Abstract
The Celera Diagnostics ViroSeq HIV-1 Genotyping System is a Food and Drug Administration-cleared, integrated system for sequence-based analysis of drug resistance mutations in subtype B human immunodeficiency virus type 1 (HIV-1) protease and reverse transcriptase (RT). We evaluated the performance of this system for the analysis of diverse HIV-1 strains. Plasma samples were obtained from 126 individuals from Uganda, Cameroon, South Africa, Argentina, Brazil, and Thailand with viral loads ranging from 2.92 to >6.0 log10 copies/ml. HIV-1 genotyping was performed with the ViroSeq system. HIV-1 subtyping was performed by using phylogenetic methods. PCR products suitable for sequencing were obtained for 125 (99%) of the 126 samples. Genotypes including protease (amino acids 1 to 99) and RT (amino acids 1 to 321) were obtained for 124 (98%) of the samples. Full bidirectional sequence data were obtained for 95 of those samples. The sequences were categorized into the following subtypes: A1/A2 (16 samples), B (12 samples), C (13 samples), D (11 samples), CRF01_AE (9 samples), F/F2 (9 samples), G (7 samples), CRF02_AG (32 samples), H (1 sample), and intersubtype recombinant (14 samples). The performances of the individual sequencing primers were examined. Genotyping of duplicate samples in a second laboratory was successful for 124 of the 126 samples. The identity level for the sequence data from two laboratories ranged from 98 to 100% (median, 99.8%). The ViroSeq system performs well for the analysis of plasma samples with diverse non-B subtypes. The availability of this genotyping system should facilitate studies of HIV-1 drug resistance in non-subtype B strains of HIV-1.
Most antiretroviral drugs approved by the U.S. Food and Drug Administration (FDA) for the treatment of human immunodeficiency virus type 1 (HIV-1) infection target the HIV-1 protease and reverse transcriptase (RT) enzymes. Unfortunately, the efficacy of these drugs is often limited by HIV-1 drug resistance, which is usually caused by mutations in the protease and RT enzymes. The U.S. Department of Health and Human Services (DHHS) and other international agencies recommend drug resistance testing for the management of patients with HIV-1 infection (9, 11; http://www.aidsinfo.nih.gov/guidelines). Sequence-based HIV-1 genotyping assays that detect drug resistance mutations in HIV-1 protease and RT are the most widely used assays for drug resistance testing.
HIV-1 strains are genetically diverse. Nine phylogenetically pure subtypes (A, B, C, D, F, G, H, J, and K) and 15 circulating recombinant forms (CRFs) are currently recognized for HIV-1 group M (HIV-1 nomenclature proposal, Los Alamos HIV Sequence Data Base [http://hiv-web.lanl.gov]). HIV-1 drug resistance mutations and their effects on drug susceptibility have been studied predominantly in patients infected with subtype B strains. Although HIV-1 subtype B causes the majority of infections in the United States and Europe, other subtypes and CRFs cause the overwhelming majority of HIV-1 infections worldwide. Analysis of drug resistance in non-subtype B HIV-1 is becoming increasingly important given the expanding availability of antiretroviral drugs throughout the world and the increasing prevalence of non-subtype B HIV-1 in regions where antiretroviral drugs are widely used (19). HIV-1 subtype can influence viral susceptibility to antiretroviral drugs (2, 6), as well as the pattern of resistance mutations that emerge after drug exposure (4, 15). HIV-1 subtype may also influence the rate at which specific resistance mutations emerge after drug exposure (7, 8, 20) and the amino acids selected at specific positions under drug pressure (14, 21). Novel, subtype-specific drug resistance mutations may also emerge in non-subtype B strains at positions not associated with drug resistance in subtype B (1). Further studies are needed to examine the emergence and fading of drug resistance mutations in individuals with non-subtype B HIV-1 and to examine genotypic correlates of drug resistance in different HIV-1 subtypes.
The ViroSeq HIV-1 Genotyping System (Celera Diagnostics, Alameda, Calif.) is an integrated system for genotyping HIV-1 protease and RT that is cleared by the FDA for clinical use. This system utilizes 9 or 10 DNA primers for analysis: 1 for reverse transcription, 2 for PCR amplification, and 6 or 7 for sequence analysis. The performance of the ViroSeq system was intensively evaluated and validated for the genotyping of HIV-1 subtype B. Subtype-specific sequence differences at the sites of primer binding may complicate the genotyping of non-subtype B HIV-1. In a previous report, a high level of performance of the ViroSeq system for analysis of 130 plasma samples with HIV-1 non-subtype B strains from Uganda (mostly subtypes A and D) was demonstrated (16). Two recent reports further evaluated the performance of the ViroSeq system for analysis of non-subtype B HIV-1 (10, 12). However, those studies used predominantly cultured isolates rather than plasma and included relatively small sample sets. In this report, we evaluated the performance of the ViroSeq system for analysis of genetically diverse strains of HIV-1 by using plasma samples obtained from 126 HIV-1-infected individuals from Brazil, Cameroon, Uganda, Thailand, Argentina, and South Africa. This analysis focused on the ability of the ViroSeq system to successfully amplify DNA for sequencing and provide complete DNA sequences of the regions of interest. The analysis included an interlaboratory comparison of assay performance.
MATERIALS AND METHODS
Specimen panel.
HIV-1-infected plasma samples were collected from 126 individual asymptomatic blood donors between 1993 and 2001 in Cameroon (61 donors), Brazil (21 donors), Uganda (17 donors), South Africa (15 donors), Thailand (9 donors), and Argentina (3 donors). Samples were collected in the presence of citrate-based anticoagulant by L. Zekeng and L. Kaptué in Cameroon; by Peter Kataaha, Nakasero Blood Bank, Kampala, Uganda; by Brooks Jackson, Department of Pathology, Johns Hopkins Medical Institutions, Baltimore, Md.; by Amilcar Tanuri, Departamento de Genetica, Instituto de Biologia, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil; by Roberto Badaro, Fundação Bahiana de Infectologia, Bahia, Brazil; by Carlos Brites, Universidade Federal da Bahia, Hospital Universitário Prof. Edgard Santos, Bahia, Brazil; by Sasitorn Bejrachandra, Sirirah Hospital, Bangkok, Thailand; and by WP Blood Transfusion Service, Cape Town, South Africa. All specimens were unlinked and collected per local regulations in the country of origin at the time of collection. This panel of specimens was likely derived from subjects who were antiretroviral-drug naive. Samples were obtained from prospective blood donors, who were anticipated to be unaware of their HIV-1 infection status, in settings where antiretroviral drugs were not widely available. All specimens tested positive for antibodies to HIV by at least one commercially available HIV-1 antibody enzyme immunoassay.
Viral load determination.
The LCx HIV RNA Quantitative Assay (Abbott Laboratories, Abbott Park, Ill.) (not licensed for use in the United States) was performed according to the manufacturer's specifications by using a sample volume of 1.0 ml. This competitive RT PCR assay targets the pol integrase region of HIV-1 (13). The upper and lower limits of quantification for the 1.0-ml sample preparation volume are 1,000,000 (6.0 log10) and 50 (1.7 log10) HIV RNA copies/ml, respectively.
HIV-1 genotyping.
HIV-1 genotyping was performed by using the Celera Diagnostics ViroSeq HIV-1 Genotyping System (version 2.0) according to the manufacturer's instructions with 0.5 ml of plasma. Analysis with this system begins with HIV-1 RNA isolation and involves high-speed centrifugation of plasma to pellet virus particles, lysis of virus particles with a chaotropic agent to release viral RNA, and isopropanol-ethanol precipitation for RNA recovery. Ten microliters of the resuspended RNA was used for the reverse transcription (65°C for 30 s, 42°C for 65 min, 99°C for 5 min) with Moloney murine leukemia virus RT followed by a single 40-cycle PCR (50°C for 10 min, 93°C for 12 min, 93°C for 20 s, 64°C for 45 s, 66°C for 3 min, 72°C for 10 min) with AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, Calif.). The PCR yielded a 1.8-kb DNA product. PCRs were performed with the Amperase uracil N-glycosylase contamination control system to reduce the risk of contaminating PCRs with products from previous amplification reactions. PCR products were purified by using spin columns and analyzed by agarose gel electrophoresis prior to sequencing. DNA sequencing was performed by using premixed BigDye terminator (Applied Biosystems) sequencing reagents with seven different primers. Conditions for cycle sequencing (25 cycles) were 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. BigDye terminator chemistry provides 99.91% accuracy at 580 bases for the ABI PRISM 3100 Genetic Analyzer (N. Marlowe, unpublished observations). Sequence data were analyzed by using Celera Diagnostics ViroSeq HIV-1 Genotyping System software (version 2.5), which assembles sequence data from the primers into a contiguous sequence that can be inspected for the identification of drug resistance mutations. Processed sequences include the region coding for protease amino acids 1 to 99 and RT amino acids 1 to 335 (full-length sequences). Corresponding genomic positions within the HXB2 reference isolate are nucleotides 2253 to 2549 for protease and 2550 to 3554 for RT. Bidirectional sequence data were obtained for protease and for RT amino acids 1 to 321. Operators at both sites (Johns Hopkins University and Celera Diagnostics) were trained and certified by the manufacturer and were proficient in running the ViroSeq assay. Each laboratory was blinded to the results obtained by the other laboratory for assay comparison.
Phylogenetic analysis.
HIV-1 sequences trimmed to 1,260 nucleotides (nt), corresponding to protease amino acids 1 to 99 and RT amino acids 1 to 321 (297 and 963 nt, respectively), were used to determine subtypes. Nucleotide alignments were obtained with the biosequence editor Seqapp v1.9a169 (obtained from http://iubio.bio.indiana.edu/soft/molbio/seqapp/). Alignments included 47 reference sequences from the Los Alamos National Laboratory (http://hiv-web.lanl.gov) recommended for HIV-1 subtyping, as well as 4 additional reference sequences, for a total of 51 sequences: A1-U455 (M62320), A1-SE7253 (AF069670), A1-92UG037 (U51190), A1-Q23-17 (AF004885), A2-94CY017-41 (AF286237), A2-97CDKTB48 (AF286238), B-HXB2 (K03455), B-JRFL (U63632), B-WEAU160 (U21135), B-RF (M17451), C-ETH2220 (U46016), C-96BW0502 (AF110967), C-95IN21068 (AF067155), C-92BR025 (U52953), D-ELI (K03454), D-NDK (M27323), D-84ZR085 (U88822), D-94UG114 (U88824), F1-VI850 (AF077336), F1-FIN9363 (AF075703), F1-93BR020 (AF005494), F2-MP255 (AJ249236), F2-MP257 (AJ249237), G-HH8793-12.1 (AF061641), G-SE6165 (AF061642), H-VI997 (AF190128), H-90CF056 (AF005496), H-VI991 (AF190127), J-SE91733 (AF082395), J-SE9280 (AF082394), K-EQTB11C (AJ249235), K-MP535 (AJ249239), CRF01_AE-93TH253 (U51189), CRF01_AE-CM240 (U54771), CRF01_AE-90CF402 (U51188), CRF02_AG-DJ263 (AF063223), CRF02_AG-DJ264 (AF063224), CRF02_AG-IBNG (L39106), CRF03_AB-KAL153-2 (AF193276), CRF03_AB-RU98001 (AF193277), CRF04_cpx-97PVCH (AF119820), CRF04_cpx-97PVMY (AF119819), CRF04_cpx-CY032 (AF049337), CRF05_DF-VI1310 (AF193253), CRF05_DF-VI961 (AF076998), CRF12_BF-A32989 (AF408630), CRF12_BF-ARMA159 (AF385936), N-YBF30 (AJ006022), SIV-CPZGAB (X52154), O-ANT70 (L20587), and O-MVP5180 (L20571). Alignment and gap stripping were performed manually. Distances between the sequences were calculated with DNADist by using the Kimura two-parameter model as an optimal substitution model with a transition-to-transversion ratio of 1.5 (PHYLIP, version 3.572, obtained from http://evolution.genetics.washington.edu/phylip.html). Neighbor-joining and consense were used to create phylogenetic trees with 500 bootstrap replications (SeqBoot). Consensus trees were displayed with TreeView. Bootstrap values of >80 were considered acceptable for subtype assignment.
Sequencing primer mismatch analysis.
Nucleotide sequences generated by the ViroSeq HIV-1 genotyping system software (version 2.5) were aligned with ViroSeq sequencing primers B, C, F, G, and H by the Clustal W method (MegAlign module, Lasergene version 5.0.1; DNASTAR Inc., Madison, Wis.) to identify nucleotide mismatches between oligonucleotide primer and target sequences. Since primers A and D are not encompassed within the processed sequence, raw sequence files generated from primer F were aligned with sequencing primers A and D.
Nucleotide sequence accession number.
GenBank accession numbers for nucleotide sequences of protease and RT are AY444180 to AY444303.
RESULTS
Analysis of HIV-1 viral load.
Plasma samples were obtained from 126 HIV-1-infected individuals from Cameroon, Brazil, Uganda, South Africa, Thailand, and Argentina. The viral loads determined by the Abbott LCx HIV assay ranged from 2.92 to more than 6 log10 RNA copies/ml, the upper limit of quantification of the assay (Table 1). The mean viral load, excluding one sample whose viral load was above the upper limit of quantification, was 4.24 log10 RNA copies/ml.
TABLE 1.
Samples used for analysis
| Country | No. of samples | Mean viral load (log10 copies/ml)a |
|---|---|---|
| Cameroon | 61 | 4.17 (2.92-5.41) |
| Brazil | 21 | 4.23 (3.04-5.18) |
| Uganda | 17 | 4.46b (3.67-6.0) |
| South Africa | 15 | 4.14 (3.10-5.62) |
| Thailand | 9 | 4.52 (3.66-5.72) |
| Argentina | 3 | 4.20 (3.69-5.15) |
| Total | 126 | 4.24b (2.92-6.0) |
Values in parentheses are ranges.
Mean excludes one sample with a viral load greater than the upper limit of quantification (>6.0 log10 copies/ml).
ViroSeq assay performance: amplification.
Analysis in the ViroSeq assay begins with RNA extraction, reverse transcription, and PCR amplification. PCR products are analyzed with agarose gel electrophoresis by visual comparison of the amount of the PCR product to a standardized size marker provided with the ViroSeq kit. Amplification was considered successful if the PCR yielded sufficient DNA for sequence analysis (assessed according to the ViroSeq product insert guidelines). To evaluate interlaboratory assay variability, the same set of samples was analyzed by investigators at Johns Hopkins University and at Celera Diagnostics. Amplification was successful for 125 (99%) of the 126 samples at both laboratories. One sample from Cameroon with a viral load of 4.69 log10 copies/ml failed to amplify at both laboratories, even when RNA was reextracted from plasma. Based on analysis of gag p24, pol integrase, and env gp41, this sample contains a subtype D strain (J. Hackett and P. Swanson, unpublished observation).
ViroSeq assay performance: sequencing.
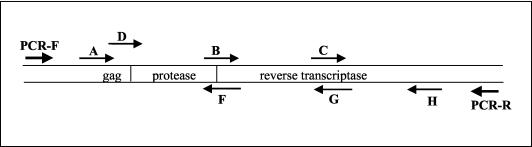
The locations and positions of sequencing primers in the ViroSeq system are shown in Fig. 1. Primers A, B, and C sequence the sense strand of the PCR product, and primers F, G, and H sequence the antisense strand of the PCR product. An analysis of the performance of individual sequencing primers was performed at Johns Hopkins University (Table 2). Full-length sequences were obtained for 124 (99%) of the 125 samples sequenced. For 95 (76%) of those samples, bidirectional sequencing data were obtained along the entire region analyzed. Sequencing was successful on the first attempt for 671 (89%) of 750 reactions with these six primers and was successful for 41 (52%) of the 79 reactions that were repeated after an initial primer failure. Sequencing was successful for 108 (86%) of 125 primer A reactions. The success rate for primer A was the lowest success rate among the six primers. Since primer A binds to the highly heterogeneous gag region of the HIV-1 genome, primer D may be used as an alternate sense primer if sequencing is unsuccessful with primer A. Sequencing with primer D was successful for 6 (30%) of 20 samples that failed to sequence with primer A; for these samples, only unidirectional sequence data were obtained for the region encoding the first 17 to 18 amino acids of protease. Sequencing was successful with either of the two alternate primers (primer A or primer D) for 114 (91%) of the samples tested and for 100, 99, 92, 98, and 94% of reactions with primers B, C, F, G, and H, respectively (Table 2). One sample failed sequencing with primers A, D, F, and G and was considered a sequencing failure. This sample also failed sequencing at Celera Diagnostics. To confirm the absence of sample cross-contamination, sequences from each sample were trimmed to 1,260 nt, aligned, and compared phylogenetically. No evidence of sample cross-contamination was observed for either site.
FIG. 1.
PCR and sequencing primers. The orientations and positions of the PCR primers (PCR-F and PCR-R) and the sequencing primers (A, B, C, D, F, G, and H) used in the ViroSeq system are shown with respect to the protease and RT coding regions.
TABLE 2.
Performance of sequencing primers by subtypea
| Subtype or CRF | n | No. (%) of successful reactions for primerb
|
No. (%) of samples with full BDSc | Primer failure(s) (single-stranded regions)d | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | D | A or D | B | C | F | G | H | ||||
| A/A2 | 16 | 14 (88) | 0/2 | 14 (88) | 16 (100) | 16 (100) | 14 (88) | 16 (100) | 15 (94) | 11 (69) | A/D (P1-R40, P1-R43), F (P1-P58, P1-P51), H (R129-R335) |
| B | 12 | 10 (83) | 2/2 | 12 (100) | 12 (100) | 12 (100) | 12 (100) | 12 (100) | 12 (100) | 12 (100) | |
| C | 13 | 12 (92) | 0/1 | 12 (92) | 13 (100) | 13 (100) | 12 (92) | 11 (85) | 13 (100) | 9 (69) | A/D (P1-R44), F (P1-P53), G (R4-R125, R4-R128) |
| D | 11 | 10 (91) | 1/1 | 11 (100) | 11 (100) | 11 (100) | 10 (91) | 11 (100) | 11 (100) | 10 (91) | F (P1-P53) |
| CRF01_AE | 9 | 5 (56) | 1/4 | 6 (67) | 9 (100) | 9 (100) | 9 (100) | 9 (100) | 9 (100) | 6 (67) | A/D (P1-R44, P1-R45, P1-R44) |
| F/F2 | 9 | 9 (100) | NT | 9 (100) | 9 (100) | 9 (100) | 8 (89) | 9 (100) | 8 (89) | 7 (78) | F (P1-P40), H (R126-R335) |
| G | 7 | 5 (71) | 0/2 | 5 (71) | 7 (100) | 7 (100) | 7 (100) | 7 (100) | 7 (100) | 5 (71) | A/D (P1-R47, P1-R48) |
| CRF02_AG | 32 | 30 (94) | 1/2 | 31 (97) | 32 (100) | 31 (97) | 30 (94) | 32 (100) | 28 (88) | 24 (75) | A/D (P1-R41), C (R223-R306), F (P1-P40, P1-P53), H (R131-R335, R133-R335, R125-R335, R124-R335) |
| H | 1 | 1 (100) | NT | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | |
| Recombinant | 14 | 12 (86) | 1/2 | 13 (93) | 14 (100) | 14 (100) | 12 (86) | 14 (100) | 13 (93) | 10 (71) | A/D (P1-R49), F (P1-P55, P1-P40), A/H (P1-P16 + R125-R335) |
| Ind | 1 | 0 | 0/1 | 0 | 1 (100) | 1 (100) | 0 | 0 | 1 (100) | 0 | ADFG (sequencing failure) |
| Total | 125 | 108 (86) | 6/20 (30) | 114 (91) | 125 (100) | 124 (99) | 115 (92) | 122 (98) | 118 (94) | 95 (76) | |
The number of samples of each subtype or CRF is shown (n). Subtype and CRF designations are based on the phylogeny of a 1,260-nt pol fragment generated by the ViroSeq system. Ind, indeterminate due to sequence failure.
Data for primer D are provided only for samples that could not be sequenced with primer A (number of successful reactions/number of reactions performed with primer D). The number of samples successfully sequenced with either of the alternate primers (A or D) is also shown. NT, not tested.
BDS, bidirectional sequences.
For each group of samples (each subtype or CRF), details are provided for samples for which one or more sequencing primers failed. The primers that failed are listed for each sample, and the regions of each sequence for which bidirectional sequence data were not obtained (single-stranded regions) are detailed in parentheses. Amino acid positions are indicated by the letters P for protease and R for RT.
Phylogenetic characterization of protease and RT.
The 124 sequences that were successfully obtained in both laboratories were first analyzed for evidence of intersubtype recombination using the Recombinant Identification program (http://hiv-web.lanl.gov) (18). Fourteen samples showed evidence of intersubtype recombination in the region analyzed. Independent phylogenetic analysis of the protease and RT coding regions was performed for the 14 putative recombinant strains. Based on this analysis, these strains included five F/B (two samples from Brazil and three samples from Argentina), and two indeterminate (Ind)/F Brazilian samples. The panel also included seven samples from Cameroon harboring recombinant strains: CRF02_AG/F (one sample), CRF02_AG/F2 (two samples), CRF02_AG/Ind (one sample), A/CRF02_AG (one sample), and Ind/Ind (two samples). Subtype assignments for the remaining 110 samples were A/A2 (16 samples), B (12 samples), C (13 samples), D (11 samples), CRF01_AE (9 samples), F/F2 (9 samples), G (7 samples), CRF02_AG (32 samples), and H (1 sample) (Table 2).
Performance of sequencing primers for different subtypes and intersubtype recombinant forms.
The performance of the ViroSeq sequencing primers at Johns Hopkins University was analyzed for samples of each subtype or CRF (Table 2). The majority of samples of all subtype categories yielded full bidirectional sequence data with no primer failures. All six primers (primers A or D, B, C, F, G, and H) were successful for over 90% of samples in all subtype categories with the following exceptions: two primer A/D failures and two primer F failures among 16 subtype A/A2 samples, two primer G failures among 13 subtype C samples, three primer A/D failures among 9 CRF01_AE samples, one primer F and one primer H failure among 9 subtype F/F2 samples, four primer H failures among 32 CRF02_AG samples, and two primer F failures among 14 intersubtype recombinant samples.
To examine nucleotide conservation at the sequencing primer binding sites, ViroSeq-generated sequences were aligned with the oligonucleotide primer sequences. The mean number and range of mismatches for the primary sequencing primers (primers A, B, C, F, G, and H) were tabulated for each subtype and CRF (Table 3). Overall, primer sites were well conserved. In most cases, the mean number of mismatches was 1 or less. The mean and range of mismatches were slightly higher for primer A with subtype F and CRF02_AG, primer B with CRF01_AE, and primer G with subtype C. The least overall conservation was observed for primer H. This sequencing primer had a mean of one mismatch or fewer with subtypes A, B, D, and H but had means of 1.1 to 2.6 mismatches with subtypes C, F, and G and with CRF01_AE, CRF02_AG, and other intersubtype recombinant strains. For primer H, the mean number of mismatches was highest (2.6) for CRF02_AG strains, ranging from 0 to 5.
TABLE 3.
Nucleotide mismatches at sequencing primer binding sitesa
| Primer | Mean no. (range) of mismatches for HIV-1 subtype or CRF
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A (n = 16) | B (n = 12) | C (n = 13) | D (n = 11) | F (n = 9) | G (n = 7) | H (n = 1) | CRF01_AE (n = 9) | CRF02_AG (n = 32) | Mosaic (n = 14) | |
| A | 0.1 (0-1) | 0.4 (0-1) | 0.4 (0-1) | 0 | 1.2 (0-5) | 1 (0-2) | 1 (1) | 0.2 (0.1) | 1.2 (0-6) | 1 (0-2) |
| B | 0.6 (0-2) | 0.2 (0-2) | 0.4 (0-2) | 0.4 (0-3) | 0.2 (0-2) | 0.7 (0-2) | 0 | 1.1 (1-2) | 0.2 (0-2) | 0.1 (0-1) |
| C | 0.8 (0-2) | 0.5 (0-1) | 0.2 (0-1) | 0.7 (0-2) | 0.4 (0-2) | 1 (1) | 0 | 0.8 (0-2) | 1.1 (1-2) | 0.6 (0-2) |
| F | 0.2 (0-2) | 0.3 (0-2) | 0.1 (0-1) | 0.8 (0-2) | 0.1 (0-1) | 0.1 (0-1) | 0 | 1 (1) | 0.1 (0-3) | 0.5 (0.1) |
| G | 0.3 (0-3) | 0.5 (0-1) | 1.6 (0-4) | 0.6 (0-2) | 0.8 (0-2) | 0.1 (0-1) | 0 (1) | 1 (1) | 0.2 (0-2) | 0.6 (0-2) |
| H | 0.6 (0-2) | 0.2 (0-1) | 2.3 (1-4) | 0.9 (0-2) | 1.2 (0-3) | 1.9 (0-3) | 0 | 2.1 (1-3) | 2.6 (0-5) | 1.1 (0-4) |
One hundred twenty-four sequences were aligned for mismatch analysis of primers B, C, F, G, and H; 115 primer F-derived sequences were analyzed for nucleotide mismatches at the primer A binding site. n, number of samples.
ViroSeq assay performance: assay reproducibility.
To evaluate interlaboratory assay variability, we compared sequencing results obtained at Johns Hopkins University with those obtained at Celera Diagnostics. Paired sequencing results were obtained for 124 of the 126 samples. The two remaining samples included one sample that failed amplification in both laboratories and one sample that failed sequencing at both laboratories. Phylogenetic analysis confirmed that the 124 paired sequences clustered together with a high degree of confidence.
The nucleotide sequence identity levels of the 124 sequence pairs ranged from 98 to 100%, with a median identity of 99.8%. Identity was >99% for 118 (95%) of the 124 paired sequences. The nucleotides identified (base calls) differed at 488 (0.3%) of the 156,364 nucleotide positions analyzed. Of these base-calling discrepancies, 479 represented the identification of a nucleotide mixture in one laboratory and the identification of one of the two corresponding nucleotides in the other laboratory (e.g., A+G = R in one laboratory and A in the other laboratory). Five of the 460 different base calls influenced the amino acids detected at positions associated with antiretroviral drug resistance: L10L/V (two base calls), L63L/P (two base calls), and Y181Y/C (one base call). Review of electropherogram data from both laboratories for these positions revealed the following: (i) both of the L10L/V mixtures were clearly detected in data from one laboratory but were not detected in data from the other laboratory, (ii) both of the L63L/P mixtures were clearly detected in data from one laboratory and were suggested by data from the second laboratory (background signal interfered with the identification of the mixtures), and (iii) the Y181C substitution was detected in one laboratory in a region where sequences from three primers overlapped. The mixture was seen in only one direction but met the criteria for mixture identification described in the ViroSeq system software manual; it was not detected in the second laboratory. Clear differences in the sequences (e.g., A in one laboratory and G in the other) were detected at only 9 (<0.01%) of the 156,364 nucleotide positions analyzed. None of these differences were at positions associated with antiretroviral drug resistance. A retrospective review of electropherograms revealed that all nine of the differences reflected the misidentification of bases at one laboratory. In six cases, misidentification may have occurred due to mobility shifts that were missed by the operator during manual inspection of the automated calls made by ViroSeq software; four of these nucleotides were in a single sequence encoding three amino acids in RT. In two cases, there was high background signal in the region.
Detection of amino acid changes at positions associated with antiretroviral drug resistance.
With the ViroSeq software, a contiguous consensus sequence derived for each sample is compared to a reference sequence, HXB-2 (GenBank accession number K03455), and differences from the reference sequence are tabulated. Non-subtype B sequences often differ from subtype B reference sequences at several positions, reflecting naturally occurring amino acid polymorphisms. The following amino acid changes were detected (the percentage of sequences with each amino acid change is shown in parentheses): in protease, M36I (90%), L63P (16%), K20R (14%), V77I (10%), L10V (9%), L10I (4%), A71T (2.4%), and M46I (one sample), and in RT, S68G (1.6%) and V118I (1.6%), plus the substitutions E44D, D67N, K70R, Y181C, T215F, K219Q, and G333E, each detected in a single sample. Four of these substitutions (D67N, K70R, T215F, and K219Q) are associated with resistance to zidovudine and were detected in a single individual from Brazil with subtype B HIV-1. The Y181C substitution is associated with high-level resistance to nevirapine; this amino acid substitution was detected in a single Cameroonian sample infected with a CRF02_AG strain.
DISCUSSION
Our previous studies using an earlier version of the ViroSeq system demonstrated excellent performance for the analysis of subtype B plasma samples (5) as well as non-subtype B plasma samples from Uganda (primarily subtypes A and D) (16). In the present report, we demonstrated that the FDA-cleared ViroSeq system (version 2.0) performs well for the analysis of genetically diverse non-subtype B and intersubtype recombinant strains. In the present study, genotypes were successfully obtained for 124 (98%) of 126 samples infected with genetically diverse HIV-1. The ViroSeq system has seven sequencing primers, which provide overlapping, bidirectional sequence data over the entire region analyzed. Bidirectional sequence data are useful for resolving sequence ambiguities and for confirming the presence of nucleotide mixtures. Nucleotide mixtures detected in genotyping assays reflect the presence of genetically diverse viral variants present in plasma samples from most HIV-1-infected individuals. The ViroSeq system is able to accurately detect specific drug resistance mutations representing 40% or more of the viral population in HIV-1 subtype B plasma samples (ViroSeq product insert). However, the detection of minority variants as mixtures may provide additional information about the presence of drug-resistant variants.
The most consistent difference in performance observed in the present study compared to previously reported results for subtype B strains (5) was in the performance of the A and D alternate primers, which bind to the highly heterogeneous gag region. For subtype B strains, sequencing was successful with the A and D primers for 92 and 88% of reactions, respectively, and at least one of the primers (either primer A or primer D) was successful with 98.4% of the samples. In contrast, for the genetically diverse strains in this study, the A and D primers were successful for 86 and 30% of the reactions, respectively. Both primers (A and D) failed for 9% of the genetically diverse samples, resulting in some regions of unidirectional sequence data.
The genetic heterogeneity of HIV-1 viruses presents a challenge for molecular assays that depend on oligonucleotide primer hybridization. Nucleotide mismatches between the primer(s) and template have the potential to reduce the efficiency of hybridization, leading to inefficient amplification or failed sequencing runs. The genetically diverse panel of samples used in the present study provided an opportunity to examine the level of nucleotide conservation within primer sites used for sequence analysis across multiple subtypes and CRFs. Consistent with the performance (>95% success rate) of the primary set of ViroSeq sequencing primers (primers A to C and F to H), a relatively high degree of nucleotide conservation was observed at the primer binding sites (Table 3). One or no mismatches were present in primer A, B, C, F, and G sites in 90% or more of the samples. For primer A, B, C, F, and G sites, two or fewer mismatches were present in 97, 99, 100, 99, and 99% of the panel members, respectively. The primer H site was less conserved, with two or fewer mismatches for 77% of this genetically diverse sample panel. Interestingly, although the level of conservation was somewhat lower in primer H, it accounted for only 20% of the failed sequencing reactions in our study. Primers A and F accounted for, respectively, 46 and 26% of the failed sequencing reactions, although their overall nucleotide conservation was higher than that for the primer H binding site. The overall level of sequence conservation within the target binding site of a primer is just one of many factors that can influence the efficiency of primer hybridization. The position and nature of the mismatch, primer length, and stringency (temperature and buffer conditions) are additional factors that contribute to mismatch tolerance. For primer F, all eight sequencing failures (reproducible at both laboratories) could be attributed to a mismatch at the 3′ terminus of the primer. The samples with primer failures included a subset (6 to 11%) of subtype A, C, D, and F and CRF02_AG specimens. In all cases, primer A provided sequence coverage on the opposite strand. Two primer G sequencing failures, both due to 3′-terminal mismatches on subtype C strains, were observed. Thus, the position of the mismatch was critical for all sequencing failures with primers F and G. Sequencing failures with primers A, C, and H were generally associated with internal mismatches near the 3′ ends of the primers or due to a higher total numbers of mismatches (three to five mismatches for primer H).
Overall, the sequencing success rates for the primers were highly concordant between laboratories. However, in five of six cases (primer F, one subtype A specimen; primer A, one subtype A, one subtype D, and two CRF01_AE specimens) in which sequencing failures were observed at Johns Hopkins University (Table 2), no mismatches were present in the primer binding sites. In the sixth case, only a single mismatch, near the 5′ end of primer A, was identified. Thus, no obvious molecular basis for failure was evident. For all six specimens, sequences were successfully generated in the second laboratory, suggesting that the failures may have resulted from technical issues.
Primer D is included in the ViroSeq kit as an accessory primer in the event of sequencing failure with primer A. Consistent with the relatively low success rate (30%) for non-subtype B strains, the primer D binding site is the least conserved of the seven ViroSeq sequencing primer binding sites. Nearly all cases of failure with primer D can be attributed to a relatively high number of total mismatches. Our data are consistent with a previous report of a low success rate for primer D with CRF01_AE strains (17). Nevertheless, primer D was successful in five cases in which primer A failed (one subtype B specimen, one subtype D specimen, two CRF02_AG specimens, and one CRF01_AE specimen) and thus is of value, even with non-subtype B strains.
The performance of the ViroSeq system (version 2.0) with non-subtype B HIV-1 was evaluated in three previous reports. Those reports also examined the performance of another commercial genotyping system, the TRUGENE HIV-1 genotyping kit (Bayer Diagnostics, Tarrytown, N.Y.), which is also cleared by the FDA for clinical use. In the first report (10), the performance of the ViroSeq system was evaluated with 15 cultured isolates of non-subtype B HIV-1 (3 subtype A, 2 subtype C, 3 subtype D, 2 subtype F, 1 subtype G, 1 subtype J, and 3 recombinant [1 A/D and 2 CRF02_A/G] isolates) and three non-subtype B plasma samples (one each of subtypes C, D, and H). All of the samples were successfully amplified and sequenced with the ViroSeq system (version 2.0). Full bidirectional sequence data were obtained for 11 (73%) of the 18 non-subtype B samples. Primer failures included both the A and D primers for three samples (subtypes F, J, and H), the F and G primers for one subtype A sample, the F primer for one subtype C sample, and the G primer for one subtype D and one CRF02_AG sample. Genotypes were also obtained for all of the non-B samples using the TRUGENE system and supplemental primers provided by the manufacturer. Samples analyzed with the TRUGENE system were noted to have more frequent sequence ambiguities.
In the second report (12), 34 cultured isolates, 27 of which were non-subtype B isolates (2 subtype A, 2 subtype A/G, 6 subtype C, 2 subtype D, 8 CRF01_AE, 4 subtype F, 2 subtype G, and 1 subtype H), were analyzed. Genotyping was successful for all of the non-subtype B samples with the ViroSeq system. Sequencing with primer A was successful for 24 (89%) of the non-subtype B samples, while sequencing with the alternate primer D was successful for only 8 (30%) of the samples. Both primers A and D failed for three isolates (of subtypes C, E, and H) and one sample for which primer H failed (a subtype C sample). Analysis with the TRUGENE system required the use of the version 1.5 primers for three (11%) of the 27 non-B samples (1 subtype C and 2 subtype G samples). Sequencing primer failures, primarily in the protease region, were described for 22 (81%) of the 27 non-subtype B samples. Samples with subtypes B and D performed better with the PR set of protease sequencing primers, whereas the P2 protease primer set was preferable for the other subtypes. The P2 primer set was successful for all samples tested but provides a shorter sequence in the protease region. Both sets of protease primers are provided with the FDA-cleared version of the TRUGENE system.
In the third report (3), genotyping was performed by using viral isolates from cell culture. Genotyping with the version 2.0 ViroSeq system was successful for 20 (80%) of 25 non-subtype B isolates. In contrast, genotyping with the TRUGENE system (version 1, without the use of supplemental primers) was successful for only 12 (50%) of 24 non-subtype B isolates. The five non-subtype B isolates that failed with the ViroSeq system included three subtype A isolates, one subtype C isolate, and one subtype J isolate. In all five cases, all seven sequencing primers failed. This type of result was not observed in the present study or in the other two studies cited. It seems unlikely that all five samples diverged genetically at the binding sites of all seven sequencing primers. Instead, these results suggest that a contaminant (e.g., from cell culture) might have interfered with the sequencing reactions. In contrast to the three previous studies described, the present study used only plasma samples for analysis.
The present report confirms the value of the ViroSeq system for the analysis of plasma samples from countries harboring diverse HIV-1 subtypes. In the present study, a high level of interlaboratory assay reproducibility was observed with this system. Further studies are needed to extend this analysis to include non-subtype B samples with low viral loads and non-subtype B samples from patients failing antiretroviral therapy. The FDA-cleared ViroSeq system offers an advantage for drug resistance monitoring in that a single set of PCR and sequencing primers can be used for the analysis of genetically diverse strains of HIV-1. The inclusion of the uracil N-glycosylase contamination control system reduces the likelihood of PCR carryover. The ViroSeq system provides a single, uninterrupted pol region sequence, and sequence data are provided in the standard FASTA format, simplifying phylogenetic analysis for subtype determination and other studies. Moreover, the ViroSeq system has been FDA-cleared for use on multiple platforms, including the high-throughput ABI Prism 3100 and 3700 DNA analyzers, making it an attractive tool for patient monitoring.
Acknowledgments
We thank Eric Shulse and the HIV Genotyping Team at Celera Diagnostics for providing reagents used in this study and Lutz Gürtler for careful review of the manuscript. ViroSeq reagents were provided by Celera Diagnostics.
This work was supported by the HIV Network for Prevention Trials (HIVNET), sponsored by the U.S. National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), DHHS, through contract N01-AI-35173 with Family Health International, contract N01-AI-45200 with Fred Hutchinson Cancer Research Center, and subcontract NOI-AI-35173-417 with Makerere Univ.; by the HIV Prevention Trials Network (HPTN), sponsored by the NIAID, National Institutes of Child Health and Human Development (NICH/HD), National Institute on Drug Abuse, National Institute of Mental Health, and Office of AIDS Research of the NIH, DHHS (contracts U01-AI-46745 and U01-AI-48054); by the Adult AIDS Clinical Trials Groups (NIH, Division of AIDS, NIAID); and by grant R01-HD-42965-01.
REFERENCES
- 1.Abecasis, A., P. Gomes, I. Derdelinckx, A. P. Carvalho, I. Diogo, F. Goncalves, J. Cabanas, M. C. Lobo, A. M. Vandamme, and R. Camacho. 2003. L89I/V: a novel mutation selected by protease inhibitor therapy in subtype G, but not in subtype B-infected patients. Antivir. Ther. 8:S140. [Google Scholar]
- 2.Apetrei, C., D. Descamps, G. Collin, I. Loussert-Ajaka, F. Damond, M. Duca, F. Simon, and F. Brun-Vezinet. 1998. Human immunodeficiency virus type 1 subtype F reverse transcriptase sequence and drug susceptibility. J. Virol. 72:3534-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beddows, S., S. Galpin, S. H. Kazmi, A. Ashraf, A. Johargy, A. J. Frater, N. White, R. Braganza, J. Clarke, M. McClure, and J. N. Weber. 2003. Performance of two commercially available sequence-based HIV-1 genotyping systems for the detection of drug resistance against HIV type 1 group M subtypes. J. Med. Virol. 70:337-342. [DOI] [PubMed] [Google Scholar]
- 4.Caride, E., K. Hertogs, B. Larder, P. Dehertogh, R. Brindeiro, E. Machado, C. A. de Sa, W. A. Eyer-Silva, F. S. Sion, L. F. Passioni, J. A. Menezes, A. R. Calazans, and A. Tanuri. 2001. Genotypic and phenotypic evidence of different drug-resistance mutation patterns between B and non-B subtype isolates of human immunodeficiency virus type 1 found in Brazilian patients failing HAART. Virus Genes 23:193-202. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham, S., B. Ank, D. Lewis, L. Wei, J. Dileanis, J. B. Jackson, P. Palumbo, P. Krogstad, and S. H. Eshleman. 2001. Performance of the Applied Biosystems ViroSeq HIV-1 Genotyping System for analysis of HIV-1 in pediatric plasma samples. J. Clin. Microbiol. 39:1254-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Descamps, D., C. Apetrei, G. Collin, F. Damond, F. Simon, and F. Brun-Vezinet. 1998. Naturally occurring decreased susceptibility of HIV-1 subtype G to protease inhibitors. AIDS 12:1109-1111. [PubMed] [Google Scholar]
- 7.Eshleman, S. H., G. Becker-Pergola, M. Deseyve, L. A. Guay, M. Mracna, T. Fleming, S. Cunningham, P. Musoke, F. Mmiro, and J. B. Jackson. 2001. Impact of HIV-1 subtype on women receiving single dose NVP prophylaxis to prevent HIV-1 vertical transmission (HIVNET 012). J. Infect. Dis. 184:914-917. [DOI] [PubMed] [Google Scholar]
- 8.Eshleman, S. H., L. A. Guay, A. Mwatha, T. Fleming, S. P. Cunningham, P. Musoke, F. Mmiro, and J. B. Jackson. 2003. Extended analysis of nevirapine (NVP) resistance in women with subtype A vs. D 6-8 weeks after single dose NVP prophylaxis: HIVNET 012. 10th Conf. Retroviruses Opportunistic Infect., abstr. 857, p. 370.
- 9.EuroGuidelines Group for HIV Resistance. 2001. Clinical and laboratory guidelines for the use of HIV-1 drug resistance testing as part of treatment management: recommendations for the European setting. AIDS 15:309-320. [DOI] [PubMed] [Google Scholar]
- 10.Fontaine, E., C. Riva, M. Peeters, J. C. Schmit, E. Delaporte, K. Van Laethem, K. Van Vaerenbergh, J. Snoeck, E. Van Wijngaerden, E. De Clercq, M. Van Ranst, and A. M. Vandamme. 2001. Evaluation of two commercial kits for the detection of genotypic drug resistance on a panel of HIV type 1 subtypes A through J. J. Acquir. Immune Defic. Syndr. 28:254-258. [DOI] [PubMed] [Google Scholar]
- 11.Hirsch, M. S., F. Brun-Vezinet, B. Clotet, B. Conway, D. R. Kuritzkes, R. T. D'Aquila, L. M. Demeter, S. M. Hammer, V. A. Johnson, C. Loveday, J. W. Mellors, D. M. Jacobsen, and D. D. Richman. 2003. Antiretroviral drug resistance testing in adults infected with human immunodeficiency virus type 1: 2003 recommendations of an International AIDS Society-USA panel. Clin. Infect. Dis. 37:113-128. [DOI] [PubMed] [Google Scholar]
- 12.Jagodzinski, L. L., J. D. Cooley, M. Weber, and N. L. Michael. 2003. Performance characteristics of human immunodeficiency virus type 1 (HIV-1) genotyping systems in sequence-based analysis of subtypes other than HIV-1 subtype B. J. Clin. Microbiol. 41:998-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johanson, J., K. Abravaya, W. Caminiti, D. Erickson, R. Flanders, G. Leckie, E. Marshal, C. Mullen, Y. Ohhashi, R. Perry, J. Ricci, J. Salituro, A. Smith, N. Tang, M. Vi, and J. Robinson. 2001. A new ultrasensitive assay for quantitation of HIV-1 RNA in plasma. J. Virol. Methods 95:81-92. [DOI] [PubMed] [Google Scholar]
- 14.Kantor, R., A. P. Carvalho, B. Wynhoven, M. A. Soares, P. Cane, J. Clarke, J. Snoeck, C. Pillay, S. Sirivichayakul, K. Ariyoshi, A. Hoguin, H. Rudich, R. Rodrigues, M. B. Bouzas, P. Cahn, L. F. Brigido, Z. Grossman, V. Soriano, W. Sugiura, P. Phanuphak, L. Morris, A. M. Vandamme, J. Weber, D. Pillay, A. Tanuri, P. R. Harrigan, R. Camacho, J. M. Schapiro, R. W. Shafer, and D. Katzenstein. 2003. Nucleic acid differences between HIV-1 non-B and B reverse transcriptase and protease sequences at drug resistance positions. Antivir. Ther. 8:S58. [Google Scholar]
- 15.Kantor, R., D. Katzenstein, R. Camacho, P. R. Harrigan, A. Tanuri, D. Pillay, A. M. Vandamme, P. Phanuphak, W. Sugiura, V. Soriano, L. Morris, A. Grossman, L. F. Brigido, J. M. Shapiro, and R. W. Shafer. 2003. Genotypic analyses of RT and protease sequences from persons infected with non-subtype B HIV-1. 10th Conf. Retroviruses Opportunistic Infect., abstr. 623, p. 281.
- 16.Mracna, M., G. Becker-Pergola, J. Dileanis, L. A. Guay, S. Cunningham, J. B. Jackson, and S. H. Eshleman. 2001. Performance of the Applied Biosystems ViroSeq HIV-1 Genotyping System for sequence-based analysis of non-subtype B HIV-1 from Uganda. J. Clin. Microbiol. 39:4323-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukaide, M., W. Sugiura, M. Matuda, S. Usuku, Y. Noguchi, K. Suzuki, K. Kawata, A. Ito, H. Sagara, K. Yamada, M. Kondo, and M. Imai. 2000. Evaluation of ViroseqTM-HIV version 2 for HIV drug resistance. Jpn. J. Infect. Dis. 53:203-205. [PubMed] [Google Scholar]
- 18.Siepel, A. C., A. L. Halpern, C. Macken, and B. T. Korber. 1995. A computer program designed to screen rapidly for HIV type 1 intersubtype recombinant sequences. AIDS Res. Hum. Retrovir. 11:1413-1416. [DOI] [PubMed] [Google Scholar]
- 19.Spira, S., M. A. Wainberg, H. Loemba, D. Turner, and B. G. Brenner. 2003. Impact of clade diversity on HIV-1 virulence, antiretroviral drug sensitivity and drug resistance. J. Antimicrob. Chemother. 51:229-240. [DOI] [PubMed] [Google Scholar]
- 20.Tanuri, A., M. A. Soares, A. T. Dumans, S. Hue, and D. Pillay. 2003. Synonymous genetic changes within subtype F may influence mutational routes to drug resistance. Antivir. Ther. 8:S142. [DOI] [PubMed] [Google Scholar]
- 21.Turner, D., B. Brenner, B. Spira, J. Schapiro, M. Songok, and M. A. Wainberg. 2003. Novel drug resistance profiles in non-B subtype HIV-1 infections. 10th Conf. Retroviruses Opportunistic Infect., abstr. 144, p. 109.