ABSTRACT
Streptococcus pneumoniae is auxotrophic for arginine, and molecular analysis of the pneumococcal genome showed that the gene encoding an arginine-ornithine antiporter (ArcD) is organized in a cluster together with the arcABC genes encoding the arginine deiminase system (ADS) of pneumococci. The ADS consists of the arginine deiminase (AD), the catabolic ornithine carbamoyltransferase (cOCT), and the carbamate kinase (CK). Pneumococcal genomes contain three ArgR-type regulators (ArgR1, ArgR2, and AhrC) that are supposed to be involved in the regulation of arginine metabolism. Here, we identified ArgR2 of TIGR4 as the regulator of the ADS and ArcD. ArgR2 binds to promoter sequences of the arc operon, and the deficiency of ArgR2 in TIGR4 abrogates expression of the ADS, including the arginine-ornithine antiporter ArcD. Intranasal infection of mice and real-time bioimaging revealed that deletion of the arcABCDT genes attenuates TIGR4. However, the acute-pneumonia model and coinfection experiments indicated that the arginine-ornithine antiporter ArcD is essential to maintain fitness, while the deficiency of ADS enzymes has a minor impact on pneumococcal fitness under in vivo conditions. Strikingly, argR2 mutant TIGR4 outcompeted the wild type in the respiratory tract, suggesting an increase in fitness and further regulatory functions of ArgR2. In contrast to TIGR4, other pneumococci, such as D39, lacking expression of ArgR2, constitutively express the ADS with a truncated nonfunctional AD. On the basis of these results, we propose that the arginine-ornithine antiporter is essential to maintain pneumococcal fitness and that the genes of the ADS cluster are positively regulated in a strain-specific manner by ArgR2.
IMPORTANCE
Pneumococci are the major etiologic agents of community-acquired pneumonia, causing more than 1.5 million deaths annually worldwide. These versatile pathogens are highly adapted to the nutrients provided by the host niches encountered. Physiological fitness is of major importance for colonization of the nasopharyngeal cavity and dissemination during invasive infections. This work identifies the regulator ArgR2 as the activator of the S. pneumoniae TIGR4 ADS and the arginine-ornithine transporter ArcD, which is needed for uptake of the essential amino acid arginine. Although ArgR2 activates ArcD expression and uptake of arginine is required to maintain pneumococcal fitness, the deficiency of ArgR2 increases TIGR4 virulence under in vivo conditions, suggesting that other factors regulated by ArgR2 counterbalance the reduced uptake of arginine by ArcD. Thus, this work illustrates that the physiological homeostasis of pneumococci is complex and that ArgR2 plays a key role in maintaining bacterial fitness.
INTRODUCTION
Streptococcus pneumoniae (pneumococcus), a member of the order Lactobacillales, is a Gram-positive bacterium residing asymptomatically as a harmless commensal in the human upper respiratory tract. However, pneumococci are also the etiologic agents of serious local and invasive infections, including otitis media, pneumonia, bacteremia, and meningitis (1–4).
During pathogenesis, pneumococci have to adapt their metabolism to different environmental conditions regarding temperature, oxygen, pH, and the availability of nutrients. Pneumococci are endowed with a large number of enzymes and transporter systems enabling them to take up and metabolize the nutrients available in their various host niches. To evade the immune defense system, they have also evolved sophisticated strategies to escape the host response and survive under harmful conditions (5).
In former studies, we demonstrated that glutamine, branched-chain amino acids, and arginine are not synthesized de novo by S. pneumoniae (6, 7). Arginine, as an essential amino acid, can be found at a concentration of about 45 µM in human plasma (8), as well as, e.g., in human cerebrospinal fluid (13 µM) (9) and muscle tissue (>1,000 µM) (8). The arginine concentration decreases during the inflammatory response (10, 11), subsequently lowering the activity of T and B cells (12). In human macrophages, arginine is important for the biosynthesis of nitric oxide, which is toxic for invasive microbes (13). In pneumococci, arginine is also a precursor for the biosynthesis of spermine, a polyamine that acts directly as a free-radical scavenger (14–16).
During evolution, organisms have developed different mechanisms for arginine catabolism. These are the arginase pathway, the arginine transaminase pathway, the arginine decarboxylase pathway, and the arginine deiminase (AD) pathway (17–21).
Various lactic acid bacteria metabolize arginine via the AD system (ADS). The ADS includes three enzymes whose genes are commonly arranged as an operon. Many ADS operons contain a fourth and a fifth gene encoding an arginine-ornithine antiporter and a putative aminopeptidase, respectively. However, the order of the genes varies among different bacteria (22–28). Overall, the ADS yields 1 mol of ornithine and CO2, 2 mol of ammonia, and 1 mol of ATP by substrate level phosphorylation per mol of arginine metabolized (17, 29). In Halobacterium salinarum, the ornithine produced by the ADS is exported in exchange for one molecule of arginine in an energy-independent manner by the membrane-bound antiporter encoded by arcD (30).
The ammonia produced by the ADS raises the cytoplasmic pH, thereby protecting the cell from potentially lethal effects of an acidic extracellular environment. This protective effect was described in oral bacteria like Streptococcus sanguis, Streptococcus gordonii, and Streptococcus rattus and partially for the pig pathogen Streptococcus suis (22, 24, 25, 31, 32).
Furthermore, 1 mol of ATP generated out of 1 mol of arginine allows the use of arginine as the only energy source as, e.g., in Pseudomonas aeruginosa. The AD pathway ArcA-ArcD is probably required for optimal anaerobic or microaerobic growth and viability of P. aeruginosa within cystic fibrosis airways (33). It was shown that the AD of S. suis, which is also present on the bacterial surface, can be induced by an increase in temperature to 42°C and a low oxygen level (34). ArcD has been characterized as an arginine transporter in several bacteria, including S. pneumoniae (35). It was shown that ArcD plays a role as an arginine uptake system and influences the structure of the pneumococcal capsule. This has an effect on the pathogenicity of different pneumococcal serotypes. The molecular basis of the effect of ArcD on the capsule is not clear (35).
Under anaerobic conditions, the expression of the ADS genes in P. aeruginosa is activated by the regulator ANR (anaerobic regulation of arginine catabolism and nitrate reduction). In the presence of arginine, the transcription of AD, induced by the ANR, is reinforced by the regulator ArgR (36). The gene encoding the ADS regulator (Crp/Fnr family), designated argR or arcR, is frequently close to the arc gene cluster (37–39). In Staphylococcus aureus, ArcR is necessary whenever arginine is the only energy source available (40). The regulation of the ADS in streptococci is highly dependent on growth conditions. Arginine induces the AD activity of S. gordonii and S. sanguis. Furthermore, AD expression is subject to carbon catabolite repression. The presence of the repressing sugar glucose lowers AD activity (37, 41). Additionally, the regulator ArgR acts as an arcABC operon expression activator in S. suis (42).
Many Gram-positive organisms, like Bacillus subtilis and Lactococcus lactis, contain a functional arginine biosynthesis pathway (43). The genome of S. pneumoniae D39 exhibits only two arg genes, namely, argG and argH, encoding argininosuccinate synthetase and argininosuccinase, respectively. These enzymes convert citrulline and aspartate to arginine (44). Remarkably, pneumococci can be positive or negative for argGH (16). Although pneumococci do not contain a complete set of arginine biosynthetic genes, three putative ArgR-type regulators (AhrC, ArgR1, and ArgR2) have been described (45). ArgR1 and AhrC form a heterohexameric complex that, at high arginine concentrations, represses five operons, including the argGH operon and the artPQ, abpA, abpB, and aliB genes, suggested to be involved in arginine uptake. In this scenario, arginine acts as an effector molecule, and under arginine limitation, the operons are derepressed (44). In the present study, we show that TIGR4, but not D39, produces the ArgR2 regulator, which activates the ADS operon in TIGR4 lacking ArgGH. In TIGR4 and other pneumococci, ArgR2 is required for optimal arginine uptake via the arginine-ornithine antiporter ArcD, and interestingly, mouse coinfections suggested that ArgR2 regulates additional genes.
RESULTS
Bioinformatic analysis of the pneumococcal AD loci and regulatory genes in pneumococci.
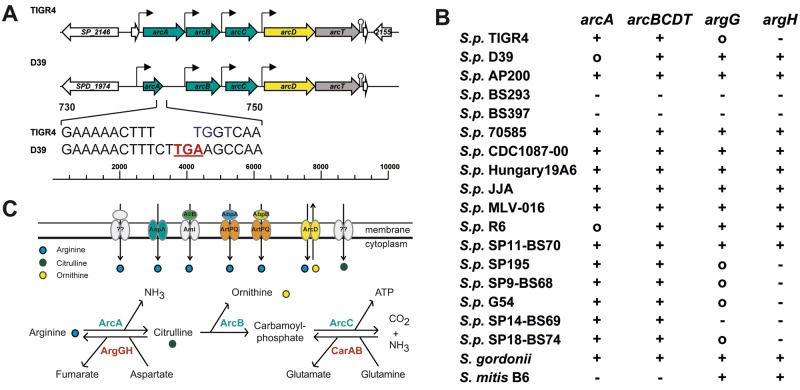
The proteins of the AD pathway in S. pneumoniae TIGR4 are encoded by the arcABCDT genes, which are located in a cluster. Bioinformatic analysis (Neural Network Promoter Prediction; http://www.fruitfly.org/seq_tools/promoter.html) showed putative promoter sequences upstream of the arcA and arcD genes, respectively. Further promoter structures were predicted upstream of arcB and arcC. A putative terminator sequence (TransTerm HP; http://transterm.cbcb.umd.edu/) was predicted downstream of arcT (Fig. 1A).
FIG 1 .
Genomic organization of ADS gene clusters in pneumococci and distribution of arc genes. (A) Genomic organization of ADS operons in S. pneumoniae TIGR4 and D39: arcA, AD; arcB, catabolic cOCT; arcC, CK; arcD, arginine-ornithine antiporter; arcT, putative aminopeptidase. The AD-encoding gene (arcA) of D39 is truncated by the insertion of a stop codon (TGA). (B) Distribution of arginine metabolism genes in different strains present in the SYBIL database. argG, argininosuccinate synthetase; argH, argininosuccinase; +, present; ○, truncated; −, absent. (C) Arginine metabolism in S. pneumoniae. CarAB, anabolic cOCT.
The deduced 409 amino acids (aa) of the AD (ArcA, SP_2148) of S. pneumoniae TIGR4 harbor conserved AD motifs, i.e., a long motif, aa 10 to 20 (SEIGKLKKVML), and three shorter conserved motifs, aa 161 to 164 (FTRD), 218 to 221 (EGGD), and 272 to 278 (MHLDTVF). All of these motifs are important for the structure and function of the AD (46). The 338 aa of the ornithine carbamoyltransferase (cOTC, ArcB, SP_2150) of TIGR4 contains the conserved carbamoylphosphate binding and catalysis (STRTR) motif (47). The amino acid sequence of the carbamate kinase (CK, ArcC, SP_2151) contains no conserved motifs (23, 48). The analysis of the arginine-ornithine antiporter (ArcD; 503 aa) shows 12 transmembrane helices (http://www.ch.embnet.org/software/TMPRED_form.html). The final gene of the ADS gene cluster, arcT (SP_2153), encodes a putative peptidase and has >60% sequence identity with ArcT of S. sanguinis (72%), S. gordonii (72%), S. suis (65%), and Streptococcus pyogenes (66%).
Comparative analyses of arc gene clusters, the anabolic argGH genes, and the genes encoding regulatory proteins ArgR1, AhrC, and ArgR2 were performed on the basis of the SYBIL database (http://strepneumo-sybil.igs.umaryland.edu/cgi-bin/current/shared/index.cgi?site=strepneumo).
Genes of the arc cluster (arcA to arcT) are present in the majority of pneumococcal genomes. In S. pneumoniae D39, R800, and R6, a stretch of 8 nucleotides (nt) starting at nt 740 differs from arcA sequences encoding a functionally active AD, most likely because of nucleotide insertions (CTTG) and a substitution of 3 nt (TGGT to AAGC) (Fig. 1A). This leads to a premature stop codon in the arcA gene, resulting in translational termination and hence in a nonfunctional AD protein. The genes arcB to arcT of R6 and D39 are identical to other pneumococcal arcB-to-arcT genes. Two other strains (S. pneumoniae BS292 and BS293) have neither the arc gene cluster nor the argGH operon (Fig. 1B). The regulatory genes (argR1, ahrC, and argR2) are annotated in all of the strains analyzed, with the exception of S. pneumoniae SP14-BS69 and SP18-BS74, which lack argR2. In addition to the genes described here, pneumococci exhibit the anabolic carbamoyl phosphate synthetase (carAB), as well as the catabolic CK (arcC) (Fig. 1C). None of the strains sequenced has all of the eight arg genes of the arginine biosynthetic pathway (argBCDEFGHJ). Interestingly, the distribution of the anabolic genes (argGH) varies among pneumococci. These genes were present in only 8 of the 33 pneumococcal strains listed in the SYBIL database, among them, S. pneumoniae D39 and R6 but not TIGR4 (Fig. 1B).
The proteins encoded by arc genes form a regulatory unit.
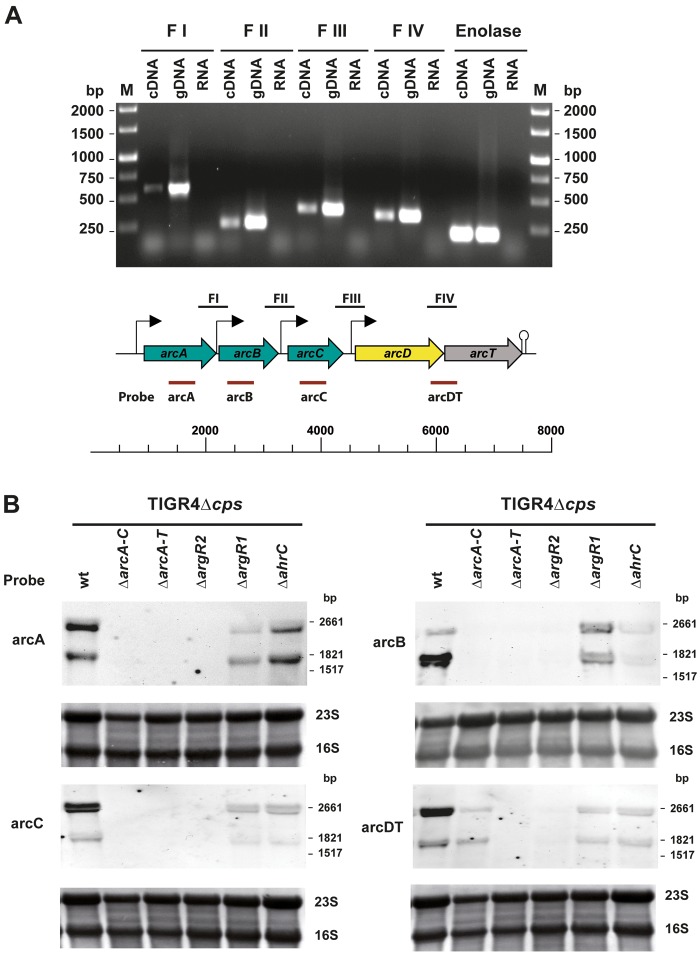
Reverse transcription (RT)-PCR analysis was performed to investigate whether the arcABCDT genes are cotranscribed. RNA isolated from S. pneumoniae TIGR4 grown in a chemically defined medium (CDM) was used to generate cDNA and PCR products representing gene-spanning sequences and intergenic regions between selected arc genes were amplified with specific primer pairs. The results suggest that the genes arcABCDT form a polycistronic operon in S. pneumoniae TIGR4 (Fig. 2A). Considering the putative promoter and terminator structures, a complete arcABCDT transcript would have a calculated length of about 6.5 kb. However, because of the predicted additional promoter and terminator structures, transcripts of 2.3 kb (arcAB), 3.4 kb (arcABC), and 3.0 kb (arcDT) are also possible. Northern blot analysis was conducted with digoxigenin (DIG)-labeled RNA probes homologous to the arcA, arcB, arcC, and arcDT mRNAs, respectively, to investigate the transcript lengths. All four probes detected specific transcripts of 2.6 and 1.6 kb in the wild-type TIGR4 strain, which are not consistent with the theoretical calculations (Fig. 2B). The ΔarcA-C, ΔarcA-T, and ΔargR2 mutants lack arcA-C-specific transcripts. Similarly, the expression of arcDT was abolished in the ΔarcA-T and ΔargR2 mutants (Fig. 2B), suggesting arcABC and arcDT regulation by ArgR2. The knockout of argR1 or ahrC also influenced arcABCDT gene expression (Fig. 2B), which suggests that ArgR1 and AhrC are involved in the complex regulation of the ADS.
FIG 2 .
Analysis of the arc gene complex expression. (A) RT-PCR analysis of arc gene cluster regions. Total mRNA was isolated from S. pneumoniae grown in THY medium, and random primers were used to amplify cDNA. Genomic DNA (gDNA) and RNA served as positive and negative controls, respectively. The following PCR fragments representing arc gene and intergenic sequences were amplified after RT: FI, arcAB intergenic region; FII, arcBC intergenic region; FIII, arcCD intergenic region; FIV, arcDT intergenic region. The enolase gene was used as a positive control. Lanes M contained molecular size markers. (B) Northern blot analysis of arcABCDT transcripts in wild-type (wt) S. pneumoniae D39 and TIGR4 and in the isogenic TIGR4 ΔarcA-C, ΔarcA-T ΔargR2, ΔargR1, and ΔahrC deletion mutants, respectively. Total RNA isolated from cultures grown in CDM to an OD600 of 0.4 was hybridized with DIG-labeled RNA probes for arcA (P704, P611), arcB (P933, P934), arcC (P935, P936), and arcDT (P705, P613). Methylene blue was used to verify the amount of RNA on hybridization membranes.
ArgR2 activates the ADS and arginine uptake in TIGR4.
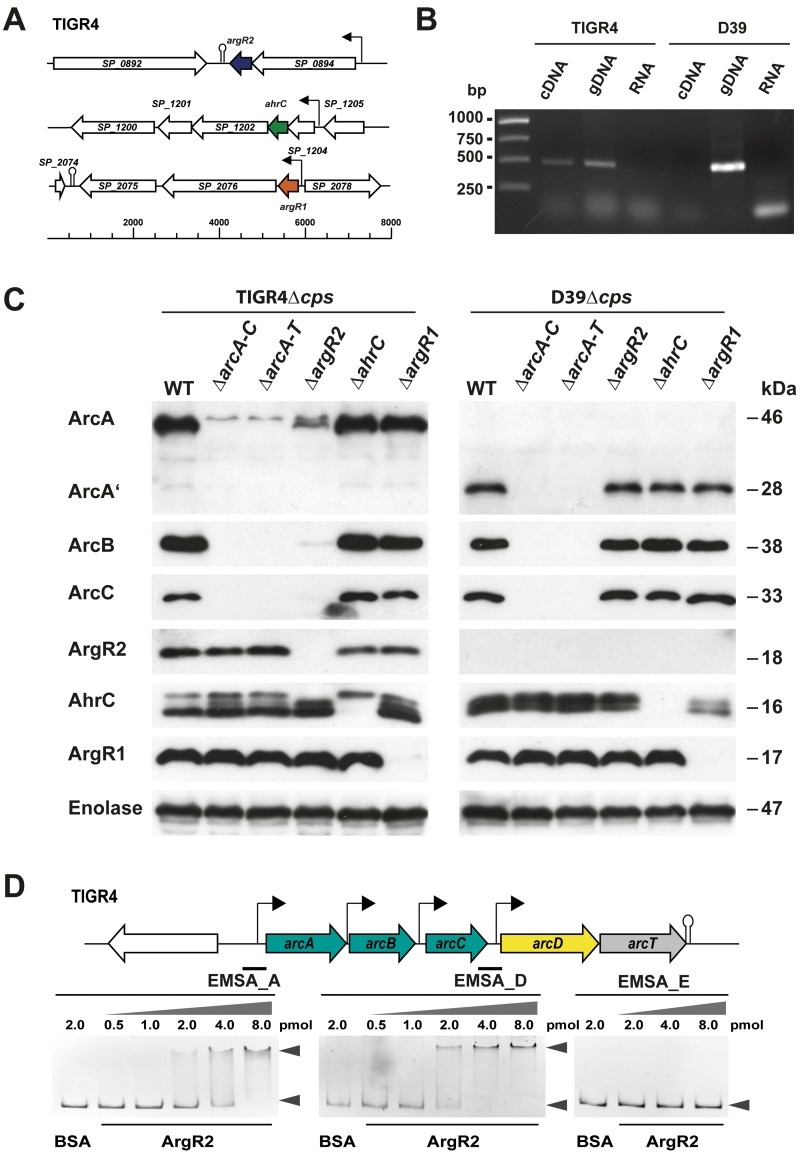
Three genes encoding ArgR-type regulatory proteins (ArgR1, ArgR2, and AhrC) are annotated in the TIGR4 genome (Fig. 3A), and if present, these genes are listed in pneumococcal genomes deposited in the SYBIL database (44). The amino acid sequence of the nutritional regulators is highly conserved (>98%) in streptococci and also shows high homology to L. lactis ArgR-type regulators. The argR2 genes of S. pneumoniae TIGR4 and D39 show a GA transition at bp 94 (TIGR4 guanine; D39 adenine), resulting in an amino acid substitution at position 31 (E31K) of ArgR2 (TIGR4 glutamate, D39 lysine). ArgR1 and AhrC form a heterohexameric complex with arginine as an effector molecule to ensure optimal uptake of arginine from the surrounding milieu (44). However, the regulator of the ADS and its impact on arginine uptake by the ArcD antiporter were unknown.
FIG 3 .
The AD gene cluster of S. pneumoniae is regulated by ArgR2. (A) Genetic regions of the three ArgR-type transcriptional regulators encoded in the TIGR4 genome. Regions: SP_0892, type I restriction enzyme; SP_0893, transcriptional regulator of arginine metabolism ArgR2; SP_0894, X-prolyl-dipeptidyl aminopeptidase gene pepX; SP_1200, GTP-binding protein LepA; SP_1201, serine/threonine protein phosphatase; SP_1202, DNA repair protein RecN; SP_1203, transcriptional regulator of arginine metabolism AhrC; SP_1204, 23S rRNA (cytidine1920-2′-O)/16S rRNA (cytidine1409-2′-O)-methyltransferase; SP_1205, geranyltranstransferase; SP_2075, ABC transporter ATP-binding protein/permease; SP_2076, pseudogene; SP_2077, transcriptional regulator of arginine metabolism ArgR1; SP_2078, arginyl-tRNA synthetase gene argRS. (B) RT-PCR analysis of argR2 expression in TIGR4 and D39. Random primers were used to amplify cDNA. Genomic DNA was used as a positive control, and RNA was used as a negative control. (C) Immunoblot analysis of ArcA, ArcB, ArcC, ArgR2, ArgR1, and AhrC synthesis in nonencapsulated pneumococcal strains (TIGR4Δcps and D39Δcps) and isogenic regulatory mutants with bacterial whole-cell cytoplasmic protein lysates. Proteins were detected with specific mouse anti-ArcA, ArcB, ArcC, ArgR2, ArgR1, or AhrC antiserum. Pneumococcal enolase was used as a loading control. The predicted molecular masses are as follows: mature ArcA in TIGR4, 46.6 kDa; fragmented ArcA′ in D39, 28.3 kDa; ArcB, 37.9 kDa; ArcC, 33.6 kDa; ArgR2, 17.8 kDa; ArgR1, 16.2 kDa; AhrC, 17.1 kDa. WT, wild type. (D) Interaction of ArgR2 with putative promoter regions of the arc operon. Purified recombinant His-ArgR2 (0.5 to 8 pmol of protein) was incubated with the 437-bp arcA promoter fragment (EMSA_A), the 391-bp arcD promoter fragment (EMSA_D), or the 307-bp enolase promoter fragment (EMSA_E, negative control; DNA, 0.2 pmol).
Immunoblot analyses were performed to assess the role of the ArgR-type regulators in the expression of proteins encoded by the AD operon. The expression of ArcA, ArcB, ArcC, and the three regulatory proteins AhrC, ArgR1, and Arg2 in pneumococcal wild-type strains TIGR4 and D39, respectively, was compared to that in the isogenic mutants by using specific mouse polyclonal antibodies generated against ArcA, ArcB, ArcC, or the regulatory proteins (Fig. 3C). The TIGR4 wild type expressed all three enzymes of the ADS, and all of the regulator proteins were detected. The mutants deficient for ArcA to ArcC and ArcA to ArcT showed no expression of the enzymes. In contrast to the argR1 and ahrC mutants, the expression of the catabolic enzymes (ArcA, ArcB, and ArcC) was diminished in the argR2 mutant, supporting the hypothesis that ArgR2 is a specific activator of the ADS and arginine uptake by ArcD.
It is noteworthy that the results of protein expression by S. pneumoniae D39 and isogenic mutants differed significantly from those of protein expression by TIGR4. Immunoblot analyses showed a truncated ArcA peptide due to the premature stop codon. The full-length ArcB and ArcC proteins were detected in the D39 wild type and, remarkably, also in the isogenic argR2 mutant. Similar to TIGR4, the deficiency of ArgR1 or AhrC had no effect on the expression of the ADS enzymes. Importantly, no protein signal could be detected for the regulator ArgR2 in D39, although the upstream sequence of argR2 is identical to the TIGR4 sequence and the only amino acid exchange (E31K) occurred at position 31. This is of particular interest because the arc genes are expressed in D39 although it lacks ArgR2, while arc gene expression in TIGR4 requires the ArgR2 regulator.
To elucidate whether the absence of ArgR2 in D39 is due to a lack of gene expression, RT-PCR analysis was performed. As depicted in Fig. 3B, a PCR fragment with intergenic argR2 primers was amplified for S. pneumoniae TIGR4, while D39 showed no PCR product, suggesting that the argR2 gene of D39 is not transcribed.
ArgR2 binds to promoter regions of the ADS.
To investigate whether ArgR2 controls the expression of the arc operon by binding to the predicted promoter regions of arcA and arcD (Fig. 1A), electrophoretic mobility shift assays (EMSAs) were performed with purified recombinant His6-ArgR2 protein. DNA-binding reactions were carried out with a 437-bp arcA or a 391-bp arcD fragment. As shown in Fig. 3D, both DNA fragments were retarded by the addition of His6-ArgR2. The amount of the DNA-protein complex increased in an ArgR2 concentration-dependent manner. These results confirm that ArgR2 acts as specific activator of the genes of the ADS.
Strain-specific expression and regulation of ADS.
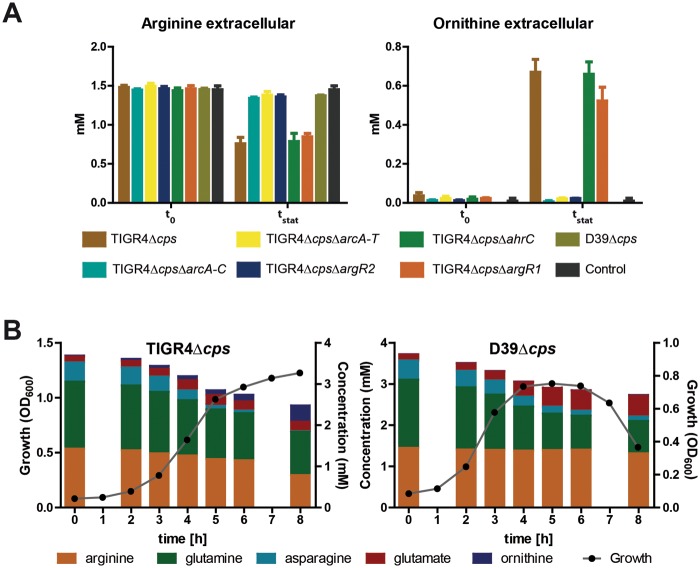
Arginine is essential for pneumococcal growth (6). 1H nuclear magnetic resonance (NMR) analysis of the growth medium indicated that the wild-type TIGR4 strain takes up arginine and releases ornithine (Fig. 4A). The analysis of the extracellular metabolites showed that the uptake of arginine lacking functional ArcD was significantly reduced in TIGR4. Moreover, the ΔarcA-C, ΔarcA-T, and ΔargR2 TIGR4 mutants showed no secretion of ornithine into the medium. Importantly, uptake of arginine by D39 was 20-fold lower than that by TIGR4 and correlated with the lower expression of the AD enzymes. Consequently, ornithine secretion was not detected in D39 (Fig. 4A). Pneumococci catabolize glutamine and asparagine as main nitrogen sources, while dispensable glutamate is secreted. Strains expressing a functional ADS, such as TIGR4, are additionally able to utilize arginine as a nitrogen source. This is accompanied by less glutamine uptake by TIGR4 than by D39 expressing a nonfunctional ADS (Fig. 4B).
FIG 4 .
Impact of the pneumococcal ADS regulator ArgR2 on consumption of nitrogen-rich arginine. (A) Uptake of arginine and export of ornithine by TIGR4Δcps and its isogenic mutants in CDM. The extracellular concentrations (millimolar) of arginine and ornithine were measured prior to the start of the cultivation (t0) and at the late exponential phase (OD600, 0.8; t1). Medium without bacterial culture was used as a control. (B) Growth curves, nitrogen-rich amino acid consumption, and end products of nitrogen metabolism formed by the TIGR4Δcps and D39Δcps strains cultured in CDM under microaerophilic conditions at 37°C. Culture supernatant samples for substrate and end product analysis by 1H-NMR were harvested at the time points indicated, and the results are represented by bars in the plots. Data from four independent experiments are shown.
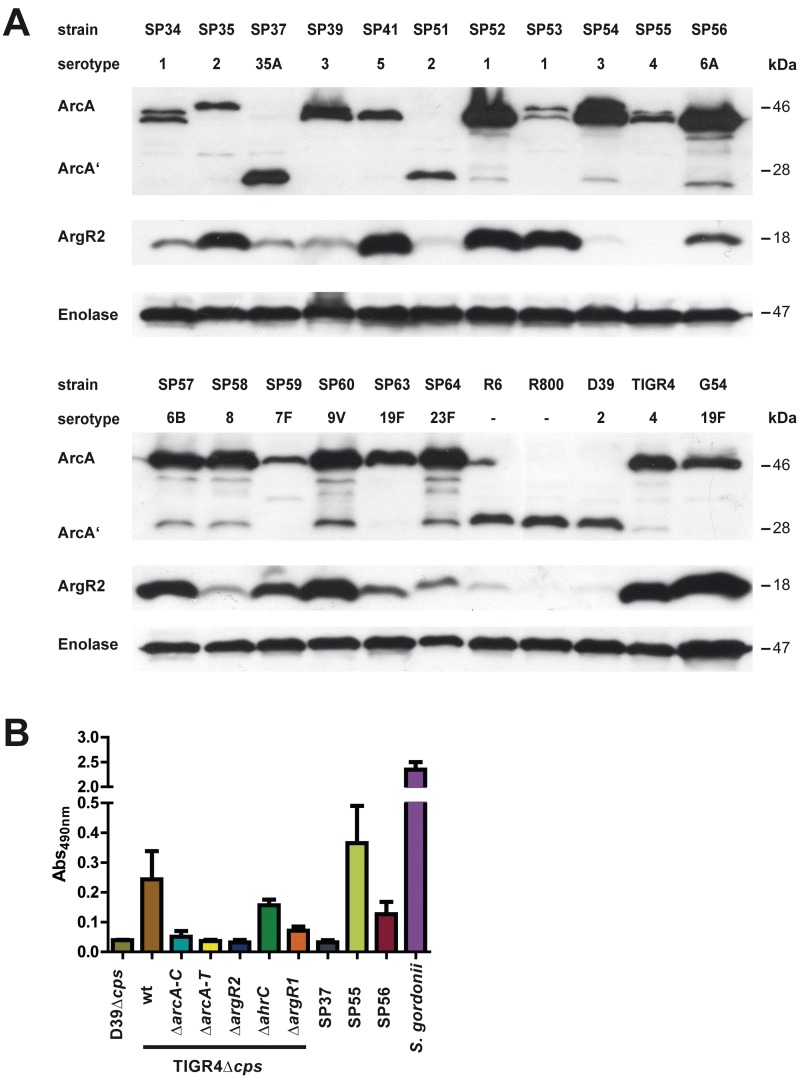
The expression of cytoplasmic ArcA and ArgR2 in 24 different pneumococcal strains and serotypes was assessed by immunoblot analysis (Fig. 5A). The results indicated the presence of ArcA in all of the strains analyzed. However, serotypes 35A (SP37) and 2 (SP51), including D39 and strains R6 and R800, produced only a truncated ArcA protein. Interestingly, the nonfunctional ArcA protein correlated with low levels or a lack of ArgR2 expression (Fig. 5B). However, ArgR2 expression was also absent from some strains expressing a full-length and functional ArcA protein (for example, SP55) (Fig. 5A and B).
FIG 5 .
Strain-specific production of ArgR2 and AD activity. (A) Immunoblot analysis of ArcA and ArgR2 expression in pneumococci. Proteins were detected with specific anti-ArcA or ArgR2 antibodies. (B) AD activity was monitored in a colorimetric enzyme assay by determination of the citrulline produced. The strains were cultivated in THY medium to an OD600 of 1.0, and 20 µg of whole protein extract was used. wt, wild type.
Enzymatic activity of the AD.
The activity of the AD was monitored in a colorimetric enzyme assay measuring the amount of produced citrulline (Fig. 5B). The results showed no enzyme activity for D39, TIGR4ΔarcAT, TIGR4ΔargR2, and serotype 35A (SP37). In contrast, AD enzyme activity was observed for TIGR4, TIGR4ΔargR1, TIGR4ΔahrC, SP55, SP56, and S. gordonii. Taken together, the immunoblot and Northern blot analyses, the enzyme activity assay, and EMSA indicate that ArgR2 is a specific positive transcriptional regulator of the pneumococcal arcABCDT operon for some S. pneumoniae strains.
Influence of the ADS, ArcD, and ArgR2 pneumococcal colonization and virulence.
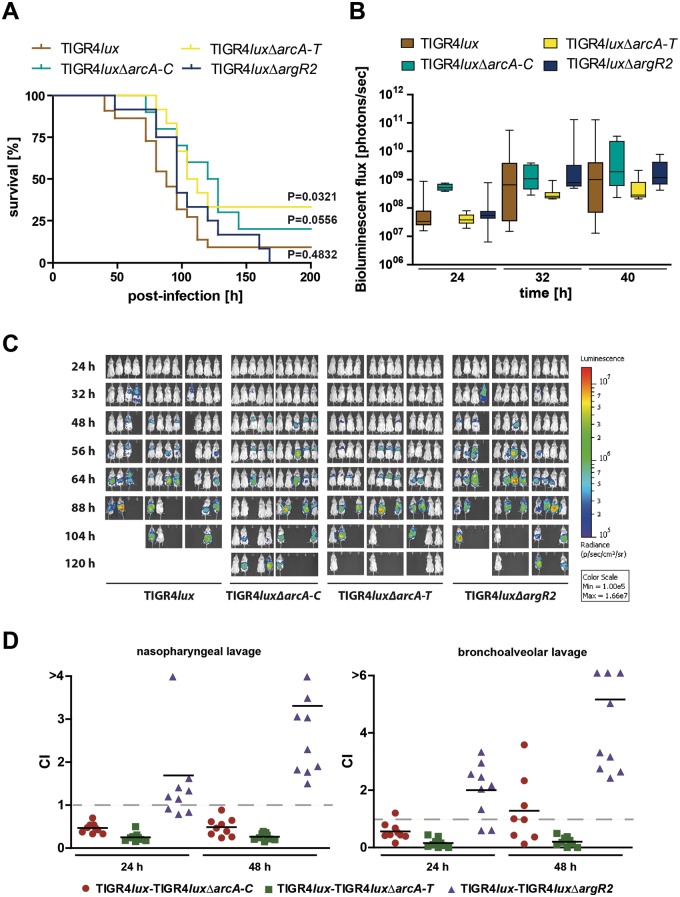
To assess the impact of the ADS and its regulator ArgR2 on TIGR4 colonization and virulence, the mouse acute-pneumonia model was used. Female CD-1 outbred mice (n = 12) were intranasally infected with 7.5 × 107 wild-type or isogenic TIGR4luxΔarcA-C, TIGR4luxΔarcA-T, and TIGR4luxΔargR2 pneumococci. The infection process was monitored in real time with an optical bioluminescence imaging system (IVIS Spectrum System). No significant differences were observed between the survival time of mice infected with wild-type or mutant pneumococci deficient for the proteins ArcA to ArcC or regulator ArgR2, while the deficiency of ArcD in the ΔarcA-T mutant attenuated pneumococci (Fig. 6A–C). To investigate the effects in more detail, coinfection experiments were conducted to decipher directly deficiencies in the in vivo bacterial fitness of the mutants and their abilities to colonize mice or to cause invasive infections. Mice (n = 9) were coinfected intranasally with equal amounts (each strain received 2.5 × 107 CFU) of the mutant strain TIGR4luxΔarcA-C, TIGR4luxΔarcA-T, or TIGR4luxΔargR2 and parental strain TIGR4lux (Fig. 6D). The ratio of wild-type to mutant bacteria was calculated 24 and 48 h postinfection by determination of the bacterial loads in the nasopharynx and airways of each individual mouse. The results of the nasopharyngeal and bronchoalveolar lavage fluid analysis showed that the ΔarcA-C and ΔarcA-T TIGR4 mutants, respectively, were outcompeted by wild-type bacteria in these host niches. In contrast, the in vivo survival rate of the TIGR4ΔargR2 mutant was obviously higher in both habitats than that of the isogenic wild-type strain. Importantly, the TIGR4 and D39 mutants deficient in genes of the ADS or the regulator ArgR2 had no growth defect in complex medium or in CDM (data not show). Hence, the in vivo experiments showed that the fitness of the TIGR4 ADS-deficient mutants was reduced under infection conditions and that arginine metabolism is coupled to virulence. Remarkably, the ArgR2-deficient mutant outcompeted the isogenic TIGR4 wild type, suggesting other, as-yet-unknown, regulatory effects of ArgR2.
FIG 6 .
Impact of the ADS enzymes, ArcD antiporter, and regulator ArgR2 on pneumococcal colonization and virulence in mice. (A) Survival of CD-1 mice after intranasal infection with pneumococci. Groups of mice (n = 12) were intranasally infected with 7.5 × 107 CFU of wild type S. pneumoniae TIGR4 or the isogenic ΔarcA-C, ΔarcA-T, or ΔargR2 mutant. (B and C) Bioluminescent optical imaging of pneumococcal dissemination after intranasal infection of CD-1 mice (n = 12). Dissemination of bioluminescent TIGR4lux, TIGR4luxΔarcA-C, TIGR4luxΔarcA-T, or TIGR4luxΔargR2 was analyzed at the time points indicated by determination of the luminescence intensity measured with the IVIS Spectrum System. The bioluminescent flux of grouped mice is represented in the box whisker graph (B). (D) Intranasal coinfection of mice with bioluminescent TIGR4lux together with TIGR4luxΔarcA-C, TIGR4luxΔarcA-T, or TIGR4luxΔargR2. Groups of CD-1 mice (n = 9) were infected with 2.5 × 107 CFU of the wild type and one of the isogenic pneumococcal mutants. At the time points indicated, mice were sacrificed and bacterial loads in the nasopharynx (nasopharyngeal lavage) and airways (bronchoalveolar lavage) were counted after the bacteria were plated on blood agar plates. CI values lower than 1 indicate higher growth of wild-type bacteria.
Influence of the ADS and regulator ArgR2 on pneumococcal phagocytosis by macrophages.
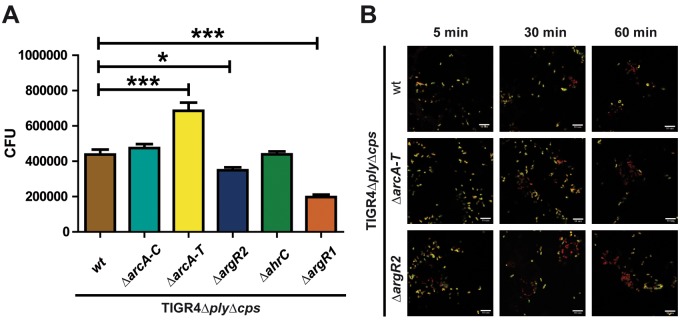
To investigate whether the ADS protects pneumococci against phagocytosis by macrophages or improves intracellular survival, nonencapsulated TIGR4Δcps pneumococci and the ΔarcA-C, ΔarcA-T, ΔargR2, ΔargR1, and ΔahrC isogenic mutants were incubated for 30 min with J774 macrophages. After the extracellular bacteria were killed, the numbers of internalized and recovered pneumococci were determined. The phagocytosis rate of TIGR4Δcps and its isogenic ΔarcA-C mutant was not significantly altered. However, the deletion of all of the genes of the AD operon (ΔarcA-T mutant) increased the internalization of pneumococci in macrophages significantly. In contrast, mutations of the regulatory proteins ArgR2 (ΔargR2) and AhrC (ΔahrC) decreased the rate of phagocytosis significantly (Fig. 7). The latter results may explain why the ArgR2-deficient mutant outcompeted the wild type under in vivo conditions.
FIG 7 .
Deficiency of arginine uptake and arginine metabolism regulator ArgR2 interferes with pneumococcal phagocytosis. J774A.1 murine macrophages were infected with a multiplicity of infection of 50 pneumococci of wild-type (wt) strain TIGR4 or its isogenic arcA-C, arcA-T, or argR2 mutant. (A) Viable and recovered intracellular pneumococci were determined by colony counting 30 min postinfection by applying the antibiotic protection assay. Shown are the mean values and standard deviations of three independent experiments performed in triplicate. *, P < 0.05; ***, P < 0.001. (B) Immunofluorescence microscopy of host cell-attached and intracellular pneumococci. J774A.1 murine macrophages were infected with pneumococcal strain TIGR4 or the ΔarcA-T or ΔargR2 mutant for 5, 30, or 60 min. Intracellular pneumococci were stained with Alexa 568 (red), while adherent bacteria appear yellow (Alexa 488 and Alexa 568). Bars represent 10 µm.
DISCUSSION
Arginine is one of the amino acids that cannot be synthesized de novo and have to be taken up by pneumococci (6, 44), since the complete biosynthetic pathway is missing from all of the pneumococcal genomes that have been sequenced. The only genes found in some pneumococcal genomes, like that of D39, encode the argininosuccinate synthetase (ArgG) and the argininosuccinase (ArgH) (16, 44). These enzymes catalyze the conversion of citrulline to arginine. Mouse models of meningitis and pneumonia showed that ArgG/ArgH-deficient pneumococci are attenuated in outgrowth in the lungs, blood, and cerebrospinal fluid (16). The catabolic ADS has been described in other streptococcal species, such as S. suis, where it covers a broad spectrum of functions. It consists of the AD, the cOCT, and the CK and catalyzes the conversion of arginine to ornithine under the release of ammonia and carbon dioxide while simultaneously generating ATP. In contrast to other prokaryotic organisms, including P. aeruginosa (33) and S. aureus (40), it seems unlikely that the main function of the ADS in group A streptococci, including S. pneumoniae, is to provide energy and metabolic intermediates. For several streptococci, such as oral bacteria and S. pyogenes, the release of ammonia plays an important role in adaptation to an acidic environment (22, 24, 31, 32, 49). Importantly, the enzymes of the ADS are still active at a very low environmental pH (50). However, pneumococci cultured under acidic conditions (pH 5.5) did not survive better than isogenic arcA-C mutants, which are unable to produce ammonia as the end product of the catabolism of arginine by the ADS (data not shown). Hence, the levels of ammonia produced are most likely not sufficient to alter the environmental pH or acid conditions in phagocytic compartments.
The regulation of arginine metabolism is generally performed by ArgR-type regulators, of which the pneumococcal genome encodes three. In S. pneumoniae D39, ArgR1 and AhrC form a heterohexameric complex that is bound to the effector molecule arginine. Under arginine-rich conditions, this complex directly represses five amino acid transport operons in a cooperative way, and it has been suggested that AbpA, AbpB, and ArtPQ, which are located in three of the five operons, form an arginine uptake unit. In addition, AliB, which is required for growth in medium with peptides (in particular, Arg-Pro-Pro and Arg-Pro-Pro-Gly-Phe) as the sole source of arginine is also controlled by ArgR1/AhrC (44). Importantly, ArgR1 and AhrC have no impact on the catabolic arcABCDT operon of the ADS of pneumococcal strain D39 (44) and differences in arginine uptake have not been measured. In L. lactis, the ArgR/AhrC complex represses in the presence of arginine the three arginine biosynthetic operons argCJDBF, gltS-argE, and argGH while activating the catabolic arcABD1C1C2TD2-yvaD operon (51–53). Also in other organisms like Escherichia coli (54), P. aeruginosa (55, 56), B. subtilis (57–59), L. plantarum (60), and Enterococcus faecalis (38), ArgR-type regulators are mediators of arginine metabolism and thus influence the expression of arginine biosynthetic and catabolic genes.
Here we show for the first time that, in S. pneumoniae TIGR4 and probably other pneumococcal strains, ArgR2 positively regulates the expression of the ADS, including the arginine-ornithine antiporter ArcD, which is shown to be essential for arginine uptake from the extracellular milieu. In addition, ArcD is essential for the full virulence of TIGR4 while the enzymes of the ADS have only a minor impact on pneumococcal fitness under in vivo conditions. Strikingly, a deficiency of ArgR2 has a detrimental effect despite downregulation of the ADS, suggesting that ArgR2 regulates other genes important for bacterial fitness or virulence and compensates for the ADS defect.
Immunoblot and Northern blot analyses demonstrated that ArgR2 is an activator of ArcABCDT expression in S. pneumoniae TIGR4 (Fig. 2B and 3C). The ArgR2-deficient TIGR4ΔargR2 mutant showed a significant decrease in arc gene and protein expression. In silico analysis and RT-PCR suggested that the arcABCDT genes form an operon. However, the transcript lengths of 2.6 and 1.6 kb observed by Northern blot analysis may point to a degradation by RNase(s) or, alternatively, that the genes of the arc operon are not transcribed as a single transcriptional unit. The phenomenon of the inconsistent size of mRNA transcripts has previously also been observed in S. suis (42; Markus Fulde personal communication). To draw reliable conclusions about transcript size, further studies are required. The results of the EMSA with fragments of the arcABC promoter and recombinant ArgR2 reveal that ArgR2 interacts with the arcABC promoter under in vitro conditions (Fig. 3D). The ArgR-type transcriptional regulators influence gene expression by binding to so-called ARG operator sites that precede target genes. ARG operator sites consist of pairs of 18-bp palindromic sequences (called ARG boxes). For E. coli, a consensus sequence was characterized (5′-TNTGNATWWWWATNCANA-3′ [conserved residues are underlined, N is any nucleotide, and W is A or T]) (54). Similar sequences have been described for other microorganisms (61–63). Even though the typical ARG box could not be determined for the promoter regions of arcA and arcD, these genes are regulated by ArgR2.
Similar to pneumococci, S. suis also contains three ArgR-type regulators. The regulator ArgR of S. suis, exhibiting high sequence homology to pneumococcal ArgR2, was found by microarray and quantitative RT-PCR analyses to regulate the S. suis arc operon (42). In streptococcal species such as S. rattus (24) and S. gordonii (37, 64), the expression of the arc genes is also ArgR2 dependent, suggesting that ArgR2 or its orthologues are specific regulators of ADS operons.
Immunoblot analyses of selected pneumococcal strains showed heterogeneity of expression for ArcA and ArgR2. In TIGR4, the arc operon is regulated by ArgR2 while in other strains the levels of ArgR2 and ArcA do not correlate, suggesting strain-specific regulation of the ADS in pneumococci. It is also still unclear whether ArgR2 regulates or whether ArgR2 itself is regulated by unknown stimuli. By comparing pneumococcal strain TIGR4 with D39, R6, SP37, SP51, and R800, respectively, several differences in the ADS and the ArgR-type regulators have been identified in this study. Although the regulator ArgR2 is the most important activator of the ADS in TIGR4, its expression could not be detected or was extremely low in D39, R6, SP37, SP51, and R800 (Fig. 3C and 5A). Despite lacking or producing low levels of the regulatory protein ArgR2, D39 and the other strains showed constitutive expression of the enzymes of the ADS (AD, OCT, CK) (Fig. 3C). In this respect, it is important to mention that the gene encoding the AD has a premature stop codon in these strains and that these strains consequently lack a functional AD (Fig. 5B). Previous transcriptome analysis demonstrated that the D39 arc operon is repressed by CcpA, which can be derepressed under carbon limitation, probably allowing the use of arginine as an alternative energy source (44, 65). However, the growth of wild-type TIGR4 and that of the isogenic ΔarcA-C mutant cultured in CDM with a low glucose concentration and in the presence of increasing arginine concentrations did not differ significantly (data not shown).
The deficiency of enzymes of the ADS and also the deficiency of ArgR2 did not impair the full virulence of TIGR4 in the mouse acute-pneumonia model (Fig. 6A), whereas the lack of a functional ArcD antiporter attenuated TIGR4. However, coinfection experiments indicated that the absence of ADS enzymes reduced the fitness of TIGR4 because the wild type outcompeted TIGR4ΔarcA-C in the nasopharynx and lungs. Similar to the acute-pneumonia model, TIGR4ΔarcA-T is significantly attenuated, highlighting the importance of the antiporter ArcD for the proper fitness and virulence of TIGR4 and probably also other pneumococcal strains. According to the metabolome analysis performed in this study and a recent study, the antiporter ArcD is the major pneumococcal arginine uptake system and is essential in strains lacking ArgGH, like TIGR4 (35). Additionally, it has to be mentioned here that the antiporter ArcD requires ornithine for its functional activity and that the ADS is the only source of ornithine in pneumococci. The lack of the ArcD transporter affected capsule expression in S. pneumoniae D39, which results in significantly enhanced phagocytosis and attenuation of D39 under in vivo conditions (35). However, when measuring the relative amount of capsular polysaccharide (CPS) by flow cytometry with anti-serotype 2- or anti-serotype 4-specific antiserum (66), the mutant D39ΔarcA-T generated in this study and lacking functional ArcD showed only a slightly smaller amount of CPS than the isogenic wild-type D39 (geometric mean fluorescence intensity [GMFI] values: D39, 389; D39ΔarcA-T, 269; D39ΔarcA-C, 327; D39ΔargR2, 408; D39Δcps, 76). Importantly, the mutant TIGR4ΔarcA-T produces amounts of CPS similar to those of isogenic wild-type TIGR4 (GMFI values: TIGR4, 342; TIGR4ΔarcA-T, 356; TIGR4ΔarcA-C, 346; TIGR4ΔargR2, 285; TIGR4Δcps, 141). Hence, the effect seems to be strain dependent and if there is any reduction of CPS, it is only to a minor degree compared to the nonencapsulated phenotypes.
The analysis of extracellular metabolites of pneumococci cultured in CDM showed that the arginine uptake of TIGR4 is 20-fold higher than that of D39, suggesting that uptake of arginine is highly reduced in pneumococci lacking a functional arginine degradation pathway, like D39. Because there is no link between arginine metabolism and alterations of cell wall composition, the results of this study and that of Gupta et al. (35) may imply that ArcD has another, not-yet-identified, function that affects the linkage of the CPS to the pneumococcal cell wall. As a secondary effect, the virulence of D39 pneumococci is impaired. The function of ArcT as an Xaa-His dipeptidase is based on in silico analysis and has not been experimentally proven. However, the arcT gene is also present in the genomes of other streptococcal species and it has been shown to be essential in a chinchilla otitis media model (35, 67).
Strikingly, ArgR2-deficient TIGR4ΔargR2 has a higher virulence potential than the wild type, which is indicated by its ability to outcompete the wild type in the nasopharynx and lungs (Fig. 6D). This suggests that ArgR2 regulates other genes encoding proteins important for bacterial fitness and virulence, respectively, and maybe compensates for the defect of the ADS and ArcD. Phagocytosis assays (Fig. 7) indicate a higher uptake of the mutant TIGR4ΔarcA-T than the isogenic wild type. In contrast, the uptake of the argR2 mutant was lower than that of TIRG4. However, accelerated or decelerated killing of intracellular pneumococci was not detected (data not shown), suggesting that the impaired colonization of the mouse lower airways by ADS-deficient pneumococci is due to their higher rates of uptake by professional phagocytes. In contrast, the argR2 mutant may overgrow the wild type because of its reduced phagocytosis. To explore a potential effect of ArgR2 deficiency on the transcriptome of TIGR4 and to identify further genes regulated by ArgR2, microarray analysis was performed. The results confirmed the downregulation of arcABCDT gene expression (mutant/wild-type ratios of 0.19, 0.17, 046, 0.42, and 0.38, respectively) and the absence of an argR2 transcript (ratio of 0.01) in the mutant TIGR4ΔargR2. Other downregulated genes were not identified. Importantly, only a few genes were upregulated in the argR2 mutant, such as the genes encoding the AdhE (alcohol dehydrogenase) or SP_1282 (ABC transporter ATP-binding protein) and the kdgA, kdgG, and gno genes encoding the enzymes of 2-keto-3-deoxygluconate metabolism. These differentially regulated genes probably do not enhance pneumococcal virulence. Hence, further studies are needed to decipher other potential targets of ArgR2.
In conclusion, this study identified ArgR2 as a key regulator of the ADS of S. pneumoniae TIGR4, and it can be assumed that this also applies to other pneumococcal strains. The regulation of the pneumococcal ADS seems to be strain dependent, as indicated by the differences between TIGR4 and D39. Importantly, ArcD, encoded by the arc operon, has been identified as a major arginine transporter that is essential for full virulence. Finally, the increased fitness of the argR2 mutant under in vivo conditions pointed to additional regulatory functions of ArgR2. The additional genes targeted by ArgR2 have to be identified in further studies to unravel the other regulatory functions of ArgR2 and understand the higher virulence potential of TIGR4ΔargR2.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. pneumoniae TIGR4 (serotype 4) and S. pneumoniae D39 (serotype 2; NCTC7466) and their isogenic mutants (see Table S1 in the supplemental material) were cultured to mid-log phase (OD600, 0.35 to 0.4) in THY medium (Todd-Hewitt broth [Oxoid, Basingstoke, England] supplemented with 0.5% yeast extract [Roth, Karlsruhe, Germany]) or CDM or grown on Columbia blood agar plates (Oxoid) at 37°C in 5% CO2. In this study, we established RPMI 1640 (PAA Laboratories) supplemented with 30.5 mM glucose, 0.65 mM uracil, 0.27 mM adenine, 1.1 mM glycine, 0.24 mM choline chloride, 1.7 mM NaH2PO4 · H2O, 3.8 mM Na2HPO4, and 27 mM NaHCO3 as CDM. The E. coli strains used in cloning procedures (see Table S1) were cultured on Luria-Bertani (LB) agar or in LB broth. Transformation of E. coli and pneumococci was conducted by standard protocols described recently (68). Pneumococcal mutants were cultured in the presence of the appropriate antibiotics, i.e., chloramphenicol (4 µg/ml), erythromycin (5 µg/ml), and/or kanamycin (150 µg/ml).
DNA techniques and sequence analysis.
For the primers and plasmids used in this study, see Tables S2 and S3 in the supplemental material. Bacterial chromosomal DNA isolation and purification were performed by a standard phenol-chloroform extraction method as described previously (69). PCR amplifications were carried out in 50-µl volumes, and reactions were subjected to denaturation at 95°C, 30 cycles, primer annealing for 0.5 min, and elongation at 72°C. The annealing temperatures depended on the primers used, and the extension time depended on the length of the expected PCR product. PCR products were purified with the Wizard SV Gel and PCR Cleanup System (Promega), and plasmids were extracted according to the Wizard Plus SV Minipreps DNA Purification System (Promega) protocol. Oligonucleotides were synthesized by Eurofins MWG Operon (Germany).
Construction of plasmids for mutagenesis and pneumococcal transformation.
Pneumococcal mutants deficient in genes of the ADS operon or regulatory genes argR1, argR2, and ahrC, respectively, were generated by insertion deletion mutagenesis as previously described (6). The genes, including upstream and downstream sequences, were amplified by PCR with the primer combinations indicated (see Table S3 in the supplemental material). The DNA fragment for arcA was amplified by PCR with TIGR4 chromosomal DNA as the template and primers M9/M10. Similarly, arcC was amplified with M17/18, arcT was amplified with M23/24, argR2 was amplified with P586/P587, ahrC was amplified with P590/591, and argR1 was amplified with M27/M28. The PCR products were cloned into the pGEM-T Easy vector (Invitrogen), resulting in plasmids pGEM695 (arcA), pGEM611 (arcC), pGEM615 (arcT), pGEM699 (argR2), pGEM703 (ahrC), and pGEM708 (argR1), respectively. The recombinant plasmids harboring the desired DNA inserts were used as the templates for inverse PCRs with primer pairs M55/56 for arcA, M59/60 for arc, M63/64 for arcT, P588/P589 for argR2, P594/595 for ahrC, and M65/P595 for argR1. The deleted gene sequences were replaced with an ermB erythromycin resistance gene cassette and amplified by PCR with primers T99 and T100 and plasmid pJDC9 (70) as the template. This resulted in plasmids pGEM696 (arcA::ermR), pGEM612 (arcC::ermR), pGEM707 (arcT::ermR), pGEM700 (argR2::ermR), pGEM704 (ahrC::ermR), and pGEM709 (argR1::ermR), which were used to transform pneumococci and generate mutants (see Table S2 in the supplemental material). The integrity of the antibiotic resistance gene cassettes in pneumococcal mutants was verified by PCR (data not shown). In addition, immunoblot assays with specific antibodies generated against recombinant proteins of the ADS or regulators indicated successful mutagenesis.
RNA preparation, Northern blot analysis, and cDNA synthesis.
Pneumococcal RNA was purified with the RNeasy minikit, including a DNase digestion step with the RNase-Free DNase Set (Qiagen), as previously described (6). RNA concentrations were determined by measuring the A280/A260 ratio with the NanoDrop ND 1000 (PeqLab). Total RNA samples (3 to 5 µg per lane) were electrophoretically separated in denaturing agarose gels (1.2%) and vacuum transferred to Hybond N+ nylon membranes (Amersham Biosciences). RNAs were cross-linked to the membranes by UV incubation, stained with methylene blue, and then hybridized with DIG-labeled RNA probes at 68°C overnight. Hybridization signals were detected with anti-DIG–alkaline phosphatase, Fab fragments, and CDP Star (Roche Applied Science). DIG-labeled RNA probes for detection of the arcA, arcB, arcC, and arcDT mRNAs were generated with the DIG RNA Labeling kit (SP6/T7) (Roche). For the oligonucleotides used to generate probes for Northern blot analysis, see Table S3 in the supplemental material. RT was performed to synthesize cDNA with 5 to 10 µg of RNA. RNA was incubated with 10 nmol of deoxynucleoside triphosphate in 20 µl of RNase-free water for 5 min at 65°C and kept on ice for 1 min, and then 4 µl of First-Strand Buffer (5×), 1 µl of random primers [pd(N)6; GE Healthcare], 1 µl of dithiothreitol (DTT; 0.1 M), 1 µl of RNasin (Promega), and 1 µl of SuperScript III reverse transcriptase (Invitrogen) were added to amplify the cDNA. The cDNA was employed as the template in the PCRs, and control PCRs were conducted with genomic DNA and total RNA as the template.
Production and purification of recombinant proteins.
Recombinant ArcA, ArcB, ArcC, ArgR1, ArgR2, and AhrC were produced as His-tagged fusion proteins in E. coli with the pET28TEV plasmid system (Novagen). Genes were amplified with deleted translation initiation codons by PCR from chromosomal DNA with specific oligonucleotide primers and incorporated restriction sites (see Table S3 in the supplemental material). PCR products were restricted by NheI and HindIII (NEB), and the fragments were cloned into similarly digested pET28TEV expression vectors (71) and verified by sequencing (Seqlab). The resulting plasmids, pET28arcA, pET28arcB, pET28arcC, pET28argR1, pET28argR2, and pET28ahrC, were transformed into E. coli strain BL21(DE3) (Novagen). For overexpression of recombinant proteins, the strains were grown in LB to an OD600 of 0.8 and induced with 1.0 mM IPTG (isopropyl-β-d-thiogalactopyranoside); the bacteria were then harvested after 2 h of induction. The N-terminally His6-tagged proteins (His6-ArcA, His6-ArcB, His6-ArcC, His6-ArgR1, His6-ArgR2, His6-AhrC) were purified by affinity chromatography with His-Trap Ni-nitrilotriacetic acid columns (GE Healthcare) according to the manufacturer’s instructions. Polyclonal mouse antisera were generated by immunization of mice with the purified protein derivative according to standard methods.
Immunoblot analysis.
Pneumococci were cultured in THY or CDM to the late exponential growth phase (OD600: THY, 1.0; CDM, 0.8), harvested by centrifugation, and washed once with phosphate-buffered saline (PBS). The bacteria were resuspended in PBS and lysed by ultrasonication. The lysate was centrifuged, and the supernatant was used for further investigation. The protein content was determined by the Bradford protein assay (Sigma-Aldrich). The protein extracts (20 µg) were mixed with an equal volume of SDS sample buffer, incubated at 95°C for 10 min, and separated by SDS–12% gel electrophoresis. The proteins were transferred to a nitrocellulose membrane with a semidry blotting system (Bio-Rad). The membranes were blocked with 10% skim milk (Roth) and incubated with mouse-derived polyclonal antiprotein antiserum (1:1,000). Goat anti-mouse IgG-peroxidase conjugate (Dianova; 1:5,000) was used as the secondary antibody, and binding activity was detected by using enhanced-chemiluminescence substrate (ECL; GE Healthcare). Rabbit-derived polyclonal antienolase antiserum and goat anti-rabbit IgG-peroxidase conjugate (Dianova; 1:5,000) was used as a loading control.
EMSA.
DNA fragments of the putative arcABCDT promoter regions were amplified by PCR with specific primer pairs (see Table S3 in the supplemental material). The promoter sequences (0.2 pmol) were incubated with different concentrations of His6-tagged ArgR2 in binding buffer (10 mM Tris/HCl [pH 7.5], 0.5 mM DTT, 5% [vol/vol] glycerol, 200 mM KCl, 5 mM MgCl2, 2.5 mM CaCl2, 10 mM arginine) for 30 min at room temperature. Protein-DNA complexes were electrophoretically separated in a native 5% polyacrylamide gel and visualized by ethidium bromide staining. Bovine serum albumin (0.2 pmol) was used as a negative control.
Determination of extracellular metabolites.
To analyze arginine uptake and ornithine secretion by pneumococci, bacteria cultured for 6 h on blood agar plates were diluted in CDM to a starting OD600 of 0.1 and incubated at 37°C. At each time point, 2 ml of medium was taken and filtered through a 0.2-µm sterile filter (Sarstaedt AG, Nürnberg, Germany) and directly frozen until analysis. 1H-NMR analysis was performed as previously described (72). In brief, 400 µl of the sample was mixed with 200 µl of sodium hydrogen phosphate buffer (0.2 mol/liter, pH 7.0), including 1 mmol/liter trimethylsilyl propanoic acid made up with 50% D2O for 1H-NMR analysis. A Bruker AVANCE-II 600 NMR spectrometer operated by TOPSPIN 3.1 software was used (both from Bruker Biospin, Rheinstetten, Germany). Qualitative and quantitative data analyses were carried out by using AMIX (Bruker Biospin, Rheinstetten, Germany).
Enzymatic activity of the AD.
S. pneumoniae was grown in THY medium at 37°C in 5% CO2. AD enzyme activity was measured by monitoring the production of citrulline from arginine as previously described (73, 74). Briefly, 100 µg of protein extract from bacteria grown in THY medium was incubated for 2 h at 37°C in 0.1 M potassium phosphate buffer containing 10 mM arginine (500 µl). After the addition of 250 µl of a mixture of H2SO4 and H3PO4 (1:3, vol/vol) and 31.3 µl of a 3% diacetyl monoxime solution, the suspension was heated for 15 min at 99°C. The relative level of citrulline, as a readout of AD activity, was determined colorimetrically at a wavelength of 490 nm.
Ethics statement.
The animal experiments conducted in this study were done in strict accordance with the guidelines of the ethics committee at The University of Greifswald, the German regulations of the Society for Laboratory Animal Science (GVSOLAS), and the European Health Law of the Federation of Laboratory Animal Science Associations (FELASA). The experiments were approved by the Landesamt für Landwirtschaft, Lebensmittelsicherheit und Fischerei Mecklenburg-Vorpommern (LALLFV M-V; Rostock, Germany) and the LALLFV M-V ethics board (LALLF M-V permits 7221.3-1.1-006/09 and 7221.3-1.1-019/11). All efforts were made to minimize suffering and ensure the highest ethical standards.
The mouse acute-pneumonia model and coinfections.
Female CD-1 outbred mice (Charles River, Sulzfeld, Germany) 8 to 10 weeks of age were used in infection experiments. Pneumococci were cultured in THY supplemented with 10% fetal bovine serum (FBS) to an OD600 of 0.35, washed once with 0.5% PBS–1% FBS, and diluted in PBS–1% FBS to get infection doses of 7.5 × 107 CFU/10 µl. Prior to intranasal infection, mice were anesthetized by intraperitoneal injections of ketamine (Ketanest S; Pfizer Pharma, Karslruhe, Germany) and xylazine (Rompun; Provet AG, Lyssach, Germany) as previously described (71, 75). Each animal was scuffed, with the nose held upright, and a 20-µl bacterial suspension (10 µl of bacteria and 10 µl of hyaluronidase [90 U]) was administered intranasally by adding a series of small droplets into the nostrils for involuntary inhalation. The infection dose was confirmed by the determination of CFU counts on blood agar plates.
In competition infection experiments, 2.5 × 107 CFU of wild-type and mutant (TIGR4luxΔarcAC, TIGR4luxΔarcAT, or TIGR4lux ΔargR2) bacteria were mixed at a 1:1 ratio. Determination of CFU counts in the nasopharynx and bronchi was performed after intranasal infection at prechosen time intervals (24 and 48 h) postinfection and as described previously (68, 76). Briefly, mice were sacrificed and their tracheas were dissected. One milliliter of sterile PBS–0.5% FBS was passed through the nasopharynx or inserted into the lung with a tracheal cannula and collected after passage (77). The output of mutant versus wild-type bacteria was determined on selective blood agar plates (Oxoid, Basingstoke, United Kingdom) containing kanamycin and/or erythromycin. The competitive index (CI) was calculated as the ratio of mutant to wild-type output CFU counts divided by the ratio of mutant to wild-type input CFU counts. A value of 1 indicates identical output CFU counts of wild-type and mutant bacteria, while a CI value lower than 1 indicates a higher output of wild-type bacteria.
Phagocytosis experiments and antibiotic protection assays.
Phagocytosis assays were conducted with J774A.1 murine macrophages (DSMZ, Braunschweig, Germany). Macrophages were incubated for 30 min with a multiplicity of infection (MOI) of 50 bacteria per cell, and the numbers of intracellular and recovered viable pneumococci were quantified by the antibiotic protection assay as described previously (7, 76). All experiments were performed at least three times in triplicate.
Cell culture adherence assays and immunofluorescence microscopy.
Pneumococcal adherence to the human lung epithelial cell line A549 (ATCC CCl-185; type II pneumocytes) was tested. The eukaryotic cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS at 37°C and 5% CO2 and infected at an MOI of 25 bacteria per cell as previously described (68, 78). Postinfection, unbound bacteria were removed and the infected host cells were fixed on glass coverslips with 3.7% paraformaldehyde. Extracellular bacteria were stained with antipneumococcal antibodies and Alexa 488-conjugated goat anti-rabbit secondary antibody, and after permeabilization with Triton X-100 (0.1%), intracellular bacteria were stained with antipneumococcal antibodies and Alexa 568- and Alexa 488-conjugated goat anti-rabbit IgG secondary antibodies, respectively, as previously described (68, 76). Immunofluorescence microscopy was performed with a fluorescence microscope (Zeiss Axio-Observer.Z1 with VisiGrid, Coolsnap HQ), and image acquisition was done with the VisiView Imaging software (Visitron Systems GmbH, Puchheim, Germany). Each bar in the images represents 10 µm. All experiments were performed at least three times with two or more replicate wells tested for each experimental setup.
SUPPLEMENTAL MATERIAL
Pneumococcal and E. coli wild-type strains used in this study and generated pneumococcal mutants employed in functional assays.
Plasmids used in this study to perform mutagenesis of pneumococci and produce recombinant proteins.
Primers used in this study to construct deletion mutants of targeted genes, perform expression cloning, EMSA or Northern blot analysis.
ACKNOWLEDGMENTS
This work was supported by a grant from the Deutsche Forschungsgemeinschaft DFG (HA 3125/3-2 to S.H. and GRK 1870 to S.H. and M.L.).
We gratefully acknowledge Gerhard Burchhardt, Kristine Sievert-Giermann, and Birgit Rietow for technical assistance, helpful discussions, and data evaluation (Department of Genetics, University of Greifswald).
Footnotes
Citation Schulz C, Gierok P, Petruschka L, Lalk M, Mäder U, Hammerschmidt S. 2014. Regulation of the arginine deiminase system by ArgR2 interferes with arginine metabolism and fitness of Streptococcus pneumoniae. mBio 5(6):e01858-14. doi:10.1128/mBio.01858-14.
REFERENCES
- 1. Bogaert D, van Belkum A, Sluijter M, Luijendijk A, de Groot R, Rümke HC, Verbrugh HA, Hermans PW. 2004. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet 363:1871–1872. 10.1016/S0140-6736(04)16357-5. [DOI] [PubMed] [Google Scholar]
- 2. Cartwright K. 2002. Pneumococcal disease in Western Europe: burden of disease, antibiotic resistance and management. Eur. J. Pediatr. 161:188–195. 10.1007/s00431-001-0907-3. [DOI] [PubMed] [Google Scholar]
- 3. Musher DM. 1992. Infections caused by Streptococcus pneumoniae: clinical spectrum, pathogenesis, immunity, and treatment. Clin. Infect. Dis. 14:801–807. 10.1093/clinids/14.4.801. [DOI] [PubMed] [Google Scholar]
- 4. Gamez G, Hammerschmidt S. 2012. Combat pneumococcal infections: adhesins as candidates for protein-based vaccine development. Curr. Drug Targets 13:323–337. 10.2174/138945012799424697. [DOI] [PubMed] [Google Scholar]
- 5. Paterson GK, Mitchell TJ. 2006. Innate immunity and the pneumococcus. Microbiology 152:285–293. 10.1099/mic.0.28551-0. [DOI] [PubMed] [Google Scholar]
- 6. Härtel T, Eylert E, Schulz C, Petruschka L, Gierok P, Grubmüller S, Lalk M, Eisenreich W, Hammerschmidt S. 2012. Characterization of central carbon metabolism of Streptococcus pneumoniae by isotopologue profiling. J. Biol. Chem. 287:4260–4274. 10.1074/jbc.M111.304311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Härtel T, Klein M, Koedel U, Rohde M, Petruschka L, Hammerschmidt S. 2011. Impact of glutamine transporters on pneumococcal fitness under infection-related conditions. Infect. Immun. 79:44–58. 10.1128/IAI.00855-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Canepa A, Filho JC, Gutierrez A, Carrea A, Forsberg AM, Nilsson E, Verrina E, Perfumo F, Bergström J. 2002. Free amino acids in plasma, red blood cells, polymorphonuclear leukocytes, and muscle in normal and uraemic children. Nephrol. Dial. Transplant. 17:413–421. 10.1093/ndt/17.3.413. [DOI] [PubMed] [Google Scholar]
- 9. Sethuraman R, Lee TL, Chui JW, Tachibana S. 2006. Changes in amino acids and nitric oxide concentration in cerebrospinal fluid during labor pain. Neurochem. Res. 31:1127–1133. 10.1007/s11064-006-9133-8. [DOI] [PubMed] [Google Scholar]
- 10. Currie GA, Gyure L, Cifuentes L. 1979. Microenvironmental arginine depletion by macrophages in vivo. Br. J. Cancer 39:613–620. 10.1038/bjc.1979.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Albina JE, Mills CD, Barbul A, Thirkill CE, Henry WL, Jr, Mastrofrancesco B, Caldwell MD. 1988. Arginine metabolism in wounds. Am. J. Physiol. 254:E459–E467. [DOI] [PubMed] [Google Scholar]
- 12. Wu G. 2009. Amino acids: metabolism, functions, and nutrition. Amino Acids 37:1–17. 10.1007/s00726-009-0269-0. [DOI] [PubMed] [Google Scholar]
- 13. Li P, Yin YL, Li D, Kim SW, Wu G. 2007. Amino acids and immune function. Br. J. Nutr. 98:237–252. 10.1017/S000711450769936X. [DOI] [PubMed] [Google Scholar]
- 14. Ha HC, Sirisoma NS, Kuppusamy P, Zweier JL, Woster PM, Casero RA., Jr. 1998. The natural polyamine spermine functions directly as a free radical scavenger. Proc. Natl. Acad. Sci. U. S. A. 95:11140–11145. 10.1073/pnas.95.19.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chattopadhyay MK, Tabor CW, Tabor H. 2003. Polyamines protect Escherichia coli cells from the toxic effect of oxygen. Proc. Natl. Acad. Sci. U. S. A. 100:2261–2265. 10.1073/pnas.2627990100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Piet JR, Geldhoff M, van Schaik BD, Brouwer MC, Valls Seron M, Jakobs ME, Schipper K, Pannekoek Y, Zwinderman AH, van der Poll T, van Kampen AH, Baas F, van der Ende A, van de Beek D. 2014. Streptococcus pneumoniae arginine synthesis genes promote growth and virulence in pneumococcal meningitis. J. Infect. Dis. 209:1781–1791. 10.1093/infdis/jit818. [DOI] [PubMed] [Google Scholar]
- 17. Abdelal AT. 1979. Arginine catabolism by microorganisms. Annu. Rev. Microbiol. 33:139–168. 10.1146/annurev.mi.33.100179.001035. [DOI] [PubMed] [Google Scholar]
- 18. Blakemore RP, Canale-Parola E. 1976. Arginine catabolism by Treponema denticola. J. Bacteriol. 128:616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Broman K, Lauwers N, Stalon V, Wiame JM. 1978. Oxygen and nitrate in utilization by Bacillus licheniformis of the arginase and arginine deiminase routes of arginine catabolism and other factors affecting their syntheses. J. Bacteriol. 135:920–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Floderus E, Linder LE, Sund ML. 1990. Arginine catabolism by strains of oral streptococci. APMIS 98:1045–1052. 10.1111/j.1699-0463.1990.tb05033.x. [DOI] [PubMed] [Google Scholar]
- 21. Mercenier A, Simon JP, Haas D, Stalon V. 1980. Catabolism of l-arginine by Pseudomonas aeruginosa. J. Gen. Microbiol. 116:381–389. [DOI] [PubMed] [Google Scholar]
- 22. Burne RA, Marquis RE. 2000. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol. Lett. 193:1–6. 10.1111/j.1574-6968.2000.tb09393.x. [DOI] [PubMed] [Google Scholar]
- 23. Dong Y, Chen YY, Snyder JA, Burne RA. 2002. Isolation and molecular analysis of the gene cluster for the arginine deiminase system from Streptococcus gordonii DL1. Appl. Environ. Microbiol. 68:5549–5553. 10.1128/AEM.68.11.5549-5553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Griswold A, Chen YY, Snyder JA, Burne RA. 2004. Characterization of the arginine deiminase operon of Streptococcus rattus FA-1. Appl. Environ. Microbiol. 70:1321–1327. 10.1128/AEM.70.3.1321-1327.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gruening P, Fulde M, Valentin-Weigand P, Goethe R. 2006. Structure, regulation, and putative function of the arginine deiminase system of Streptococcus suis. J. Bacteriol. 188:361–369. 10.1128/JB.188.2.361-369.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maghnouj A, de Sousa Cabral TF, Stalon V, Vander Wauven C. 1998. The arcABDC gene cluster, encoding the arginine deiminase pathway of Bacillus licheniformis, and its activation by the arginine repressor ArgR. J. Bacteriol. 180:6468–6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vander Wauven C, Piérard A, Kley-Raymann M, Haas D. 1984. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol. 160:928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zúñiga M, Pérez G, González-Candelas F. 2002. Evolution of arginine deiminase (ADI) pathway genes. Mol. Phylogenet. Evol. 25:429–444. 10.1016/S1055-7903(02)00277-4. [DOI] [PubMed] [Google Scholar]
- 29. Poolman B. 1993. Energy transduction in lactic acid bacteria. FEMS Microbiol. Rev. 12:125–147. 10.1111/j.1574-6976.1993.tb00015.x. [DOI] [PubMed] [Google Scholar]
- 30. Wimmer F, Oberwinkler T, Bisle B, Tittor J, Oesterhelt D. 2008. Identification of the arginine/ornithine antiporter ArcD from Halobacterium salinarum. FEBS Lett. 582:3771–3775. 10.1016/j.febslet.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 31. Casiano-Colón A, Marquis RE. 1988. Role of the arginine deiminase system in protecting oral bacteria and an enzymatic basis for acid tolerance. Appl. Environ. Microbiol. 54:1318–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nascimento MM, Gordan VV, Garvan CW, Browngardt CM, Burne RA. 2009. Correlations of oral bacterial arginine and urea catabolism with caries experience. Oral Microbiol. Immunol. 24:89–95. 10.1111/j.1399-302X.2008.00477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hogardt M, Heesemann J. 2010. Adaptation of Pseudomonas aeruginosa during persistence in the cystic fibrosis lung. Int. J. Med. Microbiol. 300:557–562. 10.1016/j.ijmm.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 34. Winterhoff N, Goethe R, Gruening P, Rohde M, Kalisz H, Smith HE, Valentin-Weigand P. 2002. Identification and characterization of two temperature-induced surface-associated proteins of Streptococcus suis with high homologies to members of the arginine deiminase system of Streptococcus pyogenes. J. Bacteriol. 184:6768–6776. 10.1128/JB.184.24.6768-6776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gupta R, Yang J, Dong Y, Swiatlo E, Zhang JR, Metzger DW, Bai G. 2013. Deletion of arcD in Streptococcus pneumoniae D39 impairs its capsule and attenuates virulence. Infect. Immun. 81:3903–3911. 10.1128/IAI.00778-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu CD, Winteler H, Abdelal A, Haas D. 1999. The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J. Bacteriol. 181:2459–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dong Y, Chen YY, Burne RA. 2004. Control of expression of the arginine deiminase operon of Streptococcus gordonii by CcpA and Flp. J. Bacteriol. 186:2511–2514. 10.1128/JB.186.8.2511-2514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barcelona-Andrés B, Marina A, Rubio V. 2002. Gene structure, organization, expression, and potential regulatory mechanisms of arginine catabolism in Enterococcus faecalis. J. Bacteriol. 184:6289–6300. 10.1128/JB.184.22.6289-6300.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maghnouj A, Abu-Bakr AA, Baumberg S, Stalon V, Vander Wauven C. 2000. Regulation of anaerobic arginine catabolism in Bacillus licheniformis by a protein of the Crp/Fnr family. FEMS Microbiol. Lett. 191:227–234. 10.1111/j.1574-6968.2000.tb09344.x. [DOI] [PubMed] [Google Scholar]
- 40. Makhlin J, Kofman T, Borovok I, Kohler C, Engelmann S, Cohen G, Aharonowitz Y. 2007. Staphylococcus aureus ArcR controls expression of the arginine deiminase operon. J. Bacteriol. 189:5976–5986. 10.1128/JB.00592-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ferro KJ, Bender GR, Marquis RE. 1983. Coordinately repressible arginine deiminase system in Streptococcus sanguis. Curr. Microbiol. 9:145–149. 10.1007/BF01567287. [DOI] [Google Scholar]
- 42. Fulde M, Willenborg J, de Greeff A, Benga L, Smith HE, Valentin-Weigand P, Goethe R. 2011. ArgR is an essential local transcriptional regulator of the arcABC operon in Streptococcus suis and is crucial for biological fitness in an acidic environment. Microbiology 157:572–582. 10.1099/mic.0.043067-0. [DOI] [PubMed] [Google Scholar]
- 43. Baumberg S, Klingel U. 1993. Biosynthesis of arginine, proline and related compounds, p 299–306 In Sonenshein AL, Hoch JA, Losick R. (ed), Bacillus subtilis and other Gram-positive bacteria. American Society for Microbiology, Washington, DC. [Google Scholar]
- 44. Kloosterman TG, Kuipers OP. 2011. Regulation of arginine acquisition and virulence gene expression in the human pathogen Streptococcus pneumoniae by transcription regulators ArgR1 and AhrC. J. Biol. Chem. 286:44594–44605. 10.1074/jbc.M111.295832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, Tettelin H, Glass JI, Winkler ME. 2007. Genome sequence of Avery’s virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 189:38–51. 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Knodler LA, Sekyere EO, Stewart TS, Schofield PJ, Edwards MR. 1998. Cloning and expression of a prokaryotic enzyme, arginine deiminase, from a primitive eukaryote Giardia intestinalis. J. Biol. Chem. 273:4470–4477. 10.1074/jbc.273.8.4470. [DOI] [PubMed] [Google Scholar]
- 47. Houghton JE, Bencini DA, O’Donovan GA, Wild JR. 1984. Protein differentiation: a comparison of aspartate transcarbamoylase and ornithine transcarbamoylase from Escherichia coli K-12. Proc. Natl. Acad. Sci. U. S. A. 81:4864–4868. 10.1073/pnas.81.15.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marina A, Uriarte M, Barcelona B, Fresquet V, Cervera J, Rubio V. 1998. Carbamate kinase from Enterococcus faecalis and Enterococcus faecium—cloning of the genes, studies on the enzyme expressed in Escherichia coli, and sequence similarity with N-acetyl-l-glutamate kinase. Eur. J. Biochem. 253:280–291. 10.1046/j.1432-1327.1998.2530280.x. [DOI] [PubMed] [Google Scholar]
- 49. Degnan BA, Fontaine MC, Doebereiner AH, Lee JJ, Mastroeni P, Dougan G, Goodacre JA, Kehoe MA. 2000. Characterization of an isogenic mutant of Streptococcus pyogenes Manfredo lacking the ability to make streptococcal acid glycoprotein. Infect. Immun. 68:2441–2448. 10.1128/IAI.68.5.2441-2448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ryan S, Begley M, Gahan CG, Hill C. 2009. Molecular characterization of the arginine deiminase system in Listeria monocytogenes: regulation and role in acid tolerance. Environ. Microbiol. 11:432–445. 10.1111/j.1462-2920.2008.01782.x. [DOI] [PubMed] [Google Scholar]
- 51. Larsen R, Buist G, Kuipers OP, Kok J. 2004. ArgR and AhrC are both required for regulation of arginine metabolism in Lactococcus lactis. J. Bacteriol. 186:1147–1157. 10.1128/JB.186.4.1147-1157.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Larsen R, Kok J, Kuipers OP. 2005. Interaction between ArgR and AhrC controls regulation of arginine metabolism in Lactococcus lactis. J. Biol. Chem. 280:19319–19330. 10.1074/jbc.M413983200. [DOI] [PubMed] [Google Scholar]
- 53. Larsen R, van Hijum SA, Martinussen J, Kuipers OP, Kok J. 2008. Transcriptome analysis of the Lactococcus lactis ArgR and AhrC regulons. Appl. Environ. Microbiol. 74:4768–4771. 10.1128/AEM.00117-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Maas WK. 1994. The arginine repressor of Escherichia coli. Microbiol. Rev. 58:631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Park SM, Lu CD, Abdelal AT. 1997. Purification and characterization of an arginine regulatory protein, ArgR, from Pseudomonas aeruginosa and its interactions with the control regions for the car, argF, and aru operons. J. Bacteriol. 179:5309–5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Park SM, Lu CD, Abdelal AT. 1997. Cloning and characterization of argR, a gene that participates in regulation of arginine biosynthesis and catabolism in Pseudomonas aeruginosa PAO1. J. Bacteriol. 179:5300–5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dennis C CA, Glykos NM, Parsons MR, Phillips SE. 2002. The structure of AhrC, the arginine repressor/activator protein from Bacillus subtilis. Acta Crystallogr. D Biol. Crystallogr. 58:421–430. 10.1107/S0907444901021692. [DOI] [PubMed] [Google Scholar]
- 58. Miller CM, Baumberg S, Stockley PG. 1997. Operator interactions by the Bacillus subtilis arginine repressor/activator, AhrC: novel positioning and DNA-mediated assembly of a transcriptional activator at catabolic sites. Mol. Microbiol. 26:37–48. 10.1046/j.1365-2958.1997.5441907.x. [DOI] [PubMed] [Google Scholar]
- 59. Gardan R, Rapoport G, Débarbouillé M. 1997. Role of the transcriptional activator RocR in the arginine-degradation pathway of Bacillus subtilis. Mol. Microbiol. 24:825–837. 10.1046/j.1365-2958.1997.3881754.x. [DOI] [PubMed] [Google Scholar]
- 60. Nicoloff H, Arsène-Ploetze F, Malandain C, Kleerebezem M, Bringel F. 2004. Two arginine repressors regulate arginine biosynthesis in Lactobacillus plantarum. J. Bacteriol. 186:6059–6069. 10.1128/JB.186.18.6059-6069.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cherney LT, Cherney MM, Garen CR, Lu GJ, James MN. 2008. Crystal structure of the arginine repressor protein in complex with the DNA operator from Mycobacterium tuberculosis. J. Mol. Biol. 384:1330–1340. 10.1016/j.jmb.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 62. Garnett JA, Marincs F, Baumberg S, Stockley PG, Phillips SE. 2008. Structure and function of the arginine repressor-operator complex from Bacillus subtilis. J. Mol. Biol. 379:284–298. 10.1016/j.jmb.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 63. Makarova KS, Mironov AA, Gelfand MS. 2001. Conservation of the binding site for the arginine repressor in all bacterial lineages. Genome Biol. 2:RESEARCH0013. 10.1186/gb-2001-2-4-research0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liu Y, Dong Y, Chen YY, Burne RA. 2008. Environmental and growth phase regulation of the Streptococcus gordonii arginine deiminase genes. Appl. Environ. Microbiol. 74:5023–5030. 10.1128/AEM.00556-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Carvalho SM, Kloosterman TG, Kuipers OP, Neves AR. 2011. CcpA ensures optimal metabolic fitness of Streptococcus pneumoniae. PLoS One 6:e26707. 10.1371/journal.pone.0026707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Abdullah MR, Gutiérrez-Fernández J, Pribyl T, Gisch N, Saleh M, Rohde M, Petruschka L, Burchhardt G, Schwudke D, Hermoso JA, Hammerschmidt S. 2014. Structure of the pneumococcal l,d-carboxypeptidase DacB and pathophysiological effects of disabled cell wall hydrolases DacA and DacB. Mol. Microbiol. 93:1183–1206. 10.1111/mmi.12729. [DOI] [PubMed] [Google Scholar]
- 67. Hitzmann A, Bergmann S, Rohde M, Chhatwal GS, Fulde M. 2013. Identification and characterization of the arginine deiminase system of Streptococcus canis. Vet. Microbiol. 162:270–277. 10.1016/j.vetmic.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 68. Jensch I, Gámez G, Rothe M, Ebert S, Fulde M, Somplatzki D, Bergmann S, Petruschka L, Rohde M, Nau R, Hammerschmidt S. 2010. PavB is a surface-exposed adhesin of Streptococcus pneumoniae contributing to nasopharyngeal colonization and airways infections. Mol. Microbiol. 77:22–43. 10.1111/j.1365-2958.2010.07189.x. [DOI] [PubMed] [Google Scholar]
- 69. Hammerschmidt S, Talay SR, Brandtzaeg P, Chhatwal GS. 1997. SpsA, a novel pneumococcal surface protein with specific binding to secretory immunoglobulin A and secretory component. Mol. Microbiol. 25:1113–1124. 10.1046/j.1365-2958.1997.5391899.x. [DOI] [PubMed] [Google Scholar]
- 70. Chen JD, Morrison DA. 1988. Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning of DNA from Streptococcus pneumoniae. Gene 64:155–164. 10.1016/0378-1119(88)90489-1. [DOI] [PubMed] [Google Scholar]
- 71. Saleh M, Bartual SG, Abdullah MR, Jensch I, Asmat TM, Petruschka L, Pribyl T, Gellert M, Lillig CH, Antelmann H, Hermoso JA, Hammerschmidt S. 2013. Molecular architecture of Streptococcus pneumoniae surface thioredoxin-fold lipoproteins crucial for extracellular oxidative stress resistance and maintenance of virulence. EMBO. Mol. Med. 5:1852–1870. 10.1002/emmm.201202435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Dörries K, Lalk M. 2013. Metabolic footprint analysis uncovers strain specific overflow metabolism and d-isoleucine production of Staphylococcus aureus COL and HG001. PLoS One 8:e81500. 10.1371/journal.pone.0081500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Oginsky EL. 1957. Isolation and determination of arginine and citrulline. Methods Enzymol. 3:639–643. 10.1016/S0076-6879(57)03434-5. [DOI] [Google Scholar]
- 74. Degnan BA, Palmer JM, Robson T, Jones CE, Fischer M, Glanville M, Mellor GD, Diamond AG, Kehoe MA, Goodacre JA. 1998. Inhibition of human peripheral blood mononuclear cell proliferation by Streptococcus pyogenes cell extract is associated with arginine deiminase activity. Infect. Immun. 66:3050–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Saleh M, Abdullah MR, Schulz C, Kohler T, Pribyl T, Jensch I, Hammerschmidt S. 2014. Following in real time the impact of pneumococcal virulence factors in an acute mouse pneumonia model using bioluminescent bacteria. J. Vis. Exp. 2014:e51174. 10.3791/51174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hermans PW, Adrian PV, Albert C, Estevão S, Hoogenboezem T, Luijendijk IH, Kamphausen T, Hammerschmidt S. 2006. The streptococcal lipoprotein rotamase A (SlrA) is a functional peptidyl-prolyl isomerase involved in pneumococcal colonization. J. Biol. Chem. 281:968–976. 10.1074/jbc.M510014200. [DOI] [PubMed] [Google Scholar]
- 77. Kerr AR, Irvine JJ, Search JJ, Gingles NA, Kadioglu A, Andrew PW, McPheat WL, Booth CG, Mitchell TJ. 2002. Role of inflammatory mediators in resistance and susceptibility to pneumococcal infection. Infect. Immun. 70:1547–1557. 10.1128/IAI.70.3.1547-1557.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bergmann S, Lang A, Rohde M, Agarwal V, Rennemeier C, Grashoff C, Preissner KT, Hammerschmidt S. 2009. Integrin-linked kinase is required for vitronectin-mediated internalization of Streptococcus pneumoniae by host cells. J. Cell Sci. 122:256–267. 10.1242/jcs.035600. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pneumococcal and E. coli wild-type strains used in this study and generated pneumococcal mutants employed in functional assays.
Plasmids used in this study to perform mutagenesis of pneumococci and produce recombinant proteins.
Primers used in this study to construct deletion mutants of targeted genes, perform expression cloning, EMSA or Northern blot analysis.