Scholar]
- Zhao J, Lambert G, Meijer AH, Rosa FM. The transcription factor Vox represses endoderm development by interacting with Casanova and Pou2. Development. 2013;140:1090–1099. doi: 10.1242/dev.082008. [DOI] [PubMed] [Google Scholar]