Abstract
Polymorphic amplified typing sequences (PATS) for Escherichia coli O157:H7 (O157) was previously based on indels containing XbaI restriction enzyme sites occurring in O-island sequences of the O157 genome. This strain-typing system, referred to as XbaI-based PATS, typed every O157 isolate tested in a reproducible, rapid, straightforward, and easy-to-interpret manner and had technical advantages over pulsed-field gel electrophoresis (PFGE). However, the system was less discriminatory than PFGE and was unable to differentiate fully between unrelated isolates. To overcome this drawback, we enhanced PATS by using another infrequently cutting restriction enzyme, AvrII (also known as BlnI), to identify additional polymorphic regions that could increase the discriminatory ability of PATS typing. Referred to as AvrII-based PATS, the system identified seven new polymorphic regions in the O157 genome. Unlike XbaI, polymorphisms involving AvrII sites were caused by both indels and single-nucleotide polymorphisms occurring in O-island and backbone sequences of the O157 genome. AvrII-based PATS by itself provided poor discrimination of the O157 isolates tested. However, when primer pairs amplifying the seven polymorphic AvrII sites were combined with those amplifying the eight polymorphic XbaI sites (combined PATS), the discriminatory power of PATS was enhanced. Combined PATS matched related O157 isolates better than PFGE while differentiating between unrelated isolates. PATS typed every O157 isolate tested and directly targeted polymorphic sequences responsible for differences in the restriction digest patterns of O157 genomic DNA, utilizing PCR rather than relying on gel electrophoresis. This enabled PATS to resolve the ambiguity in PFGE typing, including that arising from the “more distantly related” and “untypeable” profiles.
Escherichia coli O157:H7 (O157) was first identified as a human enteric pathogen in 1982 and has since been implicated in several outbreaks, as well as sporadic infections (11, 16). Presently, the pathogen ranks third after Campylobacter and Salmonella among the etiologic agents causing diarrhea in North America (10). O157 infection causes an estimated 73,000 illnesses, 2,000 hospitalizations, and 60 deaths each year in the United States (10, 16, 31; P. M. Griffin, P. S. Mead, T. van Gilder, S. B. Hunter, N. A. Strockbine, and R. V. Tauxe, Abstr. 4th Int. Symp. Workshop Shiga Toxin (Verocytotoxin)-Producing Escherichia coli Infect., p. 31, 2000). Cattle are the primary reservoir and the main source of human infection, although, sheep, deer, and goats have also been shown to transiently harbor O157 naturally (10, 16, 31). Human infection occurs through food sources that are derived from cattle (undercooked hamburger) or contaminated by cattle feces, such as salad vegetables, water, apple cider, or unpasteurized milk, or in some instances from lamb, venison, or deer jerky. In addition, infection of persons swimming in fecally contaminated lakes, as well as person-to-person and animal-to-person transmission, has also been reported (10, 39).
Outbreaks of food-borne pathogens such as O157 occur nearly simultaneously in several different states or even different countries, with varied disease manifestations. In addition, this organism has been classified as a category B bioterrorism agent by the Centers for Disease Control and Prevention (Atlanta, Ga.). Rapid typing of O157 strains would play a key role in limiting widespread outbreaks by allowing identification and withdrawal of the common source of infection. Pulsed-field gel electrophoresis (PFGE) is used by diagnostic and public health laboratories to type O157 strains. At the time PFGE was designated the “gold standard” for bacterial strain typing, it was the only technique that could provide distinctive profiles for bacterial strains in a reasonably reproducible manner (29, 38). The other methodologies available at the time included phenotypic and genotypic methods, such as biotyping, antimicrobial susceptibility patterns, bacteriophage typing, bacteriocin typing, multilocus enzyme electrophoresis patterns, whole-cell fatty acid profiles, plasmid analysis, restriction mapping, restriction fragment length polymorphism analysis, and PCR amplification patterns of repetitive, insertion, and other consensus sequences (1, 14, 24).
However, over time, certain drawbacks of PFGE have become more evident, requiring researchers to implement several modifications of the technique in order to increase sample turnaround, overcome inhibition of restriction enzymes or degradation of DNA, and minimize changes in electrophoretic patterns of DNA between gels to avoid reliance on sophisticated pattern recognition computer software to interpret the patterns generated (3, 12, 15, 23, 29). The ability of PFGE to accurately assess the genetic relatedness of O157 isolates in outbreaks was recently assessed (6). It was observed that single-restriction-enzyme PFGE gave a poor measure of genetic relatedness, as it did not resolve all the DNA fragments generated following restriction digestion and could not resolve comigrating bands (6). Assessing genetic relatedness is crucial to any epidemiological survey, and the authors suggest the use of multiple enzymes to obtain an accurate result using PFGE (6).
In previous studies, researchers tried to decipher the genetic events underlying strain variation in O157 as seen by PFGE following digestion of genomic DNA by the restriction enzyme XbaI (18, 19). XbaI is the enzyme of choice in PFGE analysis of O157 isolates, as the enzyme is an effective cutter and gives a more decipherable profile than other infrequently cutting restriction enzymes (6, 29, 38). Our analysis showed that insertions and deletions (indels) of sequences containing XbaI sites were responsible for the majority of differences between O157 strains, and based on these indels, we proposed a novel, PCR-based strain-typing tool called polymorphic amplified typing sequences (PATS, or XbaI-based PATS) (18, 19) (U.S. utility patent pending; filed 1 November 2000; PCT International patent pending; filed 1 November 2001.). XbaI-based PATS was able to type every O157 isolate tested in a reproducible manner. In contrast to PFGE, XbaI-based PATS yielded straightforward, easy-to-interpret results that correctly matched related O157 isolates (19). However, XbaI-based PATS was less discriminatory than PFGE and was unable to differentiate fully between unrelated isolates. This may have been due to our inability to utilize two repeat regions in the O157 genome in the design of the XbaI-based PATS primer pairs, as well as the occurrence of insertions and deletions in regions between XbaI sites that would have been overlooked by this typing technique.
Hence, we decided to evaluate another infrequently occurring restriction enzyme site in the O157 genome to increase the discriminatory power of PATS. The restriction enzyme AvrII (BlnI) fulfilled the criteria we had set by cutting the O157 genome infrequently, having sites within the repeat regions not targeted by XbaI-based PATS, and having only one site overlapping a region previously amplified by XbaI-based PATS. In this study, we analyzed the genetic events underlying polymorphisms at or around AvrII restriction sites in the O157 genome and used the information to enhance the discriminatory power of PATS.
MATERIALS AND METHODS
Bacteria.
Two previously characterized strains of O157, 86-24 and EDL 933, were used as controls in this study (19). In addition, 44 O157 isolates, 2 each from 22 different outbreaks collected and previously analyzed by PFGE at the Centers for Disease Control and Prevention, were also included in this study. The isolates from different outbreaks had different PFGE patterns, suggesting genetic heterogeneity among them, and showed considerable differences with XbaI-based PATS (19). The paired O157 isolates from the 22 outbreaks were given corresponding alphabetical designations, A-AA through V-VV.
Design of primer pairs amplifying O157 AvrII sites.
The sequenced EDL 933 genome (GenBank accession number AE005174) (33) was used as the prototype O157 to determine the total number of AvrII restriction sites and the DNA sequences of the regions flanking these sites. The sequences were used to design primer pairs that would yield distinct amplicons containing a single AvrII site from O157 strain EDL 933. These AvrII-based PATS primers were each assigned an IKNR prefix. We also used previously described primer pairs that amplified specific virulence genes, stx1, stx2, eae, and hlyA, and the previously identified eight polymorphic XbaI-containing sequences in the O157 genome when required (19, 30). PCR with these primer pairs was set up using previously described conditions (18, 19) that combined the hot-start and touchdown techniques (7, 8).
Primer pairs evaluating the presence of O islands 43 and 48.
O islands 43 and 48 are identical O islands encoding tellurite resistance, integrase, urease, phage proteins, and an adhesin (33). They are also referred to as the “tellurite resistance- and adherence-conferring islands” (P. I. Tarr, S. S. Bilge, J. A. Vary, N. M. Tang, M. R. Baylor, K. Potter, T. E. Besser, and S. L. Moseley, Abstr. 31st US-Japan Cholera Rel. Diarrheal Dis. Conf., 1995, p. 119-124) and are 0.5 Mb apart on the EDL 933 genome, with O island 43 inserted close to the serX tRNA gene and O island 48 inserted close to the serW tRNA gene (33). The following primer pairs used to amplify the serW and serX tRNA genes and the 3′ end of tellurite resistance- and adherence-conferring islands in a previous study (37) were included in this study: SerW (forward), 5′TCGGGGAAGGTAAGGAT3′, and SerW (reverse), 5′TTGTGATATGTATGAAGT3′; SerX (forward), 5′TTTTTCTATTGTCGATTCCTCT3′, and SerX (reverse), 5′GATCTACAAAGGCCACCAGCA3′; and Ter-island (forward), 5′GACAAACTCTCCGGGATAACTCA3′, and Ter-island (reverse), 5′TGCGGGTGCTGGTGTGGGATAA3′ (37). These primer pairs were used to determine if both or only one of the two identical O islands was present in the O157 isolates tested, as four of the AvrII-based PATS primer pairs (IKNR6A and -B, IKNR7A and -B, IKNR9A and -B, and IKNR10A and -B) mapped to these two regions. PCR was done under conditions described previously (18, 19).
Evaluation of amplicons.
PCR were initially screened for the presence or absence of amplicons. Amplicons, when present, were purified using the QIAquick PCR purification kit (Qiagen, Valencia, Calif.) and digested with the AvrII restriction enzyme (New England Biolabs, Beverly, Mass.) to confirm the presence of an AvrII restriction site within the amplicon. Undigested and digested DNA fragments were resolved on a 4% agarose gel prepared with a combination of 3% Nusieve GTG agarose (FMC BioProducts, Rockland, Maine) and 1% agarose (Shelton Scientific Inc., Shelton, Conn.) and stained with ethidium bromide.
The absence of amplicons was further investigated by PCR, Southern blotting, and sequencing (see below). Additional primer pairs were designed based on sequences upstream and downstream of AvrII sites to investigate polymorphisms resulting from indels. In addition, amplicons that showed variant digestion patterns with the AvrII restriction enzyme were evaluated by sequencing for the presence of single-nucleotide polymorphisms (SNPs) at the restriction site.
DNA extraction, sequencing, and probe labeling.
Genomic DNA was prepared using the Easy-DNA Isolation kit (Invitrogen Corp., Carlsbad, Calif.) according to the manufacturer's instructions. DNA sequencing was done at the DNA Sequencing Core Facility, Department of Molecular Biology, Massachusetts General Hospital. All DNA probes were labeled using the ECL direct nucleic acid labeling and detection system (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.).
Southern blotting.
Indels were evaluated using Southern blot hybridization. DNA was fractionated by agarose gel electrophoresis, transferred to Hybond-N+ membranes (Amersham Pharmacia Biotech, Inc.), UV cross-linked to the membrane using a Stratalinker (Stratagene, La Jolla, Calif.), and hybridized with the appropriate probes derived from O157 strain EDL 933 and labeled using the ECL direct nucleic acid labeling and detection system. Hybridization at 42°C and posthybridization washing of blots was done according to the ECL kit manual. Autoradiographs were prepared by exposure of processed blots to Scientific Imaging X-OMAT AR film (Eastman Kodak Company, Rochester, N.Y.).
Data analysis.
Dendrograms were constructed by coding molecular data as follows. The presence or absence of each of the 19 amplicons (representing eight polymorphic XbaI sites, seven polymorphic AvrII sites, and four virulence genes) was coded as a dichotomous variable. Characters representing gain or loss of a restriction site recognized by AvrII were weighted to reflect the increased probability of losing a site over gaining one; however, such weighting had no impact on the resulting dendrogram. Trees were constructed using the unweighted-pair-group method with arithmetic means option in Phylogenetic Analysis Using Parsimony software (Sinauer Associates, Inc., Sunderland, Mass.).
RESULTS
Evaluation of AvrII-based PATS primer pairs in control O157 strains.
A total of 33 AvrII restriction sites were identified in the O157 strain EDL 933 genome sequence by in silico restriction analysis. Primer pairs that would amplify ∼300- to 600-bp segments of DNA containing each AvrII restriction site were designed flanking each of these sites. The presence or absence of an amplicon with each primer pair, as well as the presence or absence of an AvrII restriction site within each amplicon, was tested by PCR, AvrII digestion, and agarose gel electrophoresis.
Thirty-three primer pairs were analyzed with the control O157 strains, EDL 933 and 86-24. With O157 strain EDL 933, 32 of the 33 primer pairs yielded the expected single amplicons containing the AvrII restriction sites. One of the primer pairs, IKNR29A and -B, consistently yielded two amplicons from both the control strains and other test O157 isolates. Sequence analysis showed that this primer pair was amplifying overlapping sequences from two repeat regions along the O157 genome. The repeat region from which the IKNR29 primers were designed was already targeted by a previously designed XbaI-based PATS primer pair, IK38A and -B (19). In addition, the second repeat region was being amplified by another AvrII-based primer pair, IKNR24A and -B, that yielded a single amplicon from that site. Hence, we decided that exclusion of IKNR29 would not compromise the typing system, and this primer pair was excluded from the final pool of 32 AvrII-based PATS primer pairs.
With O157 strain 86-24, two of the 32 AvrII-based PATS primer pairs either yielded an amplicon with an SNP (IKNR27) or failed to yield an amplicon (IKNR33) (Table 1). These polymorphic results demonstrated the ability of these primer pairs to discriminate between the two O157 control strains.
TABLE 1.
PATS profiles of E. coli O157:H7 isolates based on AvrII restriction sites and virulence genes
| PATS typea | PCR amplification and AvrII restriction digestion pattern of amplicon obtained using PATS virulence gene primer pairsb
|
PCR amplification and AvrII restriction digestion pattern of amplicon obtained using PATS virulence gene primer pairs
|
Isolate(s)c | ||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IKNR1 | IKNR2 | IKNR3 | IKNR4 | IKNR5 | IKNR6 | IKNR7 | IKNR8 | IKNR9 | IKNR10 | IKNR11 | IKNR12 | IKNR13 | IKNR14 | IKNR15 | IKNR16 | IKNR17 | IKNR18 | IKNR19 | IKNR20 | IKNR21 | IKNR22 | IKNR23 | IKNR24 | IKNR25 | IKNR26 | IKNR27 | IKNR28 | IKNR30 | IKNR31 | IKNR32 | IKNR33 | stx1 | stx2 | eae | hlyA | ||
| Control | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | E. coli O157:H7 strain EDL933 |
| Control | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 2 | 0 | 0 | 1 | 1 | 1 | E. coli O157:H7 strain 86-24 |
| 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 2 | 0 | 0 | 1 | 1 | 1 | HH, T, TT |
| 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 2 | 0 | 0 | 1 | 1 | 1 | H |
| 3 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | A, VV |
| 4 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | D, DD |
| 5 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 1 | 1 | 1 | E, EE, R, RR |
| 6 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | I, II, P, PP |
| 7 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 1 | 1 | 1 | K, KK |
| 8 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 2 | 2 | 2 | 0 | 1 | 1 | 1 | 1 | Q, QQ |
| 9 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | M, MM |
| 10 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 0 | B |
| 11 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | AA, BB, C, CC, F, FF, G, GG, J, JJ, L, LL, N, NN, O, OO, S, SS, U, UU, V |
PATS types are designated arbitrarily with different numbers.
Prefix of each PATS primer pair A-B and virulence gene primer pair F-R is indicated. 0, no amplicon; 1, amplicon without AvrII site; 2, amplicon with one AvrII site; 3, amplicon with two AvrII sites. PATS primer pairs producing polymorphic results between strains and the polymorphic scores assigned are shown in boldface.
Isolates of E. coli O157:H7 from various outbreaks that fell within a given PATS type.
Analysis of 44 O157 isolates with 32 AvrII-based PATS primer pairs.
We evaluated the discriminatory abilities of the selected 32 AvrII-based PATS primer pairs against a set of 44 O157 isolates. We also included the primer pairs amplifying the four virulence genes in the typing system as before (19). We recorded the electrophoresis results as 0, 1, 2, or 3, indicating the absence of an amplicon, the presence of an amplicon with no AvrII site, the presence of an amplicon with one AvrII site, and the presence of an amplicon with two AvrII sites, respectively. In contrast to XbaI-based PATS, AvrII-based primer pairs yielded amplicons of all types (19) (Table 1). However, amplicons obtained with the virulence gene primer pairs had a score of 0 or 1, as before (19).
Of the 32 AvrII-based PATS primer pairs, 7 produced polymorphic results and 25 produced identical results across the 44 O157 isolates tested (Table 1). The polymorphic results produced by the seven primer pairs (Table 1) ranged from the absence of an amplicon (score, 0) to the presence of an amplicon with no (score, 1) or two (score, 3) AvrII sites. Based on the scores assigned to each amplicon obtained from every isolate-primer pair combination tested, the 44 O157 isolates were separated into 11 AvrII-based PATS types (Table 1). These results were successfully reproduced in three separate analyses of the 44 O157 isolates. In addition, the absence of an amplicon was confirmed by Southern blot analysis (data not shown). The absence of the restriction site or presence of an additional restriction site was confirmed by sequencing.
DNA sequences amplified by all 32 AvrII-based PATS primer pairs were analyzed for their location in the O157 strain EDL 933 genome using the GenBank database (BLAST search program; National Center for Biotechnology Information). Of the 32 O157 AvrII-containing genome sequences, 22 occurred in sequences having homology to E. coli strain K-12 genome sequences (backbone), and 10 occurred in sequences not shared with K-12 (O islands). Among the conserved 25 AvrII-containing sequences, only 5 were in O islands. On the other hand, five of the seven polymorphic AvrII-containing sequences were in O islands, supporting a previous observation that major genomic differences between O157 strains occur in O-island sequences (Table 2) (18, 19).
TABLE 2.
Description of AvrII-containing regions polymorphic between E. coli O157:H7 isolates
| Amplicon derived from E. coli O157:H7 isolates | Region associated with amplicon in EDL 933 genomea | Length of associated O island in strain EDL933 (bp) | Description of associated O island and backbone regions |
|---|---|---|---|
| IKNR3 | O island 8 | 23,480 | Cryptic prophage CP-933H and -I |
| IKNR7 | O island 43 | 87,547 | Tellurite resistance, phage proteins, integrase, urease, adhesin |
| IKNR10 | O island 48 | 87,547 | Tellurite resistance, phage proteins, integrase, urease, adhesin |
| IKNR12 | O island 50 | 47,314 | Cryptic prophage CP-933N |
| IKNR16 | Backbone | NAb | Unknown function |
| IKNR27 | Backbone | NA | Enzyme (anaerobic respiration) |
| IKNR33 | O island 172 | 44,433 | Integrase, helicase |
Sequenced O157 strain EDL933 genome (accession no. AE005174); O island, DNA sequence unique to E. coli O157 genome; backbone, DNA sequence homologous to E. coli K-12 genome.
NA, not applicable.
Analysis of polymorphisms at AvrII restriction sites.
The seven polymorphic AvrII-containing sequences were studied in detail to ascertain the genetic nature of the differences between O157 isolates. PCR using additional primer pairs designed upstream and downstream of the polymorphic regions in the O157 strain EDL 933 genome, Southern blot hybridization, and sequencing were done. Our analysis showed that polymorphism involving amplicons IKNR12 and IKNR33 were due to deletions, while polymorphisms at IKNR3, IKNR7, IKNR10, IKNR16, and IKNR27 were due to SNPs that either deleted an AvrII site or created an additional AvrII site (Fig. 1 and 2).
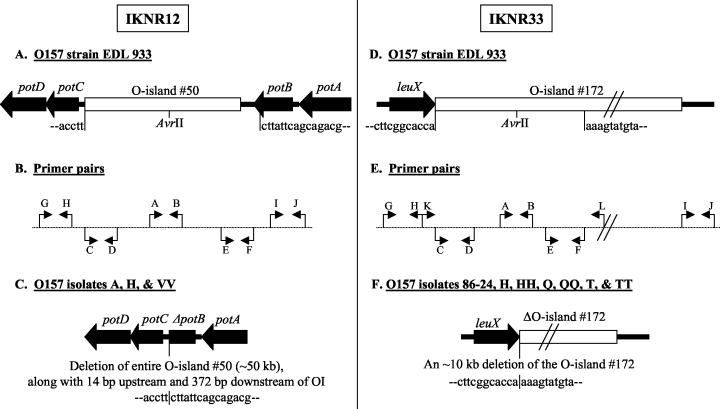
FIG. 1.
Diagrammatic representation of AvrII restriction site polymorphisms attributable to indels identified in O157 strains. (A to C) IKNR12 indel resulting from deletion of an entire O island. DNAs from O157 strain EDL 933 (A) and isolates A, H, and VV (C) are compared. Solid symbols, identical regions; open symbol, region that differs. The arrows indicate the direction of transcription of the designated genes. OI, O island. (B) Primers used to amplify segments of the DNAs. The primers are in direct alignment with the DNA segments in strain EDL 933 used to design them. (D to F) IKNR33 indel resulting from deletion of a 10-kb DNA fragment. DNAs from O157 strain EDL 933 (D) and isolates 86-24, H, HH, Q, QQ, T, and TT (F) are compared. Solid symbols, identical regions; open symbols, regions that differ. The arrows indicate the direction of transcription of the designated genes. (E) Primers used to amplify segments of the DNAs. The primers are in direct alignment with the DNA segments in strain EDL 933 used to design them.
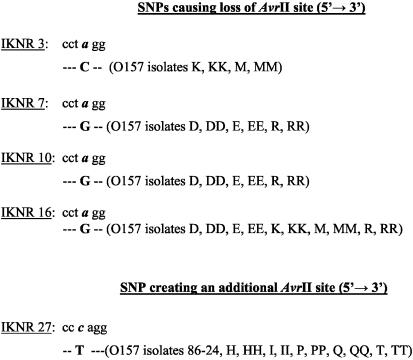
FIG. 2.
AvrII restriction site polymorphisms attributable to SNPs identified in O157 strains. SNPs causing loss or gain of an AvrII restriction site are listed with the respective O157 isolates exhibiting the polymorphism. The nucleotide altered in the wild-type sequence is shown in italics, and the replacing nucleotide is shown in uppercase.
(i) Polymorphism caused by indels: IKNR12.
The primer pair IKNR12A and -B amplified an amplicon, IKNR12, from the control O157 strain EDL 933 and most of the tested O157 isolates except isolates A, H, and VV (Table 1). This amplicon was mapped to O island 50 in O157 strain EDL933, encoding several unknown proteins and proteins associated with the cryptic prophage CP-933N (Table 2; GenBank accession number AE005174) (33). O island 50 is 47,310 bp in size and is flanked by genes involved in the spermidine-putrescine transport system, potD (1,046 bp), potC (794 bp), potB (863 bp), and potA (1,136 bp) (Fig. 1A). We designed additional primer pairs, IKNR12 C and -D, IKNR12E and -F, IKNR12G and -H, and IKNR12I and -J (Fig. 1B), and tested isolates H, A, and VV with them.
Primer pairs IKNR12A and -B, IKNR12C and -D, and IKNR12E and -F yielded amplicons of expected sizes only from EDL 933. This indicated possible deletion of sequences within, at the start of, and at the end of O island 50 in isolates A, H, and VV. Primer pairs IKNR12G and -H, and IKNR12I and -J yielded amplicons from all isolates and EDL 933, indicating the presence of DNA sequences upstream and downstream of O island 50. Primer pair IKNR12G and -J yielded amplicons only from isolates A, H, and VV, confirming a deletion of the entire O island 50 in the genomes of these isolates; this amplicon lacked an AvrII site. No amplification was obtained from EDL 933 with this primer pair, as the PCR conditions were not set to compensate for the ∼50-kb distance between the binding sites for these primers in the absence of any deletions. Sequencing and Southern blot analysis further confirmed our PCR results and observations. A 47,696-bp deletion had occurred in the genomes of O157 isolates A, H, and VV that included the entire O island 50, part of the intergenic segment between potC and O island 50 (14 bp), the entire intergenic segment between O island 50 and potB (348 bp), and part of potB itself (24 bp) (Fig. 1C). Interestingly, O island 50 has a putative insertion sequence element and a putative integrase at both ends, and an overlap in sequence was observed at the site of cleavage. These may have contributed to the excision of O island 50 in these variant isolates.
IKNR33.
Primer pairs IKNR33A and -B amplified an amplicon, IKNR33, from the control O157 strain EDL 933 and most of the tested O157 isolates except control strain 86-24 and isolates H, HH, Q, QQ, T, and TT (Table 1). This amplicon was mapped to O island 172 in O157 strain EDL933, encoding several unknown, integrase, and helicase proteins (Table 2) (GenBank accession number AE005174) (33). O island 172 is 44,433 bp in size and is flanked by the leuX (tRNA-leu) and yjhs (unknown) genes, as shown in Fig. 1D. We designed additional primer pairs, IKNR33 C and -D, IKNR33E and -F, IKNR33G and -H, IKNR33I and -J, and IKNR33K and -L (Fig. 1E), and tested the six variant isolates.
Primer pairs IKNR33A and -B, IKNR33C and -D, and IKNR33E and -F yielded amplicons of expected sizes only from EDL 933. This indicated possible deletion of sequences within and at the start of O island 172 in isolates H, HH, Q, QQ, T, and TT. Primer pairs IKNR33G and -H and IKNR33I and -J yielded amplicons from all isolates and EDL 933, indicating the conservation of DNA sequences upstream and downstream of O island 172. Primer pair IKNR33K and -L yielded amplicons only from isolates H, HH, Q, QQ, T, and TT, confirming a deletion of part of O island 172 in the genomes of these isolates; this amplicon lacked an AvrII site. No amplification was obtained from EDL 933 with this primer pair, as the PCR conditions were not set to compensate for the >10 kb distance between the binding sites for these primers in the absence of any deletions. Sequencing and Southern blot analysis confirmed that a 10,235-bp deletion had occurred in O island 172 of O157 isolates H, HH, Q, QQ, T, and TT (Fig. 1F). The deletion had occurred between two putative integrase-encoding regions, one occurring at the start of and the other within O island 172.
(ii) Polymorphism caused by SNPs.
Polymorphisms in amplicons IKNR3, IKNR7, IKNR10, IKNR16, and IKNR27 were due to SNPs occurring either at the AvrII restriction site or upstream of the site, resulting in the loss or gain of an AvrII site (Table 1 and Fig. 2), respectively. Of these five amplicons with SNPs, three were mapped to O islands and two were mapped to backbone sequences (Table 2).
Primer pairs IKNR3A and -B, IKNR7A and -B, IKNR10A and -B, and IKNR16A and -B yielded amplicons from all control and test O157 isolates. However, amplicons from some of the isolates failed to digest with the restriction enzyme AvrII. Sequence analysis indicated the presence of an SNP in the recognition sequence for this enzyme, namely, CCTAGG. The SNPs are listed in Fig. 2 as they were seen on the coding strand and were primarily transitions (adenine to guanine), with only one transversion (adenine to cytosine) seen in the IKNR3 amplicon. Similarly, primer pair IKNR27A and -B yielded amplicons from all control and test O157 isolates. However, amplicons from some of the isolates yielded three fragments upon digestion with the restriction enzyme AvrII, clearly indicating the presence of an additional site for the enzyme. Sequence analysis confirmed this observation. A transition (cytosine to thymidine on the coding strand) had occurred in the sequence upstream of the original AvrII site, creating an additional AvrII site in that region (Fig. 2).
Evaluation of O157 isolates for the presence of O islands 43 and 48.
Amplicons IKNR7 and IKNR10 mapped to the two well-characterized, identical O islands 43 and 48, which encode tellurite resistance, integrase, urease, adhesin, and other phage proteins (33, 37) (Table 2). Studies conducted by Taylor et al. have shown genomic variability in these O islands between O157 strains. Of the 16 O157 strains studied by these authors, 6 had both O islands, 6 had only one of the O islands, and 4 had neither (37). In our study, all of the O157 isolates analyzed yielded the IKNR7 and IKNR10 amplicons, clearly indicating that one or both of the O islands were always present (Table 1).
We evaluated the six O157 isolates that had SNPs in IKNR7 and IKNR10 for the presence of O islands 43 and 48. Our PCR analysis, using O157 strain EDL 933 (O island 43+ O island 48+) as a positive control, showed that all six O157 isolates contained only one O island (O island 43) inserted at serX (data not shown). Specifically, primer pairs SerW forward and reverse and Ter forward and reverse yielded amplicons of the expected sizes from all these isolates, while SerX forward and reverse failed to yield an amplicon (data not shown).
We also screened the 21 O157 isolates that grouped in the same PATS type following AvrII-based PATS analysis (Table 1) for the presence of O islands 43 and 48 to see if this would enable further differentiation of these isolates. However, the PCR analysis yielded results identical to those described above, indicating the presence of only O island 43 in these isolates as well. This eliminated the possibility of differences between the O157 isolates based on this factor.
Comparison of polymorphisms at AvrII and XbaI restriction sites.
The seven AvrII-containing regions that were polymorphic between O157 isolates occurred in O islands or backbone sequences and were caused by indels and SNPs. In contrast, as shown previously, the eight XbaI-containing regions that were polymorphic between O157 isolates were all located in O islands, and all were caused by indels (18).
One of the AvrII-containing polymorphic regions (IKNR33) and an XbaI-containing polymorphic region (IK114) could be traced to the same O island, namely, O island 172. Polymorphisms at both IKNR33 and IK114 were due to indels, suggesting that perhaps O island 172 has regions of increased recombinational activity. No overlap was observed between the remaining polymorphic regions. We also observed that some O islands contained both conserved and polymorphic regions around one or both of these restriction sites. These included O islands 8 (IK111-conserved-XbaI; IKNR3-polymorphic-AvrII), 43 (IKNR6-conserved-AvrII; IKNR7-polymorphic-AvrII), 48 (IKNR9-conserved-AvrII; IKNR10-polymorphic-AvrII), 108 (IK118-conserved-XbaI; IKB15-polymorphic-XbaI), and 172 (IK39-conserved-XbaI; IK114-polymorphic-XbaI; IKNR33-polymorphic-AvrII) (18, 19) (Tables 1 and 2).
Combined screening for polymorphic XbaI-containing and AvrII-containing regions, along with virulence genes.
In order to enhance the discriminatory ability of XbaI-based PATS, we had specifically chosen sites for the restriction enzyme AvrII, as the in silico analysis showed that some of these sites were in close proximity to some of the XbaI sites and others were fairly evenly distributed along the entire O157 genome between the XbaI sites (Genome Restriction Mapping Program; The Institute for Genomic Research Database). This would help us address two issues: (i) to be able to use the repeat regions that were excluded from XbaI-based PATS due to our inability to obtain distinct amplicons for those regions and (ii) to target polymorphisms that occurred between XbaI sites (19).
Four repeat regions, two direct repeats and two inverse repeats, were identified in the O157 genome, each of which contained a single XbaI and a single AvrII restriction enzyme site. Although the regions had >90% homology to each other, they contained a few nonoverlapping sequences. Using the nonoverlapping sequences, and the positions of the restriction enzyme sites within these repeat regions, we designed primer pairs that would yield a single amplicon from each of the repeat regions. Two of the repeat regions were successfully amplified by the XbaI-based PATS primer pairs IK38A and -B and IKB14A and -B (19), while three of the repeat regions were amplified by the AvrII-based PATS primer pairs IKNR18A and -B, IKNR24A and -B, and IKNR31A and -B (Table 1). The amplicons IKNR24 and IKNR31 amplified repeat regions that had not been targeted by XbaI-based PATS, fulfilling our goal of being able to use these regions in typing.
The distribution of the AvrII restriction enzyme sites in DNA segments between the XbaI restriction sites also helped to identify additional polymorphic regions along the O157 genome. Furthermore, the same set of 44 O157 isolates analyzed by XbaI-based PATS and AvrII-based PATS was analyzed by a combination of only those primer pairs amplifying polymorphic regions containing XbaI or AvrII restriction sites, along with the virulence gene primer pairs. The O157 isolates were differentiated into 19 groups, demonstrating that the combination of select primer pairs (combined PATS) discriminated better than the XbaI-based or AvrII-based PATS primer pairs alone (19) (Tables 1 and 3 and Fig. 3). The large clusters of isolates were split into smaller groups so that the overall distribution of the isolates was similar to that from PFGE (Fig. 3). The final PATS profiles identified the paired O157 isolates from 22 outbreaks as identical for 14 outbreaks, similar (1 amplicon difference; isolates A-AA, G-GG, H-HH, M-MM, N-NN, and P-PP) for 6 outbreaks, and different (>1 amplicon difference; isolates B-BB and V-VV) for 2 outbreaks (Table 3 and Fig. 3). The isolates recognized as different also had substantially different PFGE profiles.
TABLE 3.
PATS profiles of E. coli O157:H7 isolates based on polymorphic XbaI and AvrII restriction sites and virulence genes
| PATS typea | PCR amplification and restriction digestion pattern of amplicon obtained PATS virulence gene primer pairsb
|
Isolate(s)c | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Polymorphic XbaI sited
|
Polymorphic AvrII site
|
Virulence gene
|
||||||||||||||||||
| IK8 | IK19 | IK25 | IK114 | IK118 | IK123 | IKB3 | IKB5 | IKNR3 | IKNR7 | IKNR10 | IKNR12 | IKNR16 | IKNR27 | IKNR33 | stx1 | stx2 | eae | hlyA | ||
| Control | 0 | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | E. coli O157:H7 strain EDL933 |
| Control | 2 | 2 | 2 | 0 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 0 | 0 | 1 | 1 | 1 | E. coli O157:H7 strain 86-24 |
| 1 | 2 | 2 | 2 | 0 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 0 | 0 | 1 | 1 | 1 | HH, T, TT |
| 2 | 0 | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | AA |
| 3 | 0 | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | A |
| 4 | 2 | 2 | 2 | 0 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 2 | 3 | 0 | 0 | 1 | 1 | 1 | H |
| 5 | 0 | 2 | 0 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 0 | B |
| 6 | 2 | 2 | 0 | 2 | 2 | 2 | 0 | 0 | 1 | 2 | 2 | 2 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | M |
| 7 | 2 | 2 | 0 | 2 | 2 | 2 | 0 | 0 | 2 | 1 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | D, DD |
| 8 | 2 | 2 | 0 | 2 | 2 | 2 | 0 | 0 | 2 | 1 | 1 | 2 | 1 | 2 | 2 | 0 | 1 | 1 | 1 | E, EE, R, RR |
| 9 | 2 | 2 | 0 | 2 | 0 | 2 | 0 | 0 | 1 | 2 | 2 | 2 | 1 | 2 | 2 | 0 | 1 | 1 | 1 | K, KK |
| 10 | 2 | 0 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | GG, N |
| 11 | 0 | 2 | 0 | 2 | 2 | 0 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 1 | 1 | 1 | 1 | P |
| 12 | 2 | 2 | 0 | 2 | 2 | 0 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 1 | 1 | 1 | 1 | PP |
| 13 | 2 | 2 | 2 | 0 | 2 | 0 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 0 | 1 | 1 | 1 | 1 | Q, QQ |
| 14 | 2 | 0 | 0 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | U, UU |
| 15 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 1 | 2 | 2 | 2 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | MM |
| 16 | 2 | 2 | 0 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | VV |
| 17 | 2 | 2 | 0 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 2 | 1 | 1 | 1 | 1 | I, II |
| 18 | 2 | 2 | 0 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | L, LL, O, OO, S, SS |
| 19 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | F, FF, G, J, JJ, NN, V, C, CC, BB |
PATS types are designated arbitrarily with different numbers.
Prefix of each PATS primer pair A-B and virulence gene primer pair F-R is indicated 0, no amplicon; 1, amplicon without XbaI or AvrII site; 2, amplicon with one XbaI or AvrII site; 3, amplicon with two AvrII sites.
Isolates of E. coli O157:H7 from various outbreaks that fell within a given PATS type.
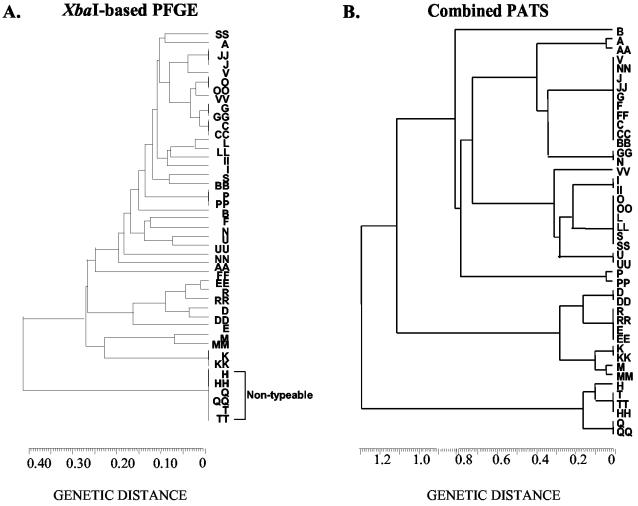
FIG. 3.
Analyses of relatedness of O157 isolates comparing XbaI-based PFGE with combined-PATS data. The dendrograms were constructed using the unweighted-pair-group method with arithmetic mean. In a previous study, PFGE gels were analyzed using Molecular Analyst Fingerprinting Plus software (Bio-Rad), and the data were exported as a band-matching table so that the two sets of data could be analyzed by the same method (18, 19).
The previously reported PFGE profiles had identified the same paired O157 isolates as identical (isolates C-CC, G-GG, J-JJ, K-KK, O-OO, and P-PP) for six outbreaks, closely related (one to three band differences; isolates A-AA, D-DD, L-LL, M-MM, N-NN, R-RR, and S-SS) for seven outbreaks, more distantly related (four to six band differences; isolates B-BB, E-EE, F-FF, I-II, U-UU, and V-VV) for six outbreaks, and untypeable (isolates H-HH, Q-QQ, and T-TT) for three outbreaks (19) (Fig. 3). As combined PATS specifically typed every O157 isolate and the system targeted specific sequences in the genome rather than relying on electrophoretic mobilities of genomic fragments, PATS was able to resolve the ambiguity in PFGE profiles. Specifically, PATS resolved the more distantly related and untypeable isolates, with B-BB and V-VV being identified as different, isolates H-HH being identified as similar, and isolates E-EE, F-FF, I-II, U-UU, Q-QQ, and T-TT being identified as identical (Table 3 and Fig. 3).
DISCUSSION
AvrII-based PATS allowed the identification of new polymorphic regions along the O157 genome in addition to those identified by XbaI-based PATS (18, 19). Specifically, 7 of the 33 AvrII restriction site-containing regions, yielding amplicons IKNR3, IKNR7, IKNR10, IKNR12, IKNR16, IKNR27, and IKNR33, targeted by AvrII-based PATS were determined to be polymorphic. The polymorphisms at these sites were found to be due to either indels or SNPs. AvrII-based PATS by itself provided poor discrimination of the 44 O157 isolates tested. However, when primer pairs amplifying the seven polymorphic AvrII sites were combined with those amplifying the eight polymorphic XbaI sites and the four virulence gene regions (combined PATS), the discriminatory power of PATS was enhanced.
PATS typed every O157 isolate tested in a reproducible manner, providing simple, accurate, and easy-to-interpret results. The consistent improvement achieved in this typing system by analyzing polymorphisms at different infrequently cutting restriction enzyme sites is reflected in the number of groups into which the 44 O157 isolates tested were distributed at each step. AvrII-based PATS was least discriminatory, dividing the O157 isolates into 11 groups. XbaI-based PATS divided the isolates into 14 groups, showing better discrimination, while combined PATS organized the isolates into 19 groups, reflecting increased discrimination (19) (Tables 1 and 3). Combined PATS was able to match the paired O157 isolates from 20 of the 22 outbreaks as identical (14 outbreaks) or closely related (6 outbreaks) while identifying the paired isolates from 2 outbreaks as different (Table 3 and Fig. 3). These O157 isolates (B-BB and V-VV), which were different by combined PATS, also had substantially different PFGE profiles (19). Likewise, most of the isolates from different outbreaks that shared the same combined PATS group also shared the same clade by PFGE (Table 3 and Fig. 3). In comparison, PFGE was able to match the paired isolates from only 13 of the 22 outbreaks as identical (6 outbreaks) or closely related (7 outbreaks); isolates from 6 outbreaks were grouped as more distantly related, while isolates from 3 outbreaks could not be typed (19) (Fig. 3). Current PFGE interpretive criteria may have overdiscriminated these isolates, as even those exhibiting a single band difference in their profiles were categorized as different. Spurious incompletely digested and/or comigrating DNA bands could have easily contributed to the incorrect assessment of isolates as being closely or more distantly related (6). In addition, we cannot rule out possible misclassification of O157 isolates to the outbreaks, as these organisms were collected at a time when subtyping was not available (19). On the other hand, PATS provided comparable, and in some instances better, discriminatory and relating abilities than PFGE. PATS, with its straightforward and less subjective results, related better to the epidemiological data than PFGE for this set of O157 isolates.
The difference between the distributions and natures of polymorphisms in XbaI and AvrII restriction sites may have contributed to the significant differences we saw in the PATS results based on these individual sites (18) (Table 1 and Fig. 3). Despite an AT-rich recognition site, only 40 XbaI restriction sites occur in the O157 strain EDL 933 genome with a 50% G+C content. Of these, 18 XbaI sites occur in backbone sequences and 22 XbaI sites occur in O-island sequences (18). All of the 18 backbone sequences and 14 of the O-island sequences containing an XbaI site are conserved among the O157 isolates tested. Polymorphic sequences containing an XbaI site are restricted to eight O islands and are all caused by indels (18). In contrast, AvrII, with a G+C-rich recognition site, has 33 sites in the O157 strain EDL 933 genome. Of these, 23 AvrII sites occur in backbone sequences and 10 occur in O-island sequences. Twenty-one backbone and five O-island sequences with AvrII sites are conserved among the O157 isolates tested, while the remaining two backbone and five O-island sequences with AvrII sites are polymorphic. These polymorphisms have been caused by both indels and SNPs. XbaI, with more sites on the O157 genome that are primarily distributed in O islands and exhibiting polymorphism caused by indels only in O islands, is probably best suited to differentiate O157 strains. Since most of the O islands are associated with phage or insertion elements capable of undergoing recombination, these may play a major role in causing polymorphisms between strains (5, 27, 36). AvrII, with fewer sites, most of which occur in backbone sequences, exhibited polymorphisms caused by SNPs in both backbone and O-island sequences that may accumulate over time, and this in turn may limit its ability to discriminate between strains (20, 26, 34).
Overall, analysis of restriction site polymorphisms in O157 indicates that twice as many indels as SNPs (10 indels and 5 SNPs) influence the process of diversification in the organism. Indels play a significant role in the evolutionary change and adaptation of bacteria, especially enterobacteria (5, 20, 26, 32, 34). Even the sequenced O157 genome contains 1.34 Mb of DNA that is not present in the K-12 genome and lacks 530 kb of DNA that is present in the K-12 genome (33). Similar indel-directed differences between the two sequenced O157 strains, EDL 933 and Sakai, can be observed (5, 13, 33). The probable existence of O157 in hypermutable states, due to defects in its methyl-directed mismatch repair system, may contribute to the large number of horizontal transfer events occurring in the genome, leading to genetic variation in the serotype and its strains (21, 22, 28). A recent study comparing closely related species and strains from six very distant systematic groups, including apes, sea urchins, bacteria, insects, nematodes, and plants, has shown that indels are responsible for many more unmatched nucleotides between closely related DNAs than are base substitutions (5).
Although most pathoadaptive mutations in bacteria that provide a selective advantage for the organism are due to indels occurring in structural and regulatory genes, SNPs, either singly or in numbers, can also influence this process (40). For instance, SNPs in the E. coli fimbrial gene can greatly influence the tropism of the organism to a variety of host cell and mucosal surfaces (40). Several SNPs have been identified in the O157 strain EDL 933 genome, especially in the backbone sequences, although the significance of each of these needs to be deciphered (33). These SNPs are primarily transitions (three transitions and one transversion). A bias toward T↔C transitions over A↔G transitions on the coding strand has been observed, with G↔T being the most common transversion (33). SNPs, primarily transitions (four transitions and one transversion), also contributed to the restriction site polymorphisms we studied. While this observation matched the trend for the O157 genome, the nucleotide bases involved in the predominant transitions and transversions were different. Three of the transitions were A↔G and one was C↔T on the coding strand. The one transversion was A↔C on the coding strand. Hence, the SNPs we observed belonged to the underrepresented group of SNPs found along the O157 genome.
PFGE is currently the standard strain-typing technique used by various public health and diagnostic laboratories to determine the relatedness of outbreak or nosocomial O157 and other bacterial isolates (2, 4, 16). Compared to the available strain-typing methodologies, PFGE provides relatively distinctive profiles for strains in several serotypes, making it a popular strain-typing tool. However, challenges in using PFGE include the fact that it is a labor-intensive and time-consuming technique that requires expensive instrumentation and software to interpret the complex electrophoretic patterns generated, and there are several factors inherent to the protocol itself that influence the patterns generated and lead to misclassification of strains (12, 15, 23). Some of the factors influencing the electrophoretic patterns of DNA in PFGE are improper storage and repeated subculturing of the isolates used to prepare the genomic DNA, which can cause spontaneous loss of plasmids or other recombinational events that alter DNA profiles; nuclease-related or electrophoresis-related degradation of DNA; methylation of DNA, causing incomplete restriction digestion of DNA; inactivation of restriction enzymes by reagents used to prepare DNA; improper resolution of smaller or larger DNA fragments, depending on the size limits of gel conditions; comigration of similar-sized DNA fragments; and nonhomologous DNA migrating as same-size bands, resulting in untypeable or incorrect profiles (3, 6, 23). In fact, PFGE fails to resolve all fragments generated after restriction digestion of the DNA on the gel under a single set of conditions, and the number of bands visualized does not match the number of sites for the restriction enzyme used to digest the DNA (3, 4, 6, 12). The choice of electrophoresis conditions, restriction enzymes, and software for the analysis of DNA patterns can alter PFGE profiles for the same strain, making it difficult to interpret and compare data (3, 6, 35). These drawbacks continue despite several measures to improve the technique by adding thiourea or HEPES to the running buffer to reduce degradation of DNA, the simplification of the protocol to make it more rapid, and the interpretation criteria designed by Gautom, Koort et al., and Tenover et al. to compensate for changes in profiles for the same strain under the conditions described above (9, 17, 38).
DNA sequence-based typing methods, such as multilocus sequence typing, are potential alternatives for PFGE, as they offer shorter assay times, less subjectivity in the interpretation of results, comparable and transferable data, and ease of automation (25). However, these techniques still rely on expensive instrumentation and software to interpret data (29). Most importantly, these typing systems are inefficient in global analysis of the genome, showing a bias toward a select set of sequences or genes that may or may not have sufficient diversity among strains of a pathogen. Also, sequencing errors may influence the results (25).
PATS provides a simple, user-friendly, straightforward, easy-to-perform and -interpret alternative to the existing techniques for bacterial strain typing. The currently standardized combined PATS is at least as good as PFGE in matching like and discriminating unlike O157 isolates. This is made possible by the ability of PATS to exploit both indels and SNPs in sequences along the bacterial genome. By doing so, the system addresses the issue of sequence variations at various sites along the bacterial genome without extensive instrumentation and helps to differentiate between homologous and nonhomologous regions. PATS is a PCR-based technique with high typeability and reproducibility, as it is not hindered by DNA methylation. The PCR format also makes it cost-effective, as it utilizes common laboratory instruments and reagents. The assay time, which is already shorter than that of PFGE or multilocus sequence typing, could be further reduced by automation if necessary. There is less subjectivity in the interpretation of results, and the data generated are easily comparable and transferable between laboratories. We are evaluating combined PATS against a new set of independent O157 isolates linked to various outbreaks to further validate our observations and to set up the interpretive criteria for the typing system. We are also working to automate the system using a DNA microarray platform and are in the process of expanding the application of PATS to other organisms.
Acknowledgments
We thank A. D. O'Brien for providing O157 strain 86-24.
This study was supported by funding from the Centers for Disease Control and Prevention-Association of Public Health Laboratories to S.B.C. and I.T.K. This work was also supported in part by the Center for Integration of Medicine and Innovative Technology (CIMIT) through U.S. Army Medical Research and Materiel Command Cooperative Agreement DAMD17-02-2-0006 awarded to S.B.C., I.T.K., and M.J.
REFERENCES
- 1.Arbeit, R. D. 1995. Laboratory procedures for the epidemiologic analysis of microorganisms, p. 190-208. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 6th ed. ASM Press, Washington, D.C.
- 2.Barrett, T. J., H. Lior, J. H. Green, R. Khakhria, J. G. Wells, B. P. Bell, K. D. Greene, J. Lewis, and P. M. Griffin. 1994. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J. Clin. Microbiol. 32:3013-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birren, B., and E. Lai. 1993. Pulsed-field gel electrophoresis—a practical guide. Academic Press, Inc., San Diego, Calif.
- 4.Bohm, H., and H. Karch. 1992. DNA fingerprinting of Escherichia coli O157:H7 strains by pulsed field gel electrophoresis. J. Clin. Microbiol. 30:2169-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Britten, R. J., L. Rowen, J. Williams, and R. A. Cameron. 2003. Majority of divergence between closely related DNA samples is due to indels. Proc. Natl. Acad. Sci. USA 100:4661-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis, M. A., D. D. Hancock, T. E. Besser, and D. R. Call. 2003. Evaluation of pulsed-field gel electrophoresis as a tool for determining the degree of genetic relatedness between strains of Escherichia coli O157:H7. J. Clin. Microbiol. 41:1843-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dieffenbach, C. W., and G. S. Dveksler. 1995. PCR primer—a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 8.Don, R. H., P. T. Cox, B. J. Wainwright, K. Baker, and J. S. Mattick. 1991. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19:4008.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gautom, R. K. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 35:727-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffin, P. M. 1995. Infections of the gastrointestinal tract, p. 739-761. Raven Press, Ltd., New York, N.Y.
- 11.Griffin, P. M., S. M. Ostroff, R. V. Tauxe, K. D. Greene, J. G. Wells, J. H. Lewis, and P. A. Blake. 1988. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann. Intern. Med. 109:705-712. [DOI] [PubMed] [Google Scholar]
- 12.Harsono, K. D., C. W. Kaspar, and J. B. Luchansky. 1993. Comparison and genomic sizing of Escherichia coli O157:H7 isolates by pulsed-field gel electrophoresis. Appl. Environ Microbiol. 59:3141-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 14.Hill, W. E., B. J. Tenge, and K. C. Jinneman. 1995. DNA subtyping and pattern analysis methods for bacterial pathogens: who's who in an outbreak. Clin. Microbiol. Newsl. 17:137-142. [Google Scholar]
- 15.Johnson, J. M., S. D. Weagant, K. C. Jinnemen, and J. L. Bryant. 1995. Use of pulsed-field gel electrophoresis for epidemiological study of Escherichia coli O157:H7 during a food-borne outbreak. Appl. Environ. Microbiol. 61:2806-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaper, J. B., and A. D. O'Brien. 1998. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. ASM Press, Washington, D.C.
- 17.Koort, J. M. K., S. Lukinmaa, M. Rantala, E. Unkila, and A. Siitonen. 2002. Technical improvement to prevent DNA degradation of enteric pathogens in pulsed-field gel electrophoresis. J. Clin. Microbiol. 40:3497-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kudva, I. T., P. S. Evans, N. T. Perna, T. J. Barrett, F. M. Ausubel, F. R. Blattner, and S. B. Calderwood. 2002. Strains of Escherichia coli O157:H7 differ primarily by insertions or deletions, not single-nucleotide polymorphisms. J. Bacteriol. 184:1873-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kudva, I. T., P. S. Evans, N. T. Perna, T. J. Barrett, G. J. DeCastro, F. M. Ausubel, F. R. Blattner, and S. B. Calderwood. 2002. Polymorphic amplified typing sequences provide a novel approach to Escherichia coli O157:H7 strain typing. J. Clin. Microbiol. 40:1152-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawrence, J. G., and H. Ochman. 1998. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. USA 95:9413-9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeClerc, J. E., B. Li, W. L. Payne, and T. A. Cebula. 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274:1208-1211. [DOI] [PubMed] [Google Scholar]
- 22.LeClerc, J. E., B. Li, W. L. Payne, and T. A. Cebula. 1999. Promiscuous origin of a chimeric sequence in the Escherichia coli O157:H7 genome. J. Bacteriol. 181:7614-7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murase, T., S. Yamai, and H. Watanabe. 1999. Changes in pulsed-field gel electrophoresis patterns in clinical isolates of enterohemorrhagic Escherichia coli O157:H7 associated with loss of Shiga toxin genes. Curr. Microbiol. 38:48-50. [DOI] [PubMed] [Google Scholar]
- 24.Musser, J. M. 1996. Molecular population genetic analysis of emerged bacterial pathogens: selected insights. Emerg. Infect. Dis. 2:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noller, A. C., M. C. McEllistrem, O. C. Stine, J. G. Morris, Jr., D. J. Boxrud, B. Dixon, and L. H. Harrison. 2003. Multilocus sequence typing reveals a lack of diversity among Escherichia coli O157:H7 isolates that are distinct by pulsed-field gel electrophoresis. J. Clin. Microbiol. 41:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ochman, H., J. G. Lawrence, and E. A. Groisman. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299-304. [DOI] [PubMed] [Google Scholar]
- 27.Ohnishi, M., C. Tanaka, S. Kuhara, K. Ishii, M. Hattori, K. Kurokawa, T. Yasunaga, K. Makino, H. Shinagawa, T. Murata, K. Nakayama, Y. Terawaki, and T. Hayashi. 1999. Chromosome of the enterohemorrhagic Escherichia coli O157:H7; comparative analysis with K-12 MG1655 revealed the acquisition of a large amount of foreign DNAs. DNA Res. 6:361-368. [DOI] [PubMed] [Google Scholar]
- 28.Ohnishi, M., J. Terajima, K. Kurokawa, K. Nakayama, T. Murata, K. Tamura, Y. Ogura, H. Watanabe, and T. Hayashi. 2002. Genomic diversity of enterohemorrhagic Escherichia coli O157 revealed by whole genome PCR scanning. Proc. Natl. Acad. Sci. USA 99:17043-17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olive, D. M., and P. Bean. 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paton, A. W., and J. C. Paton. 2002. Direct detection and characterization of Shiga toxigenic Escherichia coli by multiplex PCR for stx1, stx2, eae, ehxA, and saa. J. Clin. Microbiol. 40:271-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paulsen, I. T., L. Banerjei, G. S. A. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daughtery, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 33.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 34.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 35.Rementeria, A., L. Gallego, G. Quindos, and J. Garaizar. 2001. Comparative evaluation of three commercial software packages for analysis of DNA polymorphism patterns. Clin. Microbiol. Infect. 7:331-336. [DOI] [PubMed] [Google Scholar]
- 36.Shaikh, N., and P. I. Tarr. 2003. Escherichia coli O157:H7 Shiga toxin-encoding bacteriophages: integrations, excisions, truncations, and evolutionary implications. J. Bacteriol. 185:3596-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor, D. E., M. Rooker, M. Keelan, L. Ng, I. Martin, N. T. Perna, N. T. V. Burland, and F. R. Blattner. 2002. Genomic variability of O islands encoding tellurite resistance in enterohemorrhagic Escherichia coli O157:H7 isolates. J. Bacteriol. 184:4690-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.U.S. Department of Agriculture. 1997. An update: Escherichia coli O157:H7 in humans and cattle. Report from Centers for Epidemiology and Animal Health, p. 1-28. U.S. Department of Agriculture, Washington, D.C.
- 40.Weissman, S. J., S. L. Mosley, D. E. Dykhuizen, and E. V. Sokurenko. 2003. Enterobacterial adhesins and the case for studying SNPs in bacteria. Trends Microbiol. 11:115-117. [DOI] [PubMed] [Google Scholar]