Abstract
Candida albicans hyphae grow in a highly polarized fashion from their tips. This polarized growth requires the continuous delivery of secretory vesicles to the tip region. Vesicle delivery depends on Sec2p, the Guanine Exchange Factor (GEF) for the Rab GTPase Sec4p. GTP bound Sec4p is required for the transit of secretory vesicles from the trans-Golgi to sites of polarized growth. We previously showed that phosphorylation of Sec2p at residue S584 was necessary for Sec2p to support hyphal, but not yeast growth. Here we show that on secretory vesicles SEC2 mRNA is physically associated with Sec2p. Moreover, we show that the phosphorylation of S584 allows SEC2 mRNA to dissociate from Sec2p and we speculate that this is necessary for Sec2p function and/or translation. During hyphal extension, the growing tip may be separated from the nucleus by up to 15 μm. Transport of SEC2 mRNA on secretory vesicles to the tip localizes SEC2 translation to tip allowing a sufficient accumulation of this key protein at the site of polarized growth.
Introduction
Candida albicans hyphae show extreme polarized growth from their tip (Soll et al., 1985). This polarized growth requires a flow of post-Golgi secretory vesicles along actin cables. These deliver the additional membrane and enzymes that manufacture and re-model new cell wall required for tip extension (Sudbery, 2011; Caballero-Lima et al., 2013). Research in Saccharomyces cerevisiae has shown that at sites of polarized growth the vesicles are tethered to the exocyst, an octomeric protein complex attached to the inner surface of the plasma membrane (Terbush and Novick, 1995; Terbush et al., 1996). The vesicle-associated Rab GTPase Sec4p in its GTP-bound form is required for the late stages of the secretory pathway (Walworth et al., 1989). It mediates the tethering of vesicles with the exocyst by its interaction with the exocyst component Sec15p (Guo et al., 1999). The activating GEF of Sec4p is Sec2p, which is also vesicle associated, but released to the cytosol upon vesicle tethering to the exocyst (Walch-Solimena et al., 1997; Medkova et al., 2006; Novick et al., 2007). Thus there are vesicle-associated and cytosolic pools of Sec2p. At the tip of C. albicans hyphae both Sec2p and Sec4p are localized to a subapical spot that is similar to the Spitzenkörper of filamentous fungi (Bishop et al., 2010; Jones and Sudbery, 2010). Localization of Sec2p to this Spitzenkörper is dependent on phosphorylation at S584 by Cdk1p (Bishop et al., 2010) In contrast to the localization of Sec2p and Sec4p, the exocyst components localize to a surface crescent (Bishop et al., 2010; Jones and Sudbery, 2010).
An important aspect of protein localization in diverse cell types cells is the asymmetric localization of the encoding mRNA (Shahbabian and Chartrand, 2012; Xing and Bassell, 2013). A well-studied example in S. cerevisiae is the localization of the mRNA encoding Ash1p to daughter buds (Long et al., 1997). This process that involves four localization signals or ‘Zip codes’, three in the ORF and one in the 3′ UTR (Chartrand et al., 1999). In this case the mRNA is present in an RNA–protein complex (RNP) in association with Puf6p, Khd1p, She2p, She3p and Myo4p (Bertrand et al., 1998; Boehl et al., 2000; Long et al., 2000; Takizawa and Vale, 2000; Irie et al., 2002; Gu et al., 2004). Khd1p and Puf6p repress ASH1 translation during transport (Irie et al., 2002; Gu et al., 2004; Paquin et al., 2007). The repressive effect of Khd1p is reversed by phosphorylation by the casein kinase Yck1p. She2p is an RNA-binding protein that shuttles between the nucleus and cytoplasm. She2p is loaded co-transcriptionally onto ASH1 mRNA and forms a complex with a nuclear proteins Loc1p and Puf6p, (Shen et al., 2010; Shahbabian et al., 2014). Puf6p also binds at least 40 other transcripts in a She2p-dependent fashion, including mRNAs destined to transported to the bud. Myo4p, is a class V myosin that mediates the movement of RNPs along actin cables. She3p is the bridging protein that links She2p and Myo4p. She3p is also an RNA-binding protein so that recognition of ASH1 mRNA and its incorporation of into a ribonucleoprotein (RNP) complex requires both She2p and She3p (Müller et al., 2011). The efficient transport of ASH1 mRNA to daughter buds requires the polarized growth machinery since mutations affecting a wide range of polarity components, such as Sec4p, Cdc42p and exocyst components, reduces or abolishes ASH1 mRNA localization (Aronov and Gerst, 2004). Bud-directed RNPs associate with the cortical ER and their transport is dependent on genes required for cortical ER segregation such as MYO4 and SHE3 (Aronov et al., 2007). Consistent with the idea that RNPs are co-transported with the cortical ER to the daughter bud, She2p binds to tubular ER membranes in vitro (Shen et al., 2010; Genz et al., 2013).
Microarray experiments have demonstrated that many other mRNAs besides ASH1 can also be associated with She3p (Shepard et al., 2003). In a separate study it was shown that mRNAs that encode polarity proteins, including Sec4p are themselves transported to the sites of polarized growth in a She3p-dependent fashion (Aronov et al., 2007). These mRNAs colocalize at the tip of daughter buds with the proteins they encode. In a she3 mutant mRNA protein localization is reduced but not abolished. Thus, mRNA localization increases the efficiency of protein localization, but is not essential. ASH1 is also asymmetrically localized in C. albicans (Inglis and Johnson, 2002), and it has been shown that a number of mRNAs are asymmetrically localized in a She3p-dependent fashion (Elson et al., 2009). This She3p-dependent set of mRNAs did not include any of the proteins that have been shown to localize to the hyphal tip; and apart from ASH1, there was only limited overlap with the set of genes that is associated with She3p in S. cerevisiae. C. albicans lacks an obvious She2p homologue. In view of the finding that She3p can recognize and bind ASH1 mRNA, it is possible that She3p is solely responsible for recognizing and binding to ASH1 mRNA in C. albicans.
The tip of C. albicans hyphae can be a considerable distance from the nucleus. For example, before nuclear migration into the germ tube, which precedes mitosis, the tip may be up to 15 μm from the nucleus. In contrast the bud tip of a S. cerevisiae diploid cell is unlikely to be more than 5 μm from the nucleus. We hypothesized that it was likely that the strongly polarized distribution of proteins at the hyphal tip may be mediated by a corresponding asymmetric distribution of their encoding mRNAs. Such an idea is supported by electron micrographs in other fungal hyphae which show that the Spitzenkörper is rich in polyribosomes suggesting that it is the site of intense protein translation (Grove et al., 1970). We reasoned that the mRNAs may be transported on secretory vesicles since ASH1 mRNA transport in S. cerevisiae is dependent on components of the late secretory pathway such as Sec4p and the exocyst and localization of mRNAs encoding polarized proteins is dependent on She2p, She3p and Myo4p. We show here that mRNA is present in the immunoprecipitate of Sec2p, but surprisingly, the only mRNA present is the mRNA encoding Sec2p itself. Cell fractionation experiments shows that Sec2p mRNA co-fractionates with secretory vesicles, moreover the interaction of Sec2p with its RNA is lessened in a C-terminal truncation and is abolished by the S584E phosphomimetic substitution. Thus, secretory vesicles are not only associated with Sec2p but they also carry the mRNA that encodes Sec2p and this association is regulated by Sec2p phosphorylation at residue S584.
Results
Sec2p is associated with mRNA
To investigate whether mRNAs that encode proteins localized to the tip are themselves asymmetrically localized we used Sec2p in an RNA Immune Precipitation (RIP) experiment as an example of a protein showing localization the Spitzenkörper of C. albicans hyphae. We immune precipitated Sec2p–YFP from hyphal and yeast cells using a monoclonal antibody against GFP and Gin4p–GFP, which is not polarized to the tip, as a control. The immune precipitates (IPs) were then incubated with 32P-labelled dCTP, Oligo dT and reverse transcriptase. Figure 1A shows that the RIP from Sec2p–YFP, but not Gin4p–GFP programmed the synthesis of cDNAs. In a separate experiment we investigated whether there was a difference in hyphae compared to yeast (Fig. 1B). In this experiment we used immune precipitated YFP from cells expressing YFP from the PGK1 promoter as a control. The signal was clearly stronger from the hyphal sample compared to the yeast sample. These RIP experiments clearly show that mRNA molecules are associated with Sec2p and suggest that this association is greater in hyphal cells compared to yeast cells.
Figure 1.
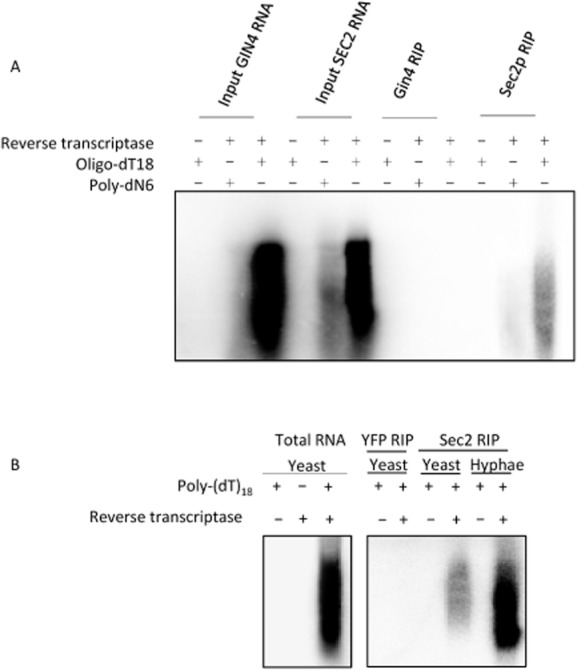
Sec2p is associated with polyA+ RNA and is more strongly associated in hyphae compared to yeast.
A. Strains expressing Sec2p–YFP or control Gin4p–GFP as indicated were used in an RNA immune precipitation experiment (RIP). Any mRNA that was present in the immune precipitates was used as a template for RT PCR using either polydT (18) or randomized polyN6 primers, 32-P dCTP and Superscipt III reverse transcriptase as described in Experimental procedures.
B. An RIP experiment using lysates from Yeast YFP expressed from the constitutive PGK1 promoter as a negative control and randomized Poly-(dT)18 as primers.
The mRNA associated with Sec2p is SEC2 mRNA
In order to identify the mRNAs associated with Sec2p we hybridized the cDNA derived from a Sec2p-3xFLAG IP to a genomic microarray using Gin4p-3xFLAG as a control. The top 10 hits from this microarray are shown in Fig. 2A. Remarkably, the top hit was Sec2p itself with a ratio compared to the Gin4p-3xFLAG control of 7.63. The next hit was HIS1 with a ratio to Gin-3xFLAG of 5.49. Since the Sec2p-3xFLAG strain was HIS1 whereas the Gin4p-3xFLAG strain was Δhis1 this result shows that the enrichment of SEC2 mRNA in the IP compared to the control is as least as great as the dynamic range of the microarray. In order to verify the presence of SEC2 mRNA and to investigate whether the remaining hits were genuine, we carried out qRT-PCR on the Sec2p-3-FLAG IP using primers to SEC2 and to a selection of five of the remaining hits. SEC2 mRNA was consistently found to be enriched in the IP (Fig. 2B). In contrast, none of the other apparent hits from the microarray showed enrichment in qRT-PCR experiments. Figure 2B also shows that the association between the SEC2 mRNA and Sec2p is dependent on Mg2++. Thus, Sec2p is associated only with its own encoding mRNA. Moreover, although there is a small enrichment in IPs from yeast cells, the enrichment in hyphae is up to three times greater, showing that the association of Sec2p mRNA with Sec2p is a feature of the strongly polarized growth of hyphae (Fig. 3A). The function of the C-terminal domain of Sec2p remains unknown. To determine if the C-terminal domain is required for binding SEC2 mRNA we repeated the RIP experiment Sec2p1–625 which lacks 126 C-terminal residues. The resulting qRT-PCR shows a significant reduction in SEC2 mRNA binding compared to control full-length Sec2p. This results show that either the C-terminal residues of Sec2p or the corresponding region of the encoding mRNA are required for binding (Fig. 3B).
Figure 2.
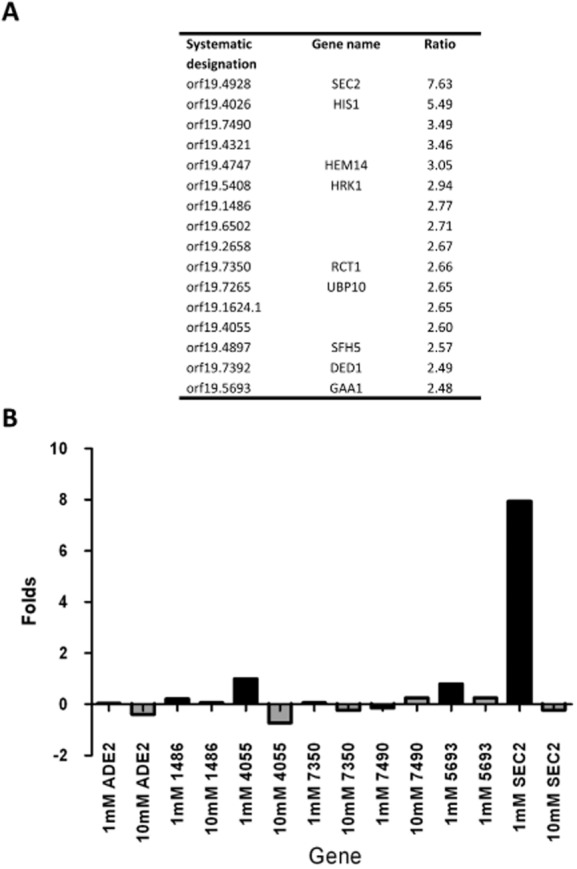
Sec2p–GFP is specifically associated with SEC2 mRNA.
A. The top 16 hits from a microarray analysis of the Sec2p-3xFLAG RIP experiment. The data are presented as the ratio of the indicated RNA to the control Gin4p-3xFLAG RIP.
B. Verification of the microarray data for SEC2 RNA and representative genes shown in panel A. qRT-PCR was performed using templates for the indicated genes when the lysis buffer contained either 1 mM EDTA or 10 mM EDTA to remove magnesium ions generally required for protein–RNA interactions. The gene names are shown in abbreviated form with the orf19 prefix not shown. The ordinate shows the fold increase relative to control Gin4p-3xFLAG RIP.
Figure 3.
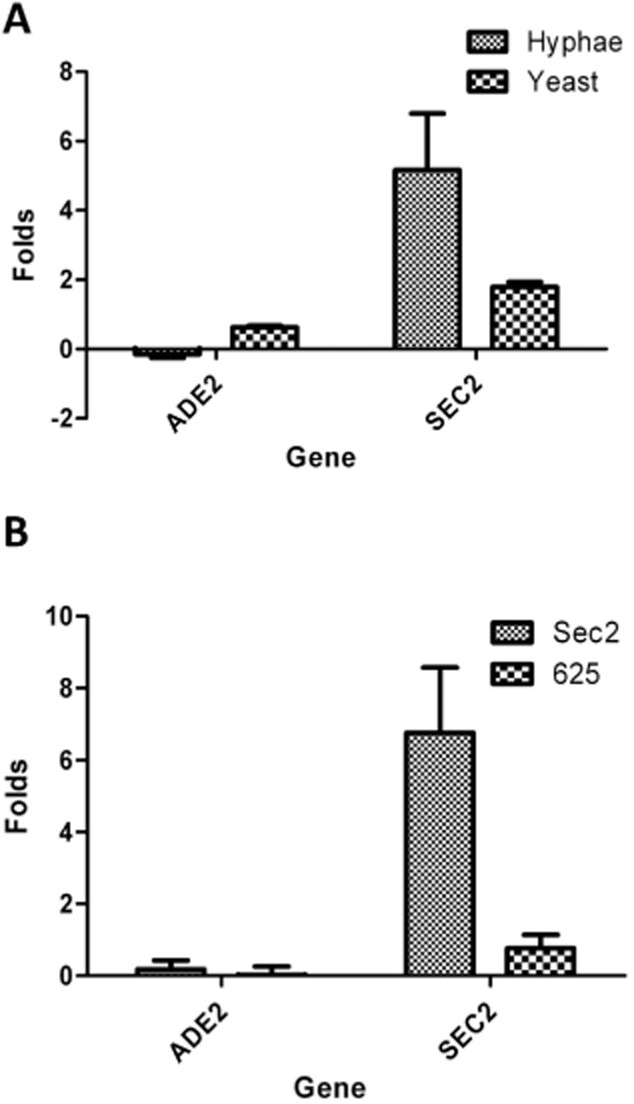
Sec2p shows a greater affinity for its mRNA in the hyphal form and requires the C-terminal domain for this affinity.
A. A RIP experiment showing the fold increase of SEC2 RNA associated with Sec2p-3xFLAG in hyphae compared to yeast.
B. A RIP experiment showing the fold increase of SEC2 RNA relative to Gin4p–GFP associated with full-length Sec2p–GFP (Sec2p1–751) compared to C-terminally truncated Sec2p (Sec2p1–625). Both experiments show the results of three biological replicates each with two technical replicates. In each case ADE2 was used as a negative control.
The association of SEC2 mRNA with Sec2p is independent of She3p-based transport mechanism
Previously a set of 40 mRNAs has been reported to be transported in a She3p-dependent manner in C. albicans hyphae (Elson et al., 2009). However, SEC2 mRNA was not a member of this set. In that study, the criteria for selecting these 40 mRNAs was that they were in the top 5% of genes showing an enrichment when the mRNAs present in a She3p RIP were analysed by a microarray experiment. It is possible that SEC2 mRNA was enriched, but at a level below this threshold. We therefore inspected the online data file of the microarray and found that SEC2 mRNA showed no enrichment. To investigate this further, we determined whether She3p and Sec2p physically associate in a reciprocal co-immunoprecipitation experiment (Fig. 4A). No association was observed. Next we determined whether Sec2p associates with SAP5 and RBT4 mRNAs that have been shown to be associated with She3p (Elson et al., 2009). We confirmed the enrichment of both of these mRNAs in a She3p IP, but there was no enrichment of these mRNAs in a Sec2p IP (Fig. 4B). We conclude that the association of SEC2 mRNA with Sec2p is independent of the She3p-based mRNA transport mechanism reported previously (Elson et al., 2009).
Figure 4.
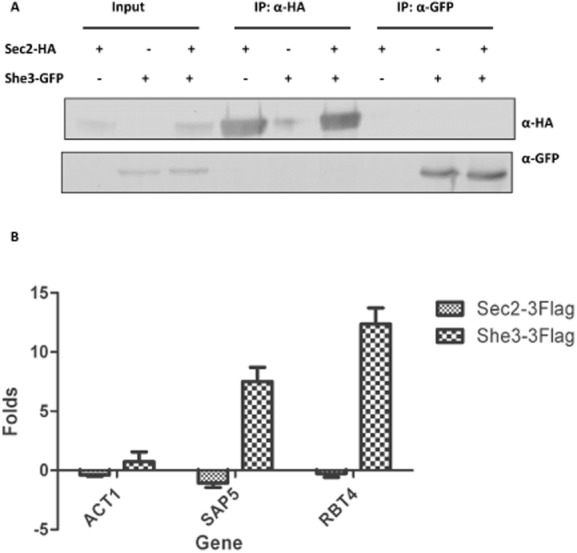
Sec2p is not associated with mRNAs transported by She3p.A.
A reciprocal immune precipitation experiment showing a lack of detectable association between She3p and Sec2p. Sec2p-HA or She3p–GFP were immune precipitated as indicated (IP). The immune precipitates were used in a Western blot experiment with monoclonal antibodies to GFP or HA as indicated.
B. RIP experiments were carried out with She3p-3xFLAG, Sec2p-3xFLAG and Gin4p-3xFLAG. RT-qPCR was carried out using primers to ACT1, SAP5 or RBT4. The chart shows the enrichment of the indicated RNAs in each immune precipitate relative to the Gin4p-3xFLAG control.
Sec2p colocalizes with SEC2 mRNA
In order to establish whether the association of Sec2p with its mRNA occurs in vivo we sought to colocalize Sec2p with its mRNA. We initially employed the MS2 system in which SEC2 mRNA was fused to a sequence containing 24 MS2 binding sites in a cell expressing MS2-binding protein fused to GFP. However, we were unable to detect a specific SEC2 mRNA signal. We next turned to a fluorescence in situ hybridization (FISH) strategy in which we colocalized the signal from Sec2p–GFP in the Spitzenkörper with the signal from the in situ hybridization of a panel of 10 oligonucleotides probes conjugated to FITC. This approach was complicated by the effect of fixation on Sec2p localization. At least in C. albicans, the Spitzenkörper is not an organelle-like structure with a fixed organization. Rather it appears to be an accumulation of secretory vesicles. Upon the fixation necessary for FISH these vesicles along with Sec2p are dispersed. To address this problem we progressively reduced both the fixation time and the concentration of the formaldehyde. We found that fixation for 5 min in 2% v/v formaldehyde resulted in the preservation of apical Sec2p localization in approximately 10–15% of the hyphae (Fig. 5A). A second problem is that the fluorescence of GFP is lost during FISH, so to localize Sec2p–GFP we used immunocytofluorescence with the secondary antibody fused to Alexofluor 633. This red fluorophore also allowed us to distinguish between the signals from FITC and GFP which would otherwise both have been in the green channel. We examined hyphae showing apical Sec2p–GFP localization to see if the mRNA signal from FISH colocalized with the Sec2p–GFP signal in the Spitzenkörper. We quantified any colocalization of the two signals using the Coloc2 plugin from the Fiji implementation of NIH ImageJ (Experimental procedures) that generates a Pearson correlation score (r), where a score of +1 indicates a perfect correlation. As a negative control we omitted the hybridization probe. Figure 5B shows the mRNA and Sec2p signals from two hyphal tips selected to show the range colocalization association observed. In the merged images the Sec2p signal is false coloured magenta so that areas of colocalization appear white. Figure 5C shows the r scores from all of the individual hyphal tips examined along with the mean and the standard deviation of the r scores. To show how the visual signals of colocalization are reflected in the r scores, the r scores from the actual hyphal tips shown in Fig. 5B are displayed with a magenta symbol in Fig. 5C. The data confirm that Sec2p–GFP colocalizes with its mRNA and that the mean of r scores is significantly different from the negative control and the non-binding Sec2p mutants described below.
Figure 5.
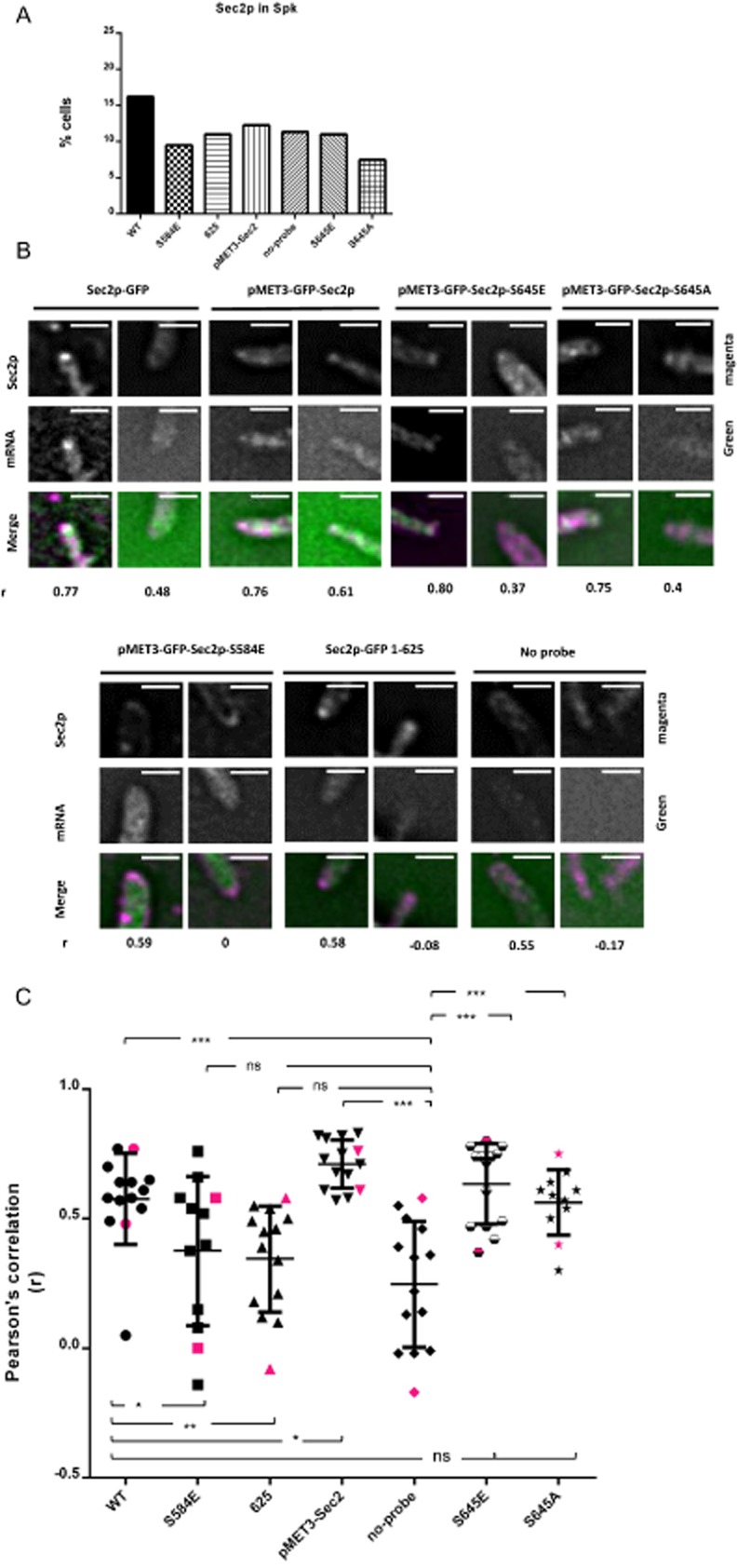
SEC2 mRNA colocalizes with Sec2p. Unbudded stationary-phase cells of the indicated genotype were induced to form hyphae for 90 min and briefly fixed with 2% v/v formaldehyde as described in the text and Experimental procedures. In the resulting hyphae Sec2p–GFP or GFP–Sec2p was visualized by immunocytofluorescence with a monoclonal anti-GFP antibody and a secondary antibody conjugated to red Alexafluor 633 (red); mRNA was visualized in the same hyphae by in situ hybridization to a panel of oligonucleotides conjugated to FITC (green).
A. Percentage hyphal tips showing apical localization of the GFP signal.
B. Two hyphal tips from each strain showing mRNA and GFP localization chosen to show the range of colocalization observed. The merged image is shown with the red signal from the GFP immunofluorescence false coloured magenta. With this green-magenta combination, which aids the colour-blind, colocalized signals appear white. The Pearson correlation coefficient (r) of colocalization is shown below each hypha. Scale bars: 5 microns.
C. Pearson correlation coefficients from all hyphae analysed in each strain. The magenta symbols indicate the correlation of the hyphae shown in panel B. Tests of significance between the data sets are shown by the horizontal brackets; *** indicates P ≤ 0.0005; ** indicates P ≤ 0.005; * indicates P ≤ 0.05; ns indicates not significant.
As well as examining the colocalization of Sec2p–GFP and its mRNA we also examined the colocalization of SEC2 mRNA in the following SEC2 alleles: First, in our original study of the role of phosphorylation in SEC2 function we constructed a number of alleles using the regulatable MET3 promoter to allow their conditional expression. We wished to test whether some of these alleles affected Sec2p binding to its mRNA. To do this it was first necessary to establish that the C-terminal GFP–Sec2p fusion bound its mRNA in the same way as the Sec2p–GFP N-terminal fusion. Figure 5B and C shows that GFP–Sec2p expressed from the MET3 promoter did indeed clearly colocalize with its mRNA. Note that this was a heterozygous construct so that there was a second wild-type SEC2 allele present. The slightly stronger and more consistent colocalization of GFP–SEC2 compared to SEC2–GFP may be due to slight overexpression of GFP–SEC2 from the MET3 promoter, and/or to the presence of two SEC2 alleles expressing mRNA in contrast to the single SEC2–GFP allele. Second, S645 is located in a region of low complexity which potentially may be part of an mRNA-binding domain. Recently it has been shown that RNA-binding proteins are enriched in low complexity regions and that phosphorylation of these regions reduces affinity of these proteins for RNA (Han et al., 2012). To test whether phosphorylation of S645 affects the affinity of Sec2p for its mRNA we used hemizygous phosphomimetic and non-phosphorylatable substitutions at this position (GFP–Sec2-S645E/sec2Δ and GFP–Sec2-S645A/sec2Δ). We found both forms of Sec2p colocalized with GFP–SEC2 mRNA in a nearly identical fashion to wild-type Sec2p–GFP (Fig. 5B and C). Importantly, because this strain was hemizygous, the only SEC2 mRNA species in these cells lacks the native 5′ UTR, thus the affinity of Sec2p for its mRNA does not require the 5′ UTR. Third, the phosphomimetic GFP–Sec2p-S584E failed to colocalize with its mRNA confirming the reduced affinity of this protein for its mRNA in the biochemical tests described above (Fig. 5B and C). Lastly, SEC21–625–GFP also failed to colocalize with its mRNA, again confirming the reduced affinity of this C-terminally truncated protein for its mRNA in the biochemical tests (Fig. 5B and C).
Sec2p mRNA is associated with secretory vesicles
One possible explanation for the association of SEC2 mRNA and its encoding protein is that IP is pulling down nascent Sec2p as it emerges from ribosomes. A precedent for this is the association of Abp140p with polyribosomes as it is translated on actin cables (Trautwein et al., 2004). Moreover, if the phenomenon was not due to the IP of nascent Sec2p, it was of interest to discover whether the association was occurring with the cytosolic pool of Sec2p or with Sec2p associated with secretory vesicles. To address both of these questions we followed the distribution of Sec2p-HA and SEC2 mRNA in a cell fractionation experiment. Cells were fractionated to produce 10 000 g, 30 000 g and 100 000 g pellets (P1–P3) and corresponding supernatants (S1–S3) (Fig. 6A). The P2 and P3 pellets were resuspended and further fractionated by Opitprep density centrifugation as previously described (Chang et al., 2008) (Fig. 6B). GFP–Arf1p was used to provide a Golgi marker [we verified that GFP–Arf1p has an intracellular distribution similar that shown for other Golgi proteins in C. albicans (Rida et al., 2006)]; Snc1p-Myc was used to provide a secretory vesicle marker; and an antibody to Rps3p was used to identify the fractions containing ribosomes (Frey et al., 2001). qRT-PCR was used to quantify SEC2 mRNA; ADE2 and ACT1 mRNAs were quantified as controls.
Figure 6.
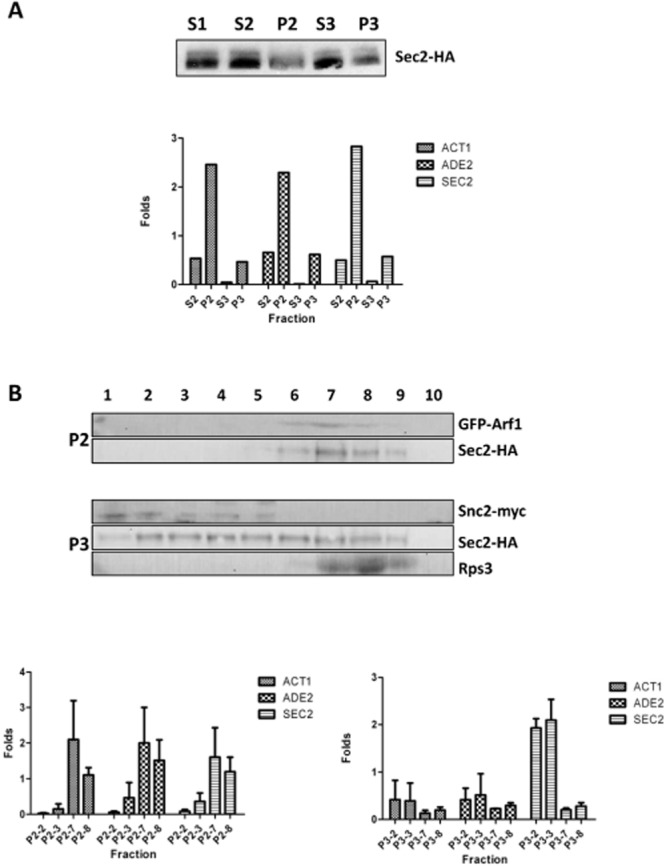
SEC2 mRNA co-fractionates with secretory vesicles.
A. Hyphal cells were harvested 90 min after induction and fractionated as described in Experimental procedures. Sec2p-HA was quantified by Western blot (top). ACT1, ADE1 and SEC2 mRNAs were quantified by qRT-PCR and fold enrichment compared to the concentration of each in the S1 supernatant. S1 = 10 000 g supernatant, S2 = 30 000 g supernatant, S3 = 100 000 g supernatant. P2 = 30 000 g pellet, P3 = 100 000 g pellet.
B. The P2 and P3 fractions were resuspended and further fractionated in an Optiprep density gradient. Fractions were collected and Arf1p–GFP, Sec2p-HA and the ribosomal protein Rps3p were detected by Western blotting. ACT1, ADE1 and SEC2 mRNAs were quantified by qRT-PCR and fold enrichment compared to the concentration of each in the S1 supernatant. Data shown are the mean and SEMs of three separate biological experiments.
The results show that Sec2p-HA is distributed in the S1–S3 supernatants and the P2 and P3 pellets (Fig. 6A). Sec2p-HA in the S3 fraction represents the cytosolic pool. SEC2 mRNA was found in both the S2, P2 and P3 fractions, but not in the S3 fraction (Fig. 6A). Thus SEC2 mRNA is not associated with the cytosolic pool of Sec2p. The density centrifugation of the resuspended P2 and P3 pellets show that Sec2p-HA co-fractionates with the Golgi marker GFP–Arf1p, the secretory vesicle marker Snc2p-myc, and to a more limited extent with the ribosomal marker Rps3p (Fig. 6B). Since secretory vesicles are derived from the Golgi, the association of Sec2p-HA with both Golgi-containing and secretory vesicle-containing fractions is to be expected. The overlapping distribution of Sec2p with ribosomal fractions may represent nascent Sec2p. The SEC2 mRNA and the control ADE2 and ACT1 mRNAs are enriched in fractions P2-7 and P2-8 which co-fractionate with the Golgi marker, Arf1p–GFP (Fig. 6B). Importantly, in three independent experiments SEC2 mRNA was consistently enriched in the P3-2 and P3-3 fractions which co-fractionate with the secretory vesicle marker Snc2p-myc, whereas there was no such enrichment of either of the control ACT1 or ADE2 mRNAs (Fig. 6B). Thus SEC2 mRNA, is associated with the secretory vesicle fraction but the control mRNAs are not. Moreover, there is no enrichment of SEC2 mRNA in the P3-7 and P3-8 fractions that overlap with the Rps3p positive fractions (Fig. 6B). Thus, the SEC2 mRNA that associates with the Sec2p is not due to the association of nascent protein with its mRNA on polyribosomes as it is translated.
Sec2p physically associates with mRNA
The association of SEC2 mRNA with secretory vesicles could be due to a direct physical association with Sec2p. Alternatively both SEC2 mRNA and Sec2p could be independently carried by secretory vesicles without any direct physical interaction. In this latter scenario the Sec2p RIP also pulls down secretory vesicles resulting in the apparent association of Sec2p and SEC2 mRNA. To address this question we carried out an RNA capture experiment. Lysates from a cell expressing either Sec2p–YFP, Sec21–625 or GFP–Sec2p S584E were UV irradiated to cross-link proteins to RNA and DNA. Cellulose-Oligo dT was then used to capture mRNA before proteins associated with the mRNA were analysed by Western blotting using a monoclonal antibody to GFP. Gin4pGFP was used as a negative control. The results show that Sec2p, but not Gin4p, directly associates with mRNA (Fig. 7). Moreover, the Sec21–625 truncation show no association, consistent with the reduction of the Sec21–625 association with SEC2 mRNA observed in RIP experiment (Fig. 3B). We previously showed that phosphorylation of a serine at residue 584 is necessary for Sec2p to support hyphal growth. GFP–Sec2p S584E showed no association with mRNA, even though it was expressed at a higher level than Sec2p which showed an association. This result suggests that the role of S584 phosphorylation is to programme the release of SEC2 mRNA.
Figure 7.
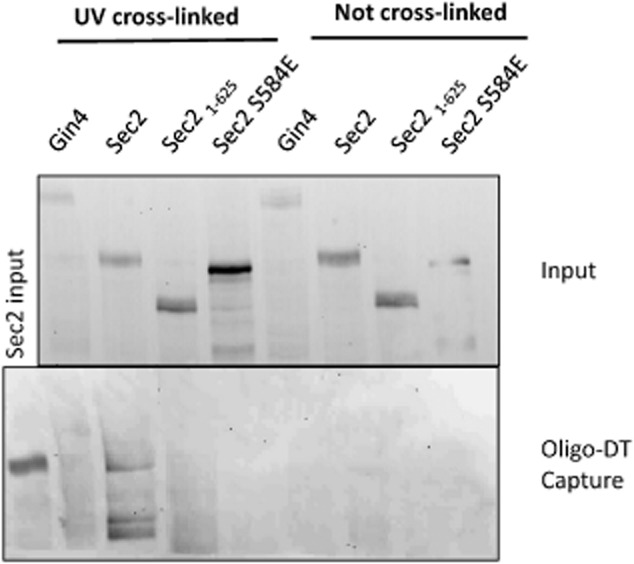
SEC2 mRNA is physically associated with Sec2p. Gin4p–GFP, Sec2p–YFP, Sec21–625 and GFP–SEC2 S584E as indicated were used in an RNA capture experiment as described in Experimental procedures. Membranes from the input and oligo-DT captured samples were probed with a monoclonal antibody to GFP.
Discussion
Previous work in S. cerevisiae has shown that mRNAs for polarity proteins are localized to the tips of small buds and colocalize with the encoded protein (Aronov et al., 2007). Moreover, this mRNA localization is depended on proteins involved in the late secretory pathway such as the GTPases Cdc42p and Rho3p. the exocyst components Sec10p and Sec15p, the Rab GTPase Sec4p and the v-SNARE components Sncp1/2 and the t-SNARE Sec9p (Aronov and Gerst, 2004). These observations led us to investigate the possibility that the mRNAs are transported on secretory vesicles. However, since polarized growth is restricted to a short period of the S. cerevisiae cell cycle, we used hyphae of the human fungal pathogen C. albicans which shows continuous polarized growth throughout the cell cycle. We immune precipitated Sec2p the GEF for Sec4p. We found evidence that mRNAs were associated with Sec2p, but remarkably, only SEC2 mRNA was present the immune precipitate and the association was independent of She3p. Sec2p is associated with vesicles, but upon vesicle tethering to the exocyst, it is thought to be displaced into the cytosol where the bulk of Sec2p is found (Medkova et al., 2006; Novick et al., 2007). Cell fractionation experiments showed that compared to control mRNAs, SEC2 mRNA is over-represented in the vesicle and Golgi fractions,. Thus, our experiments show that Sec2p is associated with its own mRNA as it travels on secretory vesicles. To our knowledge, the identification of GEF as an RNA-binding protein is unique among fungi. However, several Arf GTPases have been shown to bind mRNA in S. cerevisiae (Trautwein et al., 2004; Scherrer et al., 2010) and in mammalian cells, Arf-GEF1 was identified as a candidate RNA-binding protein in the Atlas of mRNA-binding proteins (Castello et al., 2012).
Examples of mRNA protein association in fungi
As well as the transport of ASH1 mRNA and mRNAs encoding polarity proteins by She2p/She3p/Myo4p-based process that has been extensively studied in S. cerevisiae, there are several other cases of mRNA transport that have been studied in fungi. In hyphae of the corn smut fungus Ustilago maydis, the RNA-binding protein Rrm4p has been shown to associate with a set of 63 mRNAs that are transported bidirectionally on microtubules by the motor proteins the dynein Dyn1p, and the kinesin Kin1p (König et al., 2009; Baumann et al., 2012). Rrm4p was shown to recognized CA-rich sequences in the 3′ UTR. Rrm4p RNPs colocalize with the endosomal protein Yup1 suggesting that Rrm4p RNPs are associated with endosomes which are also transported bidirectionally on microtubules by Dyn1p and Kin1p. Recently it has been shown that the early endosomes are associated with ribosomes. Significantly, the association of ribosomes with early endosomes is impaired in an rrm4 mutant and inhibitors of protein translation (Higuchi et al., 2014). Together these observations suggest that Rrm4p acts to load mRNAs onto early endosomes where they are translated. Such a mechanism may ensure that translation is equally distributed throughout the hypha. It has also been shown that septin RNA and septin protein are transported on the same endosomes, and that this is necessary for the correct delivery of septins to the cell poles (Baumann et al., 2014). This provides an interesting parallel to the present study – in both cases a protein and its encoding mRNA are transported on vesicles to their site of localization. In C. albicans 40 mRNAs have been shown to be associated with She3p (Elson et al., 2009). However, apart from ASH1, there was limited overlap with the gene set that associates with She3p in S. cerevisiae. This She3p-based mRNA transport is physiologically important as she3ΔΔ cells were defective invasive growth and epithelial cell damage. Although in situ hybridization showed that some of the She3p-associated transcripts were enriched in apical regions, none showed the specific localization to the Spitzenkörper that we have observed in the case of SEC2.
Our results show significant differences with this previous work as well as the She3p-based system in S. cerevisiae and C. albicans. First, the affinity of Sec2p for its own mRNA does not involve She2p/3p or any other previously characterized RNA-binding protein or motif in the RNA. We discuss below the way in which Sec2p may recognize its mRNA. Second, the affinity of Sec2p for its own mRNA is unusual in that it appears that only a single mRNA is bound, whereas most RNA-binding proteins recognize multiple RNAs. Third, the presence of polyribosomes and thus mRNA in the fungal Spitzenkörper has been known for some time, but our work for the first time establishes the identify of an mRNA localized in the Spitzenkörper.
How does Sec2p recognize bind to its mRNA?
In S. cerevisiae the preferred motifs for a number of RNA-binding proteins have been elucidated (Hogan et al., 2008). Many of these are located in either the 5′ or 3′ UTR. The SEC2-3xFLAG proteins we used in the RIP experiments, and the Sec2p–GFP we used in the colocalization experiments, lacked the native 3′ UTR and there was no other SEC2 mRNA species present which carried a 3′ UTR. Both of these proteins bound SEC2 mRNA, thus the interaction does not require the 3′ UTR. The colocalization experiments showed that both GFP–Sec2p S645E and GFP S645A proteins colocalized with SEC2 mRNA. Again there was no SEC2 mRNA species present which carried a 5′ UTR, thus the 5′ UTR is also not required for the interaction. The possibility that the SEC2 mRNA does not directly physically interact with Sec2p, but rather both are carried on secretory vesicles is eliminated by the RNA-capture experiments which demonstrate that Sec2p cross-links to oligo-dT purified mRNA. However, the S584E allele which mimics phosphorylation abolishes the interaction of Sec2p for its mRNA, while a C-terminal truncation Sec21–625 reduces the interaction. These observations suggest Sec2p region between 584 and 625 may be involved in the binding its mRNA.
The cellular location of the Sec2p interaction with its mRNA
It is difficult to envisage that Sec2p can shuttle between the hyphal tip and nucleus. However, recognition of cytosolic proteins for their cognate mRNAs has precedents. For example the survey of RNA binders in S. cerevisiae described above, found that 30% of RNA-binding proteins tested bound their own mRNAs, and many of these proteins were cytosolic (Scherrer et al., 2010). A similar finding was reported in survey of the RNAs bound by known RNA-binding proteins in S. cerevisiae (Hogan et al., 2008). Thus mechanisms must exist for the association of proteins and their cognate mRNAs. Many mRNAs encoding cytosolic proteins have been found to localize to the ER (Lerner et al., 2003). Moreover, the GTPase Arf1p, which is required for vesicle formation in the Golgi, has been found to associate with the mRNA-binding protein, Pab1p. This association was shown to be mRNA dependent. ASH1 was found to one of the associated mRNAs and its association was independent of She3p (Trautwein et al., 2004). However, asymmetric localization of ASH1 mRNA was abolished in an arf1 mutant. Interestingly, one of the mRNAs found to associate with Arf1p was the actin-binding protein ABP140, which was shown to localize to the distal pole of the mother cell (Kilchert and Spang, 2011). Localization of Abp140p mRNA is an interesting example of a protein that binds its own mRNA, because the first 17 amino acids of Abp140p forms an actin-binding motif; while the first 67 amino acids are sufficient to bind mRNA. Thus, it is proposed that as Abp140p is translated, the nascent polypeptide binds actin as it emerges from the polyribosome and thus tethering the polyribosome to actin cables for transport to the distal pole. Forcing the ectopic localization of Abp140p to different cellular locations resulted in the corresponding mislocalization of ABP140 mRNA. The above examples illustrate that mRNA association with proteins occurs with the ER, Golgi and on actin cables. Sec2p is a protein that cycles between the Golgi and the hyphal tip along actin cables. Figure 6 shows that SEC2 mRNA is detected in both Golgi and secretory vesicle fractions. We suggest that SEC2 mRNA is loaded onto vesicles in the Golgi before onward transport to the hyphal tip.
Possible roles of SEC2 association with its mRNA
An interesting role for the association of proteins with their cognate mRNAs has been demonstrated in the cases of mammalian thymidylate kinase (TS) and dihydofolate reductase (DHFR) which associate with their own cognate mRNAs and repress their own translation (Tai et al., 2004). In S. cerevisiae Khd1p binds to ASH1 mRNA and represses its translation (Paquin et al., 2007). Interestingly, phosphorylation of Khd1p by the casein kinase Yck1p, removes Khd1p from ASH1 mRNA and releases the translational repression. These examples suggest one possible role of Sec2p association with its mRNA is autoregulation of its translation which is relieved by phosphorylation of S584. This argument is supported by observation that the Spitzenkörper is rich in polyribosomes suggesting that it a centre for protein translation (Grove et al., 1970).
Asymmetric mRNA localization has been associated with localization of the encoded protein. Such mRNA localization mechanisms often act in parallel with other mechanisms of protein localization. For example, in S. cerevisiae proteins encoded by the mRNAs transported by the She3p system still localize in the absence of She3p, including proteins localized to sites of polarized growth (Shepard et al., 2003; Aronov et al., 2007). If the association of Sec2p with its encoding protein is promotes localization of Sec2p to the Spitzenkörper, then this may also be act in parallel with other localization mechanisms because the GFP–Sec2p_GFP 584E and Sec2p1–625–GFP still localize to the Spitzenkörper even though the mutations abolish the association with mRNA. However, the situation may be more complex than this. A C-terminal truncation, Sec2p1–583 that deletes the phosphorylation site causes mis-localization of Sec2p and abrogates hyphal growth (Bishop et al., 2010).
In S. cerevisiae it has been shown that both Ypt32p and Sec15p bind Sec2p in overlapping regions and thus must compete with each other for Sec2p binding (Ortiz et al., 2002; Medkova et al., 2006; Novick et al., 2007). Ypt32p is a Rab GTPase that acts to load Sec2p on nascent secretory vesicles in the trans-Golgi; while Sec15p is an exocyst component to which secretory vesicles are tethered prior to fusion with the plasma membrane. It has been proposed that displacement of Ypt31p by Sec15p upon vesicle docking allows Sec2p to exit the secretory vesicle upon docking to be available for another round of vesicle transport from the Golgi (Novick et al., 2007). An interesting possibility in C. albicans is that phosphorylation of Sec2p on S584 releases its binding to mRNA on the vesicles in the Spitzenkörper to allow it to be recycled to the Golgi.
Experimental procedures
Media and growth conditions
Cultures were routinely cultured on YEPD consisting of 2% glucose, 2% Bacto peptone and 1% Bacto yeast extract (Difco Becton, Dickinson and Company New Jersey) plus 80 mg l−1 uridine. SD medium consists of 0.67% w/v yeast nitrogen base (Difco, Becton, Dickinson and Company, New Jersey), 2% w/v glucose, 80 mg l−1 each of uridine or 40 mg l−1 histidine, and arginine. To induce hyphal growth cells were grown to saturation at 30°C in YEPD medium. An aliquot of the saturated culture was inoculated into fresh YEPD plus 10% calf serum (Sigma-Aldrich, St Louis, USA) so that the resultant OD600 = 0.5. The cells were then incubated at 37°C.
Strains and plasmids constructions
Strains constructed are listed in Table S1, and the oligonucleotides used are listed in Table S2. All strains were derived from BWP17 (Wilson et al., 1999). Gene deletions and C-terminal YFP and HA fusions were performed as previously described (Gola et al., 2003; Walther and Wendland, 2003; Schaub et al., 2006; Lavoie et al., 2008). All strains were checked for correct genome integration by PCR. Correct expression of protein fusion strains were also checked by Western blot. 3xFLAG fusion proteins were constructed as above using the plasmid pFA 3xFLAG Ura3 which constructed as follows: 3xFLAG was amplified by PCR from plasmid p3xFLAG-Myc-CMV26 (Sigma Aldrich) using primers 3xFLAG-F(PstI) and 3xFLAG-R. The fragment was subcloned into pGEM T easy (Promega, Madison, USA) then extracted using PstI and BamHI and cloned into pFA URA3. This plasmid was then used to create 3xFLAG fusion proteins as described above.
Protein extracts and Western blotting
Total protein extracts were prepared with modified RIPA buffer containing phosphatase inhibitors (50 mM Tris-HCl pH 7.2, 0.1% Sodium deoxycholate, 0.1% Triton X-100, 50 mM NaF, 0.2 mM sodium orthophosphate, 0.2 mM β-glycerol-phosphate, 100 mM NaCl, and protease inhibitors tablet (Roche Biosciences, Lewes, UK) and 200 μl of glass beads (0.4 mm; Sigma-Aldrich, St Louis, USA) were added. Cells were broken for 30 s in a minibeadbeater (Biospec Products, Bartlesville, USA). Soluble proteins were obtained by centrifugation of total extracts at 13 400 r.p.m. over 10 min at 4°C. For Western blots, 30 μg of protein extracts were separated on 6% SDS-PAGE, transferred to Hybond-P membranes (GE Healthcare, Chalfont St Giles, UK), and probed with anti-GFP monoclonal antibody (Roche Biosciences, Lewes, UK) anti-HA (Secondary antibodies conjugated to horseradish peroxidase were diluted 1:5000.
RNA-IP and microarray analysis
Cells from an overnight culture were diluted in YEPD and incubated for 4 h at 30°C or YEPD plus serum and incubated for 90 min at 37°C. Cells were spun down and wash once with water and once with lysis buffer (50 mM HEPES pH7.5, 100 mM NaCl, 1mMEDTA, 1 mM DTT, 10% glycerol, 0.5% Triton X-100, 6 mM MgCl2, 1 mM PMSF, plus Protease Inhibitors tablet). Cells were disrupted in lysis buffer containing 200 units RNase inhibitors (Ribosafe Bioline, London) and 0.2 μg ml−1 of heparin using a minibeadbeater (Biospec Products, as above). The lysate was cleared by centrifugation. To quantify input RNA 50 μl was removed from the lysate and analysed using RNAeasy mini kit (Qiagen, Taunton, MA ). Three millilitres containing 3 mg ml−1 of total protein was inmunoprecipitated using protein G Sepharose pre-incubated with α-FLAG monoclonal (Sigma-Aldrich) or α-GFP monoclonal antibody (Roche Biosciences, Lewes, UK). The resin was washed three times with lysis buffer without protein inhibitors. The RNA was then eluted with 0.12 μg μl−1 of Proteinase K at 37°C for 45 min. The RNA was purified using RNAqueos micro kit (Ambion, Austin, USA). Microarray analysis was carried out by NRC Biotechnology Research Institute, Montreal, Canada.
RT-qRT-PCR
Reverse transcription was carried out using Superscript III (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. The cDNA was diluted 1:5 and qRT-PCR was carried out following the manufacturer's instructions from Sensi –mix Syber green Kit (Bioline) in a Corbett Research Rotor gene qPCR machine. The primers used for the amplification are listed in Table S2. Normalization was done using the ΔΔCt method for two technical replicates and three biological replicates.
Ribonucleoparticle (mRNP) capture
An overnight culture was diluted into 400 ml of fresh medium plus 10% Calf serum (Sigma Aldrich, St Louis, USA) and incubated at 37°C. After 90 min of hyphal induction cells were harvested, washed with PBS and transferred to a 12 cm Petri dish floating in an ice-water bath and irradiated with UV light at a distance of 10 cm from the light source [Spectrolinker XL-1500 (254 nm), DOT Scientific, Burton, MI, USA] three times for 2.5 min each time. Irradiated cells were pelleted and resuspended in lysis buffer (50 mM HEPES pH 7.5, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 10% glycerol, 0.5% Triton X-100, 6 mM MgCl2, 1 mM PMSF, plus Protease Inhibitors tablet) containing 200 units RNase inhibitors (Ribosafe, Bioline), and disrupted using a minibeadbeater (Biospec Products). The lysate was cleared by centrifugation. Three milligrams of total protein was denatured by adding an equal volume of 2× binding buffer (20 mM Tris-HCl pH 7.5, 1 M NaCl, 1% SDS, 0.2 mM EDTA) and then incubated with 0.01 g of oligo-dT cellulose for 2 h at room temperature. Oligo-dT cellulose was washed three times with 50% v/v mixture lysis/binding buffer. The proteins were eluted with 60 μl of elution buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA, 50 μg ml−1 RNase) and incubated at 37°C for 30 min.
Cell fractionation
Cell fractionation and iodixanol density gradients were carried out as previously described (Chang et al., 2008) with the lysis buffer described above. Western blots were performed as described (Bishop et al., 2010). An antibody against the Rps3p ribosomal protein of S. cerevisiae was used to identify ribosomal fractions (Frey et al., 2001). Because it was not possible to combine all the epitope fusions into a single strain the experiment shown in Fig. 6 was repeated out using either a GFP–ARF1 SEC2-HA strain or a SNC2-MYC SEC2-HA strain. The data shown are a composite of the GFP–Arf1 and Sec2p-HA signals from an experiment using the GFP–ARF1 SEC2-HA strain and the Snc2-myc from an experiment using the SEC2-HA SNC2-MYC strain (where the Sec2p-HA signal was identical to that shown). An identical result was obtained when the combination of strains was reversed.
Combined immunocytochemistry and FISH
The immunocytochemistry was modified from Sudbery (2001). An overnight yeast culture was diluted into 25 ml of SD medium with 10% of calf serum to an OD600 of 0.5. Hyphal induction was carried out at 37°C for 90 min. The culture was fixed in 2% formaldehyde for 5 min. To separate cell clumps, cells were pelleted by centrifugation and washed 3× in 55 mM HCl and finally resuspended in 500 μl of 5 mg·ml−1 of pepsin in 55 mM HCl and incubated for 15 min at 37°C with gentle shaking. The cells were pelleted by centrifugation and washed 3× in buffer B (1.2 M Sorbitol, 100 mM potassium phosphate buffer pH 7.5) and finally resuspended in 1 ml spheroplasting buffer [1× Buffer B, 20 mM Vanadyl ribonucleoside complexes solution (Sigma-Aldrich), 5 μl β-Mercaptoethanol, and 1600 units of Lyticase (Sigma-Aldrich)] to digest the cell wall for 30 min at 37°C. The digested cells were washed once in buffer B and resuspended in 500 μl of buffer B. A 20 μl sample of these spheroplasts was pipetted into one well of a multi-well slide (Hendley, Essex, UK) and allowed to stand for 30 min at 4°C. All the liquid was removed by aspiration and the slides were washed in methanol at −20°C for 5 min and acetone at −20°C for 30 s. The slides were air dried. Each well was washed 10× in blocking buffer (1× PBS, 2 mg ml−1 BSA). Then, 20 μl of primary antibody (mouse anti-GFP, Roche), diluted 1:100 in blocking buffer, was added to the well. The slides were incubated overnight in a humid chamber at 4°C. The well was then washed 5× with blocking buffer plus 0.1% v/v Triton X-100, and 5× in blocking buffer. Then, 20 μl of secondary goat anti-mouse antibody conjugated with Alexafluor-633 (Life Technologies) diluted 1:250 was add to each well and incubated for 1 h at room temperature. The wells were then washed 5× in blocking buffer plus 0.1% Triton X-100, and 5× in blocking buffer. The antibodies were fixed for 5 min in blocking buffer plus 2% v/v of formaldehyde. Followed by two washes with 2× SCC for 5 min and finally 1× with 20% formamide in 2× SSC, 0.1% v/v Triton X-100 for 10 min. The FISH probe was prepared as follows: 10 ng·μl−1 of every probe (10 probes in total) labelled with Fluorescein (Sigma-Aldrich) were mixed with 1 μg·μl−1 ssDNA, 1 μg·μl−1 tRNA in solution 1 (40% Formamide, 10 mM Sodium phosphate buffer (pH 7), then denatured at 95°C for 5 min and mixed with an equal volume of solution 2 (4× SCC, 20 mM vanadyl ribonucleoside, 4 μg/μl BSA). Twenty microlitres of this mixture was added to every well and the slides were incubated in a humid chamber at 37°C for 3 h in the dark. Then, cells were washed 2× with: 20% formamide, 2× SSC for 10 min at 37°C, 1× with: 2× SSC, 0.15% Triton X-100 for 10 min at room temperature 2× with 1× SSC, for 10 min at room temperature and 2× with 1× PBS for 5 min at room temperature. Finally the slides were allowed to dry in the dark and the coverslips were mounted with mounting solution, consisting of 80% glycerol in 1× PBS containing 5 μg·ml−1 of DAPI.
Colocalization analysis
Only cells that showed at Sec2p localized to a Spitzenkorper-like structure were analysed. Colocalization of the red (protein) and green channel (mRNA) in hyphal tips was analysed using the Coloc2 plugin for the Fiji implementation of ImageJ (http://fiji.sc/Fiji) to generate a Pearson's correlation coefficient (r).
Microscopy and live cell imaging
Widefield epifluorescence microscopy and differential interference contrast (DIC) microscopy was carried out using a Delta Vision RT microscope (Applied Precision Instruments, Seattle) using an Olympus 100× UplanAPo NA 1.35 lens (Olympus Tokyo, Japan). Images were acquired and deconvolved with Softworx™ software. Images are maximum intensity projections of the deconvolved Z-stack unless otherwise stated. To visualize the nucleus DNA was stained with DAPI (Sigma-Aldrich, St Louis) To visualize the cell outline in fluorescent images, cells were counterstained with 1 μg·ml−1 Calcofluor White (Fluorescent Brightener 28, Sigma Aldrich). Where indicated cells were fixed with 2% formaldehyde and then treated with pepsin as previously described (Sudbery, 2001).
Statistical analysis
Graph pad Prism 5 was used to analyse and to represent all the data shown in this work.
Acknowledgments
This work was supported by BBSRC Project Grants BB/F007892/1 and R/130398/1. We are grateful to Andre Nantel for carrying out the microarray analysis. The authors declare they have no conflict of interest.
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher's web-site.
Supporting information
References
- Aronov S. Gerst JE. Involvement of the late secretory pathway in actin regulation and mRNA transport in yeast. J Biol Chem. 2004;279:36962–36971. doi: 10.1074/jbc.M402068200. [DOI] [PubMed] [Google Scholar]
- Aronov S, Gelin-Licht R, Zipor G, Haim L, Safran E. Gerst JE. mRNAs encoding polarity and exocytosis factors are cotransported with the cortical endoplasmic reticulum to the incipient bud in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:3441–3455. doi: 10.1128/MCB.01643-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann S, Pohlmann T, Jungbluth M, Brachmann A. Feldbrügge M. Kinesin-3 and dynein mediate microtubule-dependent co-transport of mRNPs and endosomes. J Cell Sci. 2012;125:2740–2752. doi: 10.1242/jcs.101212. [DOI] [PubMed] [Google Scholar]
- Baumann S, König J, Koepke J. Feldbrügge M. Endosomal transport of septin mRNA and protein indicates local translation on endosomes and is required for correct septin filamentation. EMBO Rep. 2014;15:94–102. doi: 10.1002/embr.201338037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH. Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- Bishop A, Lane R, Beniston R, Lazo B, Smythe C. Sudbery P. Hyphal growth in Candida albicans requires the phosphorylation of Sec2 by the Cdc28-Ccn1/Hgc1 kinase. EMBO J. 2010;29:2930–2942. doi: 10.1038/emboj.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehl F, Kruse C, Frank A, Ferring D. Jansen RP. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 2000;19:5514–5524. doi: 10.1093/emboj/19.20.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero-Lima D, Kaneva IN, Watton SP, Sudbery PE. Craven CJ. The spatial distribution of the exocyst and actin cortical patches is sufficient to organize hyphal tip growth. Eukaryot Cell. 2013;12:998–1008. doi: 10.1128/EC.00085-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann B, Strein C, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Chang W, Zaarour RF, Reck-Peterson S, Rinn J, Singer RH, Snyder M, et al. Myo2p, a class V myosin in budding yeast, associates with a large ribonucleic acid-protein complex that contains mRNAs and subunits of the RNA-processing body. RNA. 2008;14:491–502. doi: 10.1261/rna.665008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand P, Meng XH, Singer RH. Long RM. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr Biol. 1999;9:333–338. doi: 10.1016/s0960-9822(99)80144-4. [DOI] [PubMed] [Google Scholar]
- Elson SL, Noble SM, Solis NV, Filler SG. Johnson AD. An RNA transport system in Candida albicans regulates hyphal morphology and invasive growth. PLoS Genet. 2009;5:e1000664. doi: 10.1371/journal.pgen.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, Pool M. Seedorf M. Scp160p, an RNA-binding, polysome-associated protein, localizes to the endoplasmic reticulum of Saccharomyces cerevisiae in a microtubule-dependent manner. J Biol Chem. 2001;276:15905–15912. doi: 10.1074/jbc.M009430200. [DOI] [PubMed] [Google Scholar]
- Genz C, Fundakowski J, Hermesh O, Schmid M. Jansen RP. Association of the yeast RNA-binding protein She2p with the tubular endoplasmic reticulum depends on membrane curvature. J Biol Chem. 2013;288:32384–32393. doi: 10.1074/jbc.M113.486431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gola S, Martin R, Walther A, Dunkler A. Wendland J. New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast. 2003;20:1339–1347. doi: 10.1002/yea.1044. [DOI] [PubMed] [Google Scholar]
- Grove SN, Bracker CE. Morre DJ. An ultrastructural basis for hyphal tip growth in Pythium ultimum. Am J Bot. 1970;57:245–266. [Google Scholar]
- Gu W, Deng Y, Zenklusen D. Singer RH. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes Dev. 2004;18:1452–1465. doi: 10.1101/gad.1189004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Roth D, Walch-Solimena C. Novick P. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 1999;18:1071–1080. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Shi K, Wu LC, Mirzaei H, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Higuchi Y, Ashwin P, Roger Y. Steinberg G. Early endosome motility spatially organizes polysome distribution. J Cell Biol. 2014;204:343–357. doi: 10.1083/jcb.201307164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DJ, Riordan DP, Gerber AP, Herschlag D. Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis DO. Johnson AD. Ash1 protein, an asymmetrically localized transcriptional regulator, controls filamentous growth and virulence of Candida albicans. Mol Cell Biol. 2002;22:8669–8680. doi: 10.1128/MCB.22.24.8669-8680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie K, Tadauchi T, Takizawa PA, Vale RD, Matsumoto K. Herskowitz I. The Khd1 protein, which has three KH RNA-binding motifs, is required for proper localization of ASH1 mRNA in yeast. EMBO J. 2002;21:1158–1167. doi: 10.1093/emboj/21.5.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LA. Sudbery PE. Spitzenkorper, exocyst and polarisome components in Candida albicans hyphae show different patterns of localization and have distinct dynamic properties. Eukaryot Cell. 2010;9:1455–1465. doi: 10.1128/EC.00109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilchert C. Spang A. Cotranslational transport of ABP140 mRNA to the distal pole of S. cerevisiae. EMBO J. 2011;30:3567–3580. doi: 10.1038/emboj.2011.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König J, Baumann S, Koepke J, Phlmann T, Zarnack K. Feldbrügge M. The fungal RNA-binding protein Rrm4 mediates long-distance transport of ubi1 and rho3 mRNAs. EMBO J. 2009;28:1855–1866. doi: 10.1038/emboj.2009.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie H, Sellam A, Askew C, Nantel A. Whiteway M. A toolbox for epitope-tagging and genome-wide location analysis in Candida albicans. BMC Genomics. 2008;9:578–591. doi: 10.1186/1471-2164-9-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner RS, Seiser RM, Zheng TL, Lager PJ, Reedy MC, Keene JD. Nicchitta CV. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA. 2003;9:1123–1137. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long RM, Singer RH, Meng XH, Gonzalez I, Nasmyth K. Jansen RP. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- Long RM, Gu W, Lorimer E, Singer RH. Chartrand P. She2p is a novel RNA-binding protein that recruits the Myo4p–She3p complex to ASH1 mRNA. EMBO J. 2000;19:6592–6601. doi: 10.1093/emboj/19.23.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medkova M, France YE, Coleman J. Novick P. The rab exchange factor Sec2p reversibly associates with the exocyst. Mol Biol Cell. 2006;17:2757–2769. doi: 10.1091/mbc.E05-10-0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Heym RG, Mayer A, Kramer K, Schmid MA, Cramer P, et al. A cytoplasmic complex mediates specific mRNA recognition and localization in yeast. PLoS Biol. 2011;9:e1000611. doi: 10.1371/journal.pbio.1000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Medkova M, Dong G, Hutagalung A, Reinisch K. Grosshans B. Interactions between Rabs, tethers, SNAREs and their regulators in exocytosis. Biochem Soc Trans. 2007;34:683–686. doi: 10.1042/BST0340683. [DOI] [PubMed] [Google Scholar]
- Ortiz D, Medkova M, Walch-Solimena C. Novick P. Ypt32 recruits the Sec4p guanine nucleotide exchange factor, Sec2p, to secretory vesicles; evidence for a Rab cascade in yeast. J Cell Biol. 2002;157:1005–1015. doi: 10.1083/jcb.200201003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin N, Ménade M, Poirier G, Donato D, Drouet E. Chartrand P. Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol Cell. 2007;26:795–809. doi: 10.1016/j.molcel.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Rida PCG, Nishikawa A, Won GY. Dean N. Yeast-to-hyphal transition triggers formin-dependent Golgi localization to the growing tip in Candida albicans. Mol Biol Cell. 2006;17:4364–4378. doi: 10.1091/mbc.E06-02-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub Y, Dunkler A, Walther A. Wendland J. New pFA-cassettes for PCR-based gene manipulation in Candida albicans. J Basic Microbiol. 2006;46:416–429. doi: 10.1002/jobm.200510133. [DOI] [PubMed] [Google Scholar]
- Scherrer T, Mittal N, Janga SC. Gerber P. A screen for RNA-binding proteins in yeast indicates dual functions for many enzymes. PLoS ONE. 2010;5:e15499. doi: 10.1371/journal.pone.0015499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbabian K. Chartrand P. Control of cytoplasmic mRNA localization. Cell Mol Life Sci. 2012;69:535–552. doi: 10.1007/s00018-011-0814-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbabian K, Jeronimo C, Forget A, Robert F. Chartrand P. Co-transcriptional recruitment of Puf6 by She2 couples translational repression to mRNA localization. Nucleic Acids Res. 2014;42:8692–8704. doi: 10.1093/nar/gku597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, St-Denis A. Chartrand P. Cotranscriptional recruitment of She2p by RNA pol II elongation factor Spt4-Spt5/DSIF promotes mRNA localization to the yeast bud. Genes Dev. 2010;24:1914–1926. doi: 10.1101/gad.1937510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard KA, Gerber AP, Jambhekar A, Takizawa PA, Brown PO, Herschlag D, et al. Widespread cytoplasmic mRNA transport in yeast: Identification of 22 bud-localized transcripts using DNA microarray analysis. Proc Natl Acad Sci USA. 2003;100:11429–11434. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll DR, Herman MA. Staebell MA. The involvement of cell wall expansion in the two modes of mycelium formation of Candida albicans. J Gen Microbiol. 1985;131:2367–2375. doi: 10.1099/00221287-131-9-2367. [DOI] [PubMed] [Google Scholar]
- Sudbery PE. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localisation. Mol Microbiol. 2001;41:19–31. doi: 10.1046/j.1365-2958.2001.02459.x. [DOI] [PubMed] [Google Scholar]
- Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- Tai NW, Schmitz JC, Liu J, Lin XK, Bailly M, Chen TM. Chu E. Translational autoregulation of thymidylate synthase and dihydrofolate reductase. Front Biosci. 2004;9:2521–2526. doi: 10.2741/1413. [DOI] [PubMed] [Google Scholar]
- Takizawa PA. Vale RD. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc Natl Acad Sci USA. 2000;97:5273–5278. doi: 10.1073/pnas.080585897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terbush DR. Novick P. Sec6, Sec8, and Sec15 are components of a multisubunit complex which localizes to small bud tips in Saccharomyces cerevisiae. J Cell Biol. 1995;130:299–312. doi: 10.1083/jcb.130.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terbush DR, Maurice T, Roth D. Novick P. The exocyst is a multi-protein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Trautwein M, Dengjel J, Schirle M. Spang A. Arf1p provides an unexpected link between COPI vesicles and mRNA in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:5021–5037. doi: 10.1091/mbc.E04-05-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena C, Collins RN. Novick PJ. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J Cell Biol. 1997;137:1495–1509. doi: 10.1083/jcb.137.7.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther A. Wendland J. An improved transformation protocol for the human fungal pathogen Candida albicans. Curr Genet. 2003;42:339–343. doi: 10.1007/s00294-002-0349-0. [DOI] [PubMed] [Google Scholar]
- Walworth NC, Goud B, Kabcenell AK. Novick PJ. Mutational analysis of Sec4 suggests a cyclical mechanism for the regulation of vesicular traffic. EMBO J. 1989;8:1685–1693. doi: 10.1002/j.1460-2075.1989.tb03560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson B, Davis D. Mitchell AP. Rapid hypothesis testing in Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L. Bassell GJ. mRNA localization: an orchestration of assembly, traffic and synthesis. Traffic. 2013;14:2–14. doi: 10.1111/tra.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information


