Abstract
From November 2000 to October 2001, a reverse transcription-PCR using primers directed to the norovirus RNA polymerase coding region was included in a viral and bacterial routine screening to diagnose sporadic cases of acute gastroenteritis among children in Asturias, Spain. The role of noroviruses (8.6% of the positively diagnosed cases) as the cause of sporadic pediatric gastroenteritis was evaluated with respect to the detection rates of other gastroenteritis-associated viruses and bacteria. The results indicated that noroviruses were less common than rotaviruses (36.9%), Campylobacter spp. (28.8%), and Salmonella spp. (18.4%) but more frequent than astroviruses (4.3%), adenoviruses (3.8%), and Yersinia spp. (2.2%). Mixed infections involving noroviruses were rarely observed (0.5%). The presence of a norovirus-associated pediatric gastroenteritis peak in summer, as well as the complete absence of norovirus-associated cases in colder months, challenges the view that norovirus infections exclusively have wintertime seasonality. On the other hand, phylogenetic analysis of the amplified fragments showed that the norovirus strains responsible were closely related. A further study using the full-length capsid region showed that these strains could be included into genogroup II, Bristol/Lorsdale cluster, and were closely related to the 1995 and 1996 U.S. subset of strains associated with outbreaks recorded worldwide between 1995 and 1996.
Gastroenteritis remains a major public health problem worldwide, especially among children. More than 700 million cases of acute gastroenteritis are estimated to occur annually in children less than 5 years old. The mortality associated with gastroenteritis has been estimated to be 3.5 to 5 million per year (14, 33).
Many different pathogens have been found in the stools of children with gastroenteritis. From them, bacteria, such as Salmonella spp., Shigella spp., Campylobacter spp., and Yersinia spp. among others and viruses, such as rotaviruses, adenoviruses, and astroviruses, have been clearly established as etiologic agents of gastroenteritis in children (8, 21, 28).
Recently, noroviruses (formerly known as “Norwalk-like viruses”) have been included as a common cause of outbreaks and sporadic cases of gastroenteritis worldwide in individuals of all ages. Their role as the major cause of viral gastroenteritis outbreaks has been recently reported (6, 18, 39). Nevertheless, few studies have been carried out to evaluate the relative contribution of noroviruses to pediatric sporadic gastroenteritis with respect to other classic gastroenteritis-associated viruses and bacteria.
The Norovirus genus within the family Caliciviridae includes single-stranded positive-sense RNA viruses. They have been divided into two distinct genogroups, GGI and GGII, and further subdivided into several clusters or genotypes based on genetic divergence in regions of the RNA-dependent RNA polymerase and the major viral capsid protein (VP60) (38). Thus, GGI includes Norwalk, Southampton, Chiba, and Desert Shield viruses, whereas GGII includes Hawaii, Snow Mountain Agent, Toronto, and Bristol viruses, among others. Recently, a genetic classification system for the noroviruses was proposed on the basis of the complete capsid protein sequence (3). According to this system, GGI and GGII include 7 and 10 clusters, respectively. Because of the high genetic diversity shown by this group of viruses, the molecular characterization of the genotypes is essential to understand the epidemiology of the strains causing pediatric gastroenteritis.
The aim of this study was to evaluate the relative contribution of noroviruses to sporadic cases of pediatric gastroenteritis over an extended time period and to investigate the genotype of the strains involved.
MATERIALS AND METHODS
Stool samples.
Between November 2000 and October 2001, a total of 363 children with clinical symptoms of gastroenteritis were seen in the Emergency Department of Hospital Universitario Central de Asturias. A patient with gastroenteritis was defined as a patient with one or more of the following symptoms: nausea, vomiting, and/or diarrhea. Stool specimens were collected during the acute phase of infection. All samples were obtained by direct deposition in a sterile container and were transported the same day to our laboratory, where they were stored at 4°C until they were processed. Specimens for bacteriological culture were inoculated into media on the day of collection. Specimens for virus were prepared as 20% homogenates in phosphate-buffered saline and stored at 4°C for up 1 week until they were tested. The study included only fecal specimens from sporadic cases. Although all the children were under 14 years old, the exact age of 54 of them was unknown. To analyze the effect of the age, the remaining 309 patients (mean age, 2.6 years; age range, 1 month to 14 years) were grouped into two categories on the basis of logical and comparable cutoff points: less than 5 years old (79.6% of all patients) and 5 to 14 years old (20.4%).
Laboratory diagnosis.
Fecal specimens were routinely screened for etiologic agents of diarrhea. Conventional bacterial culture procedures to isolate gastroenteritis-associated bacteria, such as Salmonella, Campylobacter, Shigella, Yersinia, Vibrio, Plesiomonas, and Aeromonas species, were carried out. On the other hand, commercial immunoassays IDEIA Rotavirus, IDEIA Adenovirus, and IDEIA Astrovirus (Dako Ltd., Ely, United Kingdom) were used according to manufacturer's instructions to detect specific antigens from group A rotaviruses, adenoviruses type 40 and 41, and astroviruses, respectively.
RNA extraction and RT-PCR.
Viral RNA from noroviruses was purified from 20% stool suspensions in phosphate-buffered saline by using the guanidine isothiocyanate method. Briefly, 200 μl of guanidine isothiocyanate was added to 50 μl of the stool suspension and the mixture incubated at 60°C for 10 min. Nucleic acids were precipitated by addition of 250 μl of isopropanol and further centrifugation at 14,000 rpm for 15 min. The pellet was washed with 1 ml of 75% ethanol, dried, resuspended in 50 μl of diethyl pyrocarbonate-treated water, and stored at −20°C. To detect noroviruses, reverse transcription (RT)-PCRs were carried out using the Titan One Tube RT-PCR System (Roche Corp., Indianapolis Ind.) and primers NV1 and NV2, which were deduced from the RNA-dependent RNA polymerase coding region of the Norwalk virus, the prototype strain of noroviruses (GI/1, according to Ando's nomenclature) (Table 1). Five microliters of extracted RNA was added to 20 μl of an RT-PCR mixture according to the manufacturer's instructions. Amplification was performed using a GeneAmp PCR system 9600 thermal cycler (Applied Biosystems, Foster City, Calif.) using the following conditions: retrotranscription at 48°C for 45 min; denaturation at 94°C for 2 min; 40 cycles at 94°C for 30 s, 55°C for 30 s, and 68°C for 30 s; and one final cycle at 68°C for 10 min. RT-PCR products were analyzed using 2% agarose-Tris-borate-EDTA gel electrophoresis. The primer pair used is expected to produce a 306-bp RT-PCR fragment.
TABLE 1.
In-house PCR primers used for RT-PCR and sequencing analysis
| Name | Sequence (5′-3′) | Sense | Position |
|---|---|---|---|
| NV1a | GAAGATGGCCCCCTCATCT | + | 4511-4529c |
| NV2a | GGCCTCAGACAGTGCACAGA | − | 4717-4816c |
| NV9b | ACCCAGAAAAATTGACAG | + | 4632-4649d |
| NV14b | ACCCATCAGATGGGTTGGC | − | 5106-5124d |
| NV12b | TGGTGAGGACTCGACAAG | − | 5680-5697d |
| NV10b | GCTTTGGCCAGCTTCAAG | − | 6207-6224d |
| NVdT5b | TGTTTGAGTGCAAGCTTC | + | 6538-6555d |
| NV6b | GCAAAGAAAGCTCCAGCCA | − | 6705-6723d |
| NVdT3a | (T)17AAAGACACTAAAG | − | 7543-7572d |
To amplify a 3,335-nt region from the 3′ end of the viral genome, viral RNA from RT-PCR positive samples was extracted using the QIAmp RNA extraction kit (Qiagen GmbH, Hilden, Germany). The RT-PCRs were performed using primers NV1 and NVdT3 (Table 1). The amplification was carried out using the following conditions: retrotranscription at 48°C for 45 min; denaturation at 94°C for 2 min; 30 cycles at 94°C for 1 min, 55°C for 1 min, and 68°C for 2 min; and one final cycle at 68°C for 10 min. The PCR products were analyzed by electrophoresis in 1% agarose-Tris-borate-EDTA.
To avoid cross-contamination in RT-PCR, procedures of extraction of viral RNA were carried out in a room physically separated from that for performing RT-PCRs. Furthermore, positive and negative controls were included in all PCR assays.
Cloning and sequence analysis.
The PCR products were excised from agarose gels and purified by adsorption onto silica beads using the DNA purification kit Ultraclean 15 (MO BIO Laboratories, Solana Beach, Calif.). Purified DNA fragments were cloned into pGEM-T (Promega Corp., Madison, Wis.) by using a protocol described by the manufacturer. Positive clones were identified by minipreparation of plasmid DNA followed by restriction enzyme analysis. The 306-bp DNA inserts were fully sequenced in both directions using T7 and SP6 primers. These primers, as well as those described in Table 1 were used to investigate the full nucleotide sequence of the larger 3,335-bp insert DNA. All sequencing reactions were performed by the chain-termination method on an automated sequencer (ABI PRISM 3100 Genetic Analyzer; Applied Biosystems). The translation products encoded by the cloned cDNAs were deduced using the DM5 program (24). The nucleotide or deduced amino acid sequences were aligned using Clustal W using the program default parameters (34). The phylogenetic trees were plotted using the program TreeView (version 1.6.6) (26).
Statistical analyses.
Chi-squared tests were performed using GraphPad InStat version 3.00 for Windows 95 (GraphPad Software).
Nucleotide sequence accession number.
The nucleotide sequence of the 3′ end of the Ast6139/2001/SP isolate viral genome has been deposited in GenBank under accession number AJ583672.
RESULTS
Etiology of sporadic cases of acute gastroenteritis.
Among the 363 stool specimens studied from children with gastroenteritis, 179 cases could not be related to a known pathogen (49.3%), whereas 184 stool samples (50.7%) contained at least one identifiable agent. As summarized in Table 2, individual enteropathogens were found in 170 of the samples (92.4% of the total positive identifications) by isolating a bacterium from 88 of the fecal samples (47.8%) or detecting a virus in 82 cases (44.5%). Dual infections were found in only 14 cases (7.6%). Of the 184 cases associated to one or two types of pathogens, group A rotaviruses were the most frequently detected (36.9% of the positive cases). The other enteric viruses found were noroviruses (8.6%), astroviruses (4.3%), and adenoviruses type 40 or 41 (3.8%). Among the bacteria, Campylobacter jejuni was the one most frequently isolated (28.8%), followed by Salmonella enterica serovar Enteritidis (18.4%), S. enterica serovar Typhimurium (4.3%), and Yersinia enterocolitica (2.2%) (Table 2). On the other hand, the main type of association detected in dual infections was virus-bacterium (4.9% of the total positive identifications) with rotaviruses and C. jejuni as the most common pathogens implicated (Table 2). Noroviruses were detected in a single dual infection (6.3% of all norovirus-associated cases), whereas astroviruses were present in three mixed infections (37.5% of all astrovirus-associated cases).
TABLE 2.
Enteropathogens identified associated to sporadic cases of gastroenteritis in Asturias, Spain
| Type of infectionand pathogen | No. of cases (%a) |
|---|---|
| Single | 170 (92.4) |
| Viruses | |
| All | 82 (44.5) |
| Rotavirus | 57 (31.0) |
| Norovirus | 15 (8.1) |
| Astrovirus | 5 (2.7) |
| Adenovirus | 5 (2.7) |
| Bacteria | |
| All | 88 (47.8) |
| C. jejuni | 45 (24.5) |
| S. enterica serovar Enteritidis | 31 (16.8) |
| S. enterica serovar Typhimurium | 8 (4.3) |
| Y. enterocolitica | 4 (2.2) |
| Dual | |
| All | 14 (7.6) |
| Rotavirus + C. jejuni | 5 (2.7) |
| Rotavirus + S. enterica serovar Enteritidis | 2 (1.1) |
| Rotavirus + adenovirus | 2 (1.1) |
| Rotavirus + norovirus | 1 (0.5) |
| Rotavirus + astrovirus | 1 (0.5) |
| C. jejuni + astrovirus | 2 (1.1) |
| C. jejuni + S. enterica serovar Enteritidis | 1 (0.5) |
Percentage with respect to the 184 cases with positive pathogen identification.
The seasonal distribution of the episodes of acute gastroenteritis per enteropathogen showed that diarrheal episodes caused by viruses were distributed throughout the year with one rotavirus-associated peak from March to June (58,8% of all rotavirus-associated cases) and, surprisingly, a norovirus-associated peak in June-July (50% of all norovirus-associated cases). Furthermore, no noroviruses were detected in winter months. No seasonal distribution was observed in the diarrheal episodes caused by bacteria.
The age of the individuals was one of the factors analyzed. In children less than 5 years old, viruses and bacteria were detected with a similar frequency (30.4 versus 25.2% of the patients in this category, respectively), while in children 5 to 14 years old bacteria caused more infections than viruses (34.9 versus 6.3%; P < 0.0001). On the other hand, viruses were more common among children under 5 years than in those 5 to 14 years old (30.4 versus 6.3%; P < 0.0001). In fact, all the viruses were clearly more frequently detected in this age group (94.6% of all rotavirus-, 92.3% of all norovirus-, and 100% of all astrovirus- and adenovirus-associated cases, respectively). Although, bacteria were isolated with similar frequencies among children under 5 years or from those 5 to 14 years old (25.2 versus 34.9%), S. enterica serovar Enteritidis was more frequently found in children 5 to 14 years old (20.6 versus 6.5%; P = 0.0062).
To investigate the effect of the fecal specimen consistency, stool samples were classed as liquid, semisolid, or solid (18.9, 61.6, and 19.5%, respectively). Considering the 363 specimens processed, a bacterium was isolated from 45.3% of the liquid, 24.1% of the semisolid, and 11.4% of the solid specimens (P = 0,001), while a virus was detected in 17.1% of the liquid, 27.9% of the semisolid, and 17.1% of the solid specimens, respectively. An analysis of the type of enteropathogen involved showed that S. enterica serovar Enteritidis was isolated from 28.1% of the liquid, 5.7% of the semisolid, and 1.4% of the solid specimens (P < 0.0001). No other correlation between consistency of the stool and the other pathogens was found. Dual infections were excluded from the analysis.
Molecular epidemiology of Noroviruses.
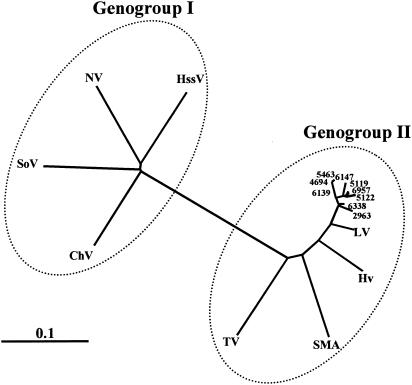
To investigate the variability of the strains detected over the 24-month period, the 306-bp RT-PCR products from 12 of the 16 isolates characterized in this study were purified and sequenced. The alignment of the amplicon nucleotide sequences revealed that four of the isolates were identical to the consensus sequence, whereas the remaining eight differed by only 2 to 8 bp (Fig. 1). In addition, a phylogenetic analysis of the RNA polymerase region from the nine different isolates described in this study and eight of the most representative members of genogroup I and II noroviruses was carried out, showing that all the Asturias isolates described in this work clustered together and were closely related to genogroup II, Bristol/Lorsdale cluster (Fig. 2).
FIG. 1.
Alignment of norovirus sequences within a conserved region of the RNA polymerase region, amplified using NV1 and NV2 primers. Nucleotide residue changes with respect to the consensus sequence are indicated. Numbers on the left correspond to the isolate name. The amplicon nucleotide residue numbers are indicated on the right. Primer sequences are excluded.
FIG. 2.
Phylogenetic analysis of the nine different norovirus isolates obtained in the present study. The nucleotide sequences of the 267-bp amplicons derived from the RNA polymerase coding regions were compared to representative genogroup I and II norovirus sequences. Abbreviations: NV, Norwalk virus, M87611; SoV, Southampton virus, L07418; DSV, Desert Shield virus, U04469; ChV, Chiba virus, AB042808; HssV, Hesse virus, AF093797; TV, Toronto virus, U02030; SMA, Snow Mountain Agent, L23831; HV, Hawaii virus, U07611; LV, Lorsdale virus, X86557.
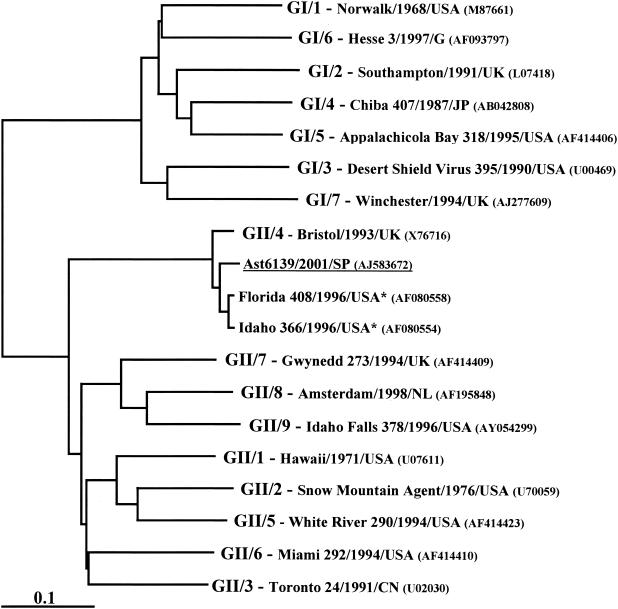
To further characterize the strains responsible of sporadic gastroenteritis in our survey, according to Ando's classification, the full-length capsid region of the representative isolate Ast6139/2001/SP was amplified, cloned and sequenced. Phylogenetic analysis using the deduced VP60 amino acid sequence from Ast6139/2001/SP isolate and the full-length capsid sequences of representative norovirus strains of the genetic clusters proposed by Ando et al. (3) confirmed that Ast6139/2001/SP was a member of the Bristol/Lorsdale cluster (GII/4, in the Ando's classification) (Fig. 3). Furthermore, a close relationship to the 95/96-US subset, a group of GII/4 strains associated with outbreaks of gastroenteritis in several countries from five continents, was also found.
FIG. 3.
Phylogenetic analysis of capsid amino acid sequences of Ast6139/2001/SP (underlined), two representative strains of the 95/96-US subset (asterisks), and representative norovirus strains of the genetic clusters proposed by Ando et al. (3).
DISCUSSION
Etiology of sporadic cases of acute gastroenteritis.
The role of rotaviruses, Campylobacter, and Salmonella as the most frequent causes of gastroenteritis in children has been reported by several authors (4, 8, 12, 28). Recently, the importance of noroviruses as the second cause of viral gastroenteritis has been reported (5, 6, 10), but bacterial agents were excluded by these authors. To our knowledge, the present work constitutes the first study designed to investigate the relative contribution of noroviruses to sporadic cases of pediatric gastroenteritis in Spain with respect to other classic gastroenteritis-associated viruses and bacteria. A first finding of the present report is that only half of the cases of acute pediatric gastroenteritis studied could be related to the presence of a known pathogen. Similar identification rates have been described in previous studies (7, 17, 21) supporting the existence of a large number of undiagnosed pathogens causing gastroenteritis, despite the significant advances made in diagnostic technology.
In our study, rotaviruses were the most frequent agents detected, confirming their role in mild diarrhea during childhood. The other main causes of sporadic cases of gastroenteritis in children were C. jejuni and Salmonella spp., followed by astroviruses, adenoviruses, and Y. enterocolitica, which were only occasionally detected. The development of an in-house RT-PCR to detect noroviruses has let us to include them in our study and evaluate their relative detection rate with respect to other gastroenteritis-associated viruses and bacteria. Our data supported the relevance of noroviruses as an under-appreciated cause of diarrhea in children from Asturias. Their role as the second cause of sporadic cases of viral gastroenteritis has also been reported by several studies carried out in other European countries, such as The Netherlands (16), France (Rouen) (22), Ireland (13), and Spain (Madrid) (6). Nevertheless, these studies have reported that noroviruses are responsible for 10 to 15% of all gastroenteritis cases. The detection of only a single genotype during a 1-year period could indicate the inability to detect some of the other norovirus clusters by our assay. The use of nondegenerate primers and/or a relatively high annealing temperature used in the RT-PCRs could explain this limitation and be the cause of the lower detection rate found in this study.
Viral and bacterial intestinal pathogens could affect either the same or different regions of the gut, and their effects would be enhanced (11). Mixed infections have been described previously in children with acute gastroenteritis, but few data on the role of noroviruses in dual infections are available. Barnes et al. (4) found 1.6% of mixed infections. The main type of association detected was virus-bacteria (80%), with rotaviruses followed by Salmonella as the most common pathogens, but neither noroviruses nor astroviruses were studied. Román et al. (29) detected 5% of mixed infections, the majority of these being combinations of rotavirus with astrovirus or adenovirus, and astrovirus with Salmonella, although no attempts to identify noroviruses were made. Bon et al. (5) found 16.7% of dual viral infections with rotaviruses as the most common agent implicated. In this study approximately half of the norovirus infections were mixed infections. Our results show the relative importance of virus-bacteria associations, being rotaviruses and Campylobacter the most common pathogens implicated. Nevertheless, noroviruses were not frequently implicated in associations with bacteria or other viruses. It is worth noting that a high percentage of cases of gastroenteritis associated with astroviruses were mixed infections (29).
Although several studies have described a traditional rotavirus seasonal peak in winter (15, 36), other reports have shown a peak in the late winter and early spring and even in late spring and summer (9). Gastroenteritis caused by norovirus infection has been described as a highly seasonal syndrome, often referred to as “winter vomiting disease.” Nevertheless, a recent study has reported a midsummer peak in 2002 in United Kingdom (19). The rotavirus- and norovirus-associated peaks found in the present report, as well as the lack of norovirus-associated cases in colder months support the relevance of these viruses as the cause of gastroenteritis in spring and summer and challenges the view that rotavirus and norovirus infections exclusively have wintertime seasonality. On the other hand, our data support the previously reported not seasonal distribution of diarrheal episodes caused by bacteria (8, 27).
Several authors have shown that bacteria caused more infections than viruses among children 5 to 14 years old (10). The low detection rate of viruses, as well as the high incidence of S. enterica serovar Enteritidis found in the present study further supported this idea. The high incidence of S. enterica serovar Enteritidis described in the present work, the only enteropathogen most frequently isolated among children in this group of age, is in clear controversy with previous data reported in other countries, where this bacterium was highly prevalent among patients less than 5 years old, although the difference between the proportion of patients less than 5 years old who tested positive for Salmonella and the proportion of older patients who tested positive was not found statistically significant (10, 30). On the other hand, viruses are considered the most common enteropathogens in children less than 5 years old. In spite of the high detection rate of viruses in our study, bacteria and viruses were detected with a similar frequency. The reason for this discrepancy is that we have found that C. jejuni is an important cause of diarrhea in children less than 5 years of age (84.1% of all C. jejuni-associated cases). In fact, in most developed countries, infection with Campylobacter species is associated with two age peaks, one in children under 5 years and another in young adults (15 to 19 years old) (1, 32). While, rotaviruses have been described as the primary cause of gastroenteritis in children less than 5 years old, the high prevalence of noroviruses has been also reported (5, 10, 35). Our results supported the importance of these viral pathogens in this group of age, suggesting the existence of factors that affect this predominance. Thus, the high consultation rate (79.6% of all patients belong to this group of age in our study), the close contact at day care centers, and the lack of knowledge on personal hygiene might have effects on viral infections (20). Furthermore, the existence of a built-up immunity that prevents symptomatic rotavirus infections at an older age has been previously described (27). Few data about the pathogenesis of norovirus infection has been reported, although the low detection rate of norovirus among older children (only one case in our study) suggests a similar built-up immunity.
The effect of the fecal specimen consistency on the diagnostic yield was also investigated. The dogma that gastroenteritis viruses are more likely to be found in liquid than in solid fecal specimens is commonly accepted. However, a recent report suggested that solid fecal specimens at the end of an episode of diarrhea would have a high viral diagnostic yield (23). Our study has shown a lack of correlation between stool consistency and the gastroenteritis-associated viruses found. On the other hand, the fact that bacteria are more likely to be found in liquid fecal specimens has been described (31). Our results support the argument that this is only the case for S. enterica serovar Enteritidis. These findings might have additional implications for those involved in establishing diagnostic algorithms for the investigation of viral gastroenteritis.
Molecular epidemiology of noroviruses.
Because of the high genetic diversity of noroviruses, molecular characterization of the genotypes of the strains causing pediatric gastroenteritis is essential to understand the epidemiology of noroviruses. Antigen detection enzyme-linked immunosorbent assays, based on the use of hyperimmune antisera or monoclonal antibodies to numerous recombinant GI and GII noroviruses capsid proteins, have been recently developed. Although, these assays are highly sensitive, their use in diagnostic laboratories has been limited by their narrow specificity, they are able to recognize the capsid protein of the immunizing virus but not those from other noroviruses (2, 18). Taking into account that there is a logical correlation between antigenic grouping based on the use of antibodies to recombinant capsid proteins and genomic grouping based on capsid amino acid sequences, Ando et al. (3) have proposed a classification scheme of noroviruses based on the ORF2 amino acid sequences, which would facilitate the designation of new strains, as well as the evaluation and standardization of enzyme immunoassays. The characterization of the norovirus strains detected in our study has shown that they belong to Bristol/Lorsdale cluster (GII/4, in Ando's classification). Similar studies carried out by other research groups worldwide have also reported the prevalence of Bristol-like viruses in sporadic cases and outbreaks (6, 13, 37). The high prevalence of the GII/4 strains worldwide is supported by the detection of the 95/96-US subset, a group of GII/4 strains associated with outbreaks of gastroenteritis in several countries from five continents (25, 39). Our results indicate that these viruses, responsible for large outbreaks of gastroenteritis, are also represented in sporadic community cases. The reasons for the high prevalence of these strains are unknown, but the identification of viral determinants involved in adaptation and virulence may be useful in defining why the GII/4 strains have become prevalent globally.
The high detection rate of noroviruses as the cause of diarrhea in children reported in this study supports their inclusion in routine screenings to diagnose sporadic cases of acute gastroenteritis. On the other hand, the genetic classification of these viruses will help us to improve our knowledge about the most prevalent strains.
Acknowledgments
This work was supported by project grants 01/0976 and 01/3139 from Fondo de Investigaciones Sanitarias, Madrid, Spain.
REFERENCES
- 1.Adak, G. K., J. M. Cowden, S. Nicholas, and H. S. Evans. 1995. The public health laboratory service national case-control study of primary indigenous sporadic cases of campylobacter infection. Epidemiol. Infect. 115:15-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altmar, R. L., and M. K. Estes. 2001. Diagnosis of noncultivable gastroenteritis viruses, the human caliciviruses. Clin. Microbiol. Rev. 14:15-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of “Norwalk-like viruses.” J. Infect. Dis. 181:S336-S348. [DOI] [PubMed] [Google Scholar]
- 4.Barnes, G. L., E. Uren, K. B. Stevens, and R. F. Bishop. 1998. Etiology of acute gastroenteritis in hospitalized children in Melbourne, Australia, from April 1980 to March 1993. J. Clin. Microbiol. 36:133-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bon, F., P. Fascia, M. Dauvergne, D. Tenenbaum, H. Planson, A. M. Petion, P. Pothier, and E. Kohli. 1999. Prevalence of group A rotavirus, human calicivirus, astrovirus, and adenovirus type 40 and 41 infections among children with acute gastroenteritis in Dijon, France. J. Clin. Microbiol. 37:3055-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buesa, J., B. Collado, P. López-Andújar, R. Abu-Mallouh, J. Rodríguez Díaz, A. García Díaz, J. Prat, S. Guix, T. Llovet, G. Prats, and A. Bosch. 2002. Molecular epidemiology of caliciviruses causing outbreaks and sporadic cases of acute gastroenteritis in Spain. J. Clin. Microbiol. 40:2854-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caeiro, J. P., J. J. Mathewson, M. A. Smith, Z. D. Jiang, M. A. Kaplan, and H. L. Dupont. 1999. Aetiology of outpatient paediatric nondysenteric diarrhoea: a multicenter study in the United States. Pediatr. Infect. Dis. J. 18:94-97. [DOI] [PubMed] [Google Scholar]
- 8.Caprioli, A., C. Pezzella, R. Morelli, A. Giammanco, S. Arista, D. Crotti, M. Facchini, P. Guglielmetti, C. Piersimoni, and I. Luzzi. 1996. Enteropathogens associated with childhood diarrhea in Italy. The Italian Study Group on Gastrointestinal Infections. Pediatr. Infect. Dis. J. 15:876-883. [DOI] [PubMed] [Google Scholar]
- 9.Chiu, T. F., C. N. Lee, P. I. Lee, C. L. Kao, H. C. Lin, C. Y. Lu, H. Y. Tseng, H. L. Hsu, C. Y. Lee, and L. M. Huang. 2000. Rotavirus gastroenteritis in children: 5-year experience in a medical center. J. Microbiol. Immunol. Infect. 33:181-186. [PubMed] [Google Scholar]
- 10.de Wit, M. A., M. P. Koopmans, L. M. Kortbeek, N. J. van Leeuwen, J. Vinje, and Y. T. van Duynhoven. 2001. Etiology of gastroenteritis in sentinel general practices in the Netherlands. Clin. Infect. Dis. 33:280-288. [DOI] [PubMed] [Google Scholar]
- 11.di Biase, A. M., G. Petrone, M. P. Conte, L. Seganti, M. G. Ammendolia, A. Tinari, F. Iosi, M. Marchetti, and F. Superti. 2000. Infection of human enterocyte-like cells with rotavirus enhances invasiveness of Yersinia enterocolitica and Y. pseudotuberculosis. J. Med. Microbiol. 49:897-904. [DOI] [PubMed] [Google Scholar]
- 12.Durepaire, N., M. P. Pradie, M. C. Ploy, M. Mounier, S. Ranger-Rogez, C. Martin, and F. Denis. 1995. Adenoviruses from stool samples in hospital units. Comparison with main pathogens in gastroenteritis (Rotavirus, Campylobacter, Salmonella). Pathol. Biol. 43:601-610. [PubMed] [Google Scholar]
- 13.Foley, B., J. O'Mahony, C. Hill, and J. G. Morgan. 2001. Molecular detection and sequencing of “Norwalk-like viruses” in outbreaks and sporadic cases of gastroenteritis in Ireland. J. Med. Virol. 65:388-394. [DOI] [PubMed] [Google Scholar]
- 14.Guerrant, R. L., J. M. Hughes, N. L. Lima, and J. Crane. 1990. Diarrhoea in developed and developing countries: magnitude, special settings, and etiologies. Rev. Infect. Dis. 12:S41-S50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koopmans, M., and D. Brown. 1999. Seasonality and diversity of group A rotaviruses in Europe. Acta Paediatr. Suppl. 88:14-19. [DOI] [PubMed] [Google Scholar]
- 16.Koopmans, M., J. Vinje, M. de Wit, I. Leenen, W. Poel, and Y. Duynhonven. 2000. Molecular epidemiology of human enteric caliciviruses in The Netherlands. J. Infect. Dis. 181:S262-S269. [DOI] [PubMed] [Google Scholar]
- 17.Kotloff, K. L., S. S. Wasserman, and J. Y. Steciak. 1988. Acute diarrhoea in Baltimore children attending an outpatient clinic. Pediatr. Infect. Dis. J. 7:753-759. [DOI] [PubMed] [Google Scholar]
- 18.Lopman, B. A., D. W. Brown, and M. Koopmans. 2002. Human caliciviruses in Europe. J. Clin. Virol. 24:137-160. [DOI] [PubMed] [Google Scholar]
- 19.Lopman, B. A., M. Reacher, C. Gallimore, G. K. Adak, J. J. Gray, and D. W. Brown. 2003. A summertime peak of “winter vomiting disease”: surveillance of Noroviruses in England and Wales, 1995 to 2002. BMC Public Health 24:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louhiala, P. J., N. Jaakkola, R. Ruotsalainen, and J. J. Jaakkola. 1997. Day-care centers and diarrhea: a public health perspective. J. Pediatr. 131:476-479. [DOI] [PubMed] [Google Scholar]
- 21.Maltezou, H. C., A. Zafiropoulou, M. Mavrikou, E. Bozavoutoglou, G. Liapi, M. Foustoukou, and D. A. Kafetzis. 2001. Acute diarrhoea in children treated in an outpatient setting in Athens, Greece. J. Infect. 43:122-127. [DOI] [PubMed] [Google Scholar]
- 22.Marine-Cardine, A., K. Gourlain, O. Mouterde, N. Castignolles, M. F. Hellot, E. Mallet, and C. Buffet-Janvresse. 2002. Epidemiology of acute viral gastroenteritis in children hospitalized in Rouen, France. Clin. Infect. Dis. 34:1170-1178. [DOI] [PubMed] [Google Scholar]
- 23.McCaughey, C., H. J. O'Neill, D. E. Wyatt, S. N. Christie, P. T. Jackson, and P. V. Coyle. 1998. Effect of faecal consistency on virological diagnosis. J. Infect. 36:145-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mount, D. W., and B. Conrad. 1986. Improved programs for DNA and protein sequence analysis on the IBM personal computer and other standard computer systems. Nucleic Acids Res. 14:443-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noel, J. S., R. L. Fankhauser, T. Ando, S. S. Monroe, and R. I. Glass. 1999. Identification of a distinct common strain of “Norwalk-like viruses” having a global distribution. J. Infect. Dis. 179:1334-1444. [DOI] [PubMed] [Google Scholar]
- 26.Page, R. D. M. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 27.Parashar, U. D., R. C. Holman, J. S. Bresee, M. J. Clarke, P. H. Rhodes, R. L. Davis, R. S. Thompson, J. P. Mullooly, S. B. Black, H. R. Shinefield, S. M. Marcy, C. M. Vadheim, J. I. Ward, R. T. Chen, R. I. Glass, et al. 1998. Epidemiology of diarrheal disease among children enrolled in four West Coast health maintenance organizations. Pediatr. Infect. Dis. J. 17:605-611. [DOI] [PubMed] [Google Scholar]
- 28.Prats, G., T. Llovet, C. Munoz, R. Sole, B. Mirelis, C. Izquierdo, P. Rodriguez, M. E. Sabanes, N. Rabella, R. Pericas, F. Sanchez, N. Margall, F. Navarro, and P. Coll. 1997. Etiology of enteritis in a university general hospital in Barcelona (1992-1995). Enferm. Infecc. Microbiol. Clin. 15:349-356. [PubMed] [Google Scholar]
- 29.Román, E., I. Wilhelmi, J. Colomina, J. Villar, M. L. Cilleruelo, V. Nebreda, M. Del Alamo, and A. Sanchez-Fauquier. 2003. Acute viral gastroenteritis: proportion and clinical relevance of multiple infections in Spanish children. J. Med. Microbiol. 52:435-440. [DOI] [PubMed] [Google Scholar]
- 30.Saeed, A. M., et al. (ed.). 1999. Salmonella enterica serovar enteriditis in humans and animals: epidemiology, pathogenesis and control. Iowa State University Press, Ames.
- 31.Singh, T., M. Verma, J. Chhatwal, B. Chacko, H. Kaur, and H. Prabhakar. 1995. Predictive utility of clinical and stool parameters in bacterial diarrhoea in children. Indian J. Med. Sci. 49:285-290. [PubMed] [Google Scholar]
- 32.Skirrow, M. B. 1991. Epidemiology of campylobacter enteritis. Int. J. Food Microbiol. 12:9-16. [DOI] [PubMed] [Google Scholar]
- 33.Snyder, J. D., and M. H. Merson. 1982. The magnitude of the global problem of acute diarrhoeal disease: a review of active surveillance data. Bull W. H. O. 60:605-613. [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multisequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tompkins, D. S., M. J. Hudson, H. R. Smith, R. P. Eglin, J. G. Wheeler, M. M. Brett, R. J. Owen, J. S. Brazier, P. Cumberland, V. King, and P. E. Cook. 1999. A study of infectious intestinal disease in England: microbiological findings in cases and controls. Commun. Dis. Public Health. 2:108-113. [PubMed] [Google Scholar]
- 36.van der Donck, I., L. van Hoovels, K. de Leener, T. Goegebuer, L. Vanderwegen, J. Frans, M. Rahman, and M. van Ranst. 2003. Severe diarrhea due to rotavirus infection in a Belgian hospital 1981-2002. Acta Clin. Belg. 58:12-18. [DOI] [PubMed] [Google Scholar]
- 37.Vinje, J., S. A. Altena, and M. P. Koopmans. 1997. The incidence and genetic variability of small round-structured viruses in outbreaks of gastroenteritis in The Netherlands. J. Infect. Dis. 176:1374-1378. [DOI] [PubMed] [Google Scholar]
- 38.Wang, J., X. Jiang, H. P. Madore, J. Gray, U. Desselberger, T. Ando, Y. Seto, I. Oishi, J. F. Lew, and K. Y. Green. 1994. Sequence diversity of small, round-structured viruses in the Norwalk virus group. J. Virol. 68:5982-5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White, P. A., G. S. Hansman, A. Li, J. Dable, M. Isaacs, M. Ferson, C. J. McIver, and W. D. Rawlinson. 2002. Norwalk-like virus 95/96-US strain is a major cause of gastroenteritis outbreaks in Australia. J. Med. Virol. 68:113-118. [DOI] [PubMed] [Google Scholar]