ABSTRACT
The plant pathogen Ralstonia solanacearum possesses two genes encoding a trehalose-6-phosphate synthase (TPS), an enzyme of the trehalose biosynthetic pathway. One of these genes, named ripTPS, was found to encode a protein with an additional N-terminal domain which directs its translocation into host plant cells through the type 3 secretion system. RipTPS is a conserved effector in the R. solanacearum species complex, and homologues were also detected in other bacterial plant pathogens. Functional analysis of RipTPS demonstrated that this type 3 effector synthesizes trehalose-6-phosphate and identified residues essential for this enzymatic activity. Although trehalose-6-phosphate is a key signal molecule in plants that regulates sugar status and carbon assimilation, the disruption of ripTPS did not alter the virulence of R. solanacearum on plants. However, heterologous expression assays showed that this effector specifically elicits a hypersensitive-like response on tobacco that is independent of its enzymatic activity and is triggered by the C-terminal half of the protein. Recognition of this effector by the plant immune system is suggestive of a role during the infectious process.
IMPORTANCE
Ralstonia solanacearum, the causal agent of bacterial wilt disease, infects more than two hundred plant species, including economically important crops. The type III secretion system plays a major role in the pathogenicity of this bacterium, and approximately 70 effector proteins have been shown to be translocated into host plant cells. This study provides the first description of a type III effector endowed with a trehalose-6-phosphate synthase enzymatic activity and illustrates a new mechanism by which the bacteria may manipulate the plant metabolism upon infection. In recent years, trehalose-6-phosphate has emerged as an essential signal molecule in plants, connecting plant metabolism and development. The finding that a bacterial pathogen could induce the production of trehalose-6-phosphate in plant cells further highlights the importance of this metabolite in multiple aspects of the molecular physiology of plants.
INTRODUCTION
Ralstonia solanacearum is a soilborne plant-pathogenic betaproteobacterium with a wide host range and a wide geographic distribution (1). Plants from more than 50 botanical families, including major agricultural crops, are affected by this bacterial wilt disease. As a root and vascular pathogen, R. solanacearum is a model system to investigate the molecular mechanisms of bacterial pathogenicity (2, 3). Among the many virulence determinants that have been identified, the type III secretion system (T3SS) is essential to R. solanacearum pathogenesis. This T3SS machinery was shown to deliver a large set of 70 to 75 effector proteins directly into the cytoplasm of plant cells (4–7). A classification of type III effector (T3E) genes found in the R. solanacearum species complex using a unified nomenclature was recently proposed (4). The expression of the T3SS and T3E genes is under the transcriptional control of the hrpB regulatory gene (8). Excluding rare cases on specific hosts (9, 10) disruption of single T3E genes does not significantly alter disease symptom development, and this certainly reflects the synergistic and overlapping functions of the T3Es in R. solanacearum pathogenicity. Cumulative disruption of T3E could indeed lead to disease symptom reduction (9, 11) or reduced multiplication in planta (12). Some single T3E mutant strains were also reported to have reduced levels of multiplication in planta compared to their multiplication in the wild type when used in competitive assays (13).
T3Es can also betray the pathogen by triggering a defense reaction from the plant immune system if the plant possesses a resistance protein able to specifically recognize one of these T3Es (14). Several of these T3Es, formerly called “avirulence” determinants, have been characterized in R. solanacearum (3). RipP1 (formerly known as PopP1), RipP2 (PopP2), and RipAX2 (Rip36) are avirulence determinants triggering resistance on petunia, Arabidopsis, and eggplant lines, respectively (15–17). On tobacco species, two T3Es in R. solanacearum strain GMI1000, RipAA (AvrA) and RipP1, elicit a hypersensitive reaction (HR), a localized and programmed cell death at the sites of infection. The double inactivation of ripAA and ripP1 is sufficient to restore pathogenicity, showing that both determinants restrict the host range of R. solanacearum on Nicotiana spp. (18).
The molecular function of most R. solanacearum T3Es once inside plant cells remains elusive. Many T3Es do not have homology with proteins of known function, and relatively few functional studies have been conducted (10–12). There is evidence that RipP2 displays acetyltransferase activity (19), whereas the RipG (GALA) effector family was shown to interact with plant SKP1-like proteins, presumably acting as active components of the host ubiquitination machinery to subvert plant immunity (9, 20, 21). In addition, R. solanacearum possesses a transcription activator-like effector named RipTAL that is likely to promote disease through the transcriptional activation of host genes, as shown for Xanthomonas spp. (22, 23).
In this study, we identified and characterized a novel T3E from R. solanacearum strain GMI1000 that displays homology to trehalose-6-phosphate synthases, a class of enzymes that are ubiquitous in prokaryotes and eukaryotes. Complementation assays in yeast were used to demonstrate its biochemical activity and identify essential residues for enzymatic function. Interestingly, trehalose-6-phosphate has recently emerged as an essential signal molecule in plants, connecting plant metabolism and development (24). Heterologous expression assays also show that this T3E is specifically inducing an HR-like response in tobacco.
RESULTS
RipTPS encodes an effector protein homologous to trehalose phosphate synthases and is translocated into plant cells by the T3SS.
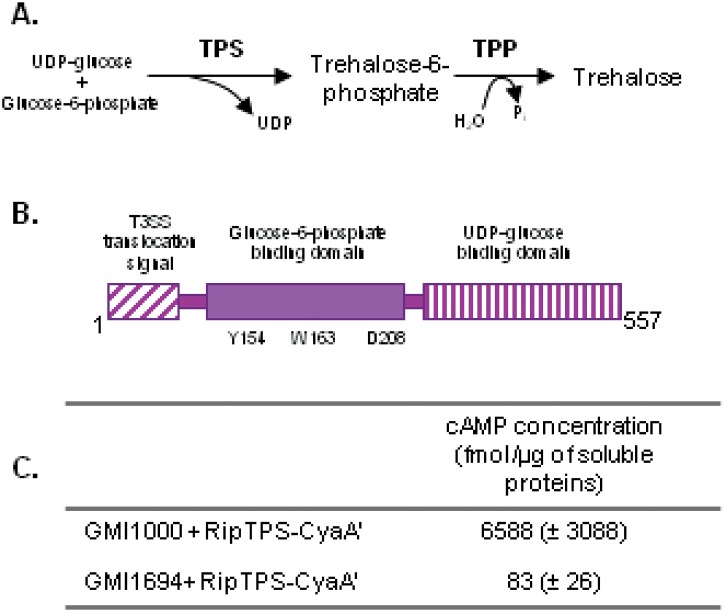
The RSp0731 gene product displays homology with proteins of the glycosyltransferase superfamily that possess a trehalose-6-phosphate synthase (TPS) activity. TPS catalyzes the transfer of glucose from UDP-glucose to glucose-6-phosphate, forming trehalose-6-phosphate (Tre6P) and UDP (Fig. 1). RSp0731 encodes a 557-amino acid (aa) protein that has 39 and 45% identity with OtsA from Escherichia coli and R. solanacearum, respectively. E. coli OtsA is a TPS enzyme for which a tridimensional structure was determined and on which residues required for its TPS activity have been identified (25, 26). All of the latter key residues required for substrate binding are conserved in RSp0731 (see Fig. S1 in the supplemental material). A striking difference between the RSp0731 protein and other bacterial TPS enzymes is that it carries a 77-aa N-terminal extension (Fig. S1) harboring the typical amino acid composition bias that characterizes effectors translocated through the T3SS (8). In order to demonstrate that this N-terminal domain directs the translocation of RSp0731 into plant cells through the T3SS, we used the adenylate cyclase (CyaA) assay already used in R. solanacearum (5, 6) that relies on the CyaA domain of the Bordetella pertussis cyclolysin (27). A chimeric protein fusion consisting of the 83 N-terminal residues from RSp0731 fused to CyaA was engineered on a plasmid vector and introduced into the wild-type strain GMI1000 and the T3SS (hrcV) mutant derivative GMI1694. The cyclic AMP (cAMP) concentration was monitored in tobacco plant tissue following infiltration with these strains, and the results shown in Fig. 1C indicate that this value was 73-fold higher in the wild-type strain than in the T3SS mutant. This shows that the RSp0731 N-terminal domain allows T3SS-dependent translocation into host cells; RSp0731 was therefore renamed RipTPS, in agreement with the new nomenclature (4).
FIG 1 .
ripTPS encodes a T3E homologous to trehalose phosphate synthases. (A) Enzymatic reactions catalyzed by trehalose-6-phosphate synthase (TPS) and trehalose-6-phosphate phosphatase (TPP) in the trehalose biosynthetic pathway. (B) Schematic representation of RipTPS functional domains and some of the conserved residues predicted to be essential for activity. (C) The concentrations of cyclic AMP (cAMP) were measured in N. tabacum leaves after infiltration of R. solanacearum strains with or without a functional T3SS (GMI1000 and hrcV, respectively) expressing the RipTPS N-terminal domain (first 88 amino acids) fused to the calmodulin-dependent adenylate cyclase. Standard errors of the results of three independent experiments are shown in brackets.
ripTPS is a conserved effector in the R. Solanacearum species complex, and homologues are present in other bacterial plant pathogens.
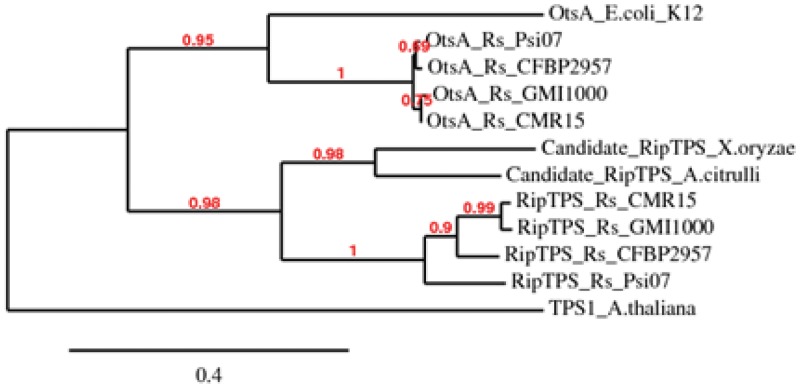
Because the RipTPS effector is translocated into plant cells, we investigated the phylogenetic relationship of the protein with other bacterial TPS sequences. As the plant TPS is clearly an outgroup in this phylogenetic reconstruction (Fig. 2), it seems that the R. solanacearum ripTPS most probably originated from a duplication of a bacterial otsA gene. Further comparative genomic analyses show that ripTPS is widely conserved in strains of the R. solanacearum species complex. Indeed, it is present in each of the four main phylotypes, with the exception of Ralstonia syzygii and blood disease bacterium strains, which are undergoing a reductive genomic evolution (28) and presumably have lost the gene. All these RipTPS homologues display a high level of protein sequence identity (from 76 to 99%) among the various strains. Interestingly, in most of the strains, ripTPS is physically associated on the genome with another type III effector gene (ripAV), contrary to the other (otsA) TPS genes, which are physically associated with a trehalose-6-phosphate phosphatase (otsB)-related gene. In order to identify RipTPS effector homologues in other bacterial pathogens, we performed BLAST searches and detected proteins in Xanthomonas oryzae pv. oryzae and Acidovorax citrulli with a RipTPS architecture, i.e., a TPS domain and an approximately 100-aa N-terminal extension potentially involved in T3SS export (Fig. 2; also Fig. S1 in the supplemental material). This suggested that RipTPS-like effectors may be present in other vascular plant pathogens.
FIG 2 .
Bacterial TPS phylogeny. The accession numbers of the proteins aligned in this phylogeny can be found in the legend to Fig. S1 in the supplemental material. The outgroup Arabidopsis thaliana TPS1 (accession number NP_177979) was added in this analysis. Sequences were aligned with MUSCLE and curated with GBlocks, and the phylogeny was reconstructed with PhyML. All analysis was run using the http://phylogeny.fr webserver (58). Bootstrap values are indicated in red. Scale is number of substitutions per site.
Functional complementation of a yeast tps-defective mutant by R. solanacearum ripTPS.
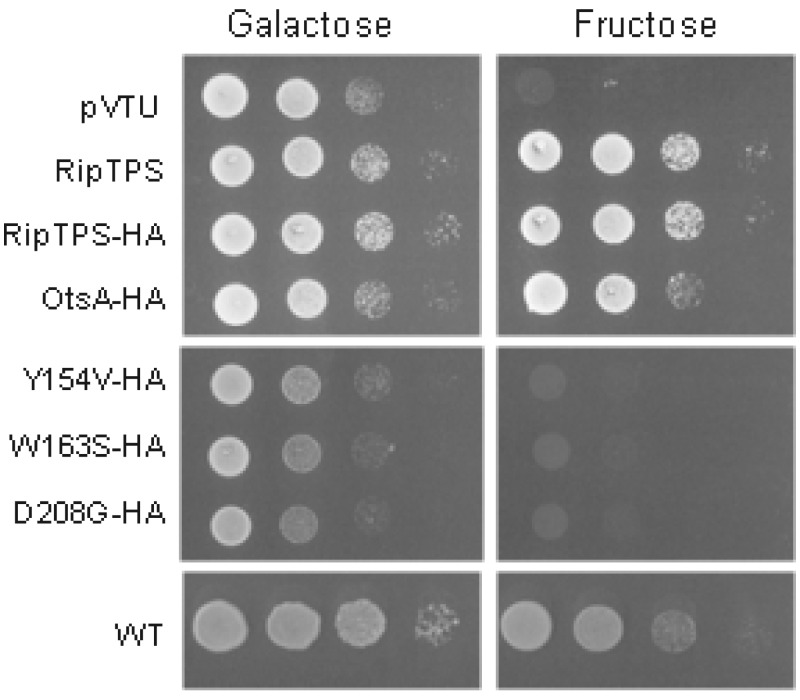
In Saccharomyces cerevisiae, trehalose is synthesized by a TPS enzyme encoded by tps1 and a trehalose-6-phosphate phosphatase (TPP) encoded by tps2. Inactivation of tps1 causes an absence of trehalose accumulation but also a growth arrest when glucose or fructose that is rapidly fermented is used as a carbon source (24). The expression of the E. coli otsA gene was shown to partially restore the growth of a yeast tps1 mutant in the presence of fermentable sugars (29). Since we also wanted to quantify Tre6P production, we decided to test for complementation by ripTPS using the tps1 tps2 double mutant strain, which has the same growth defect on fructose as the single tps1 mutant, i.e., it is unable to grow on fructose but remains able to grow on galactose (30). When the tps1 tps2 mutant strain was transformed with ripTPS, growth on fructose was restored to the wild-type level (Fig. 3). Similar results were observed with the expression of the E. coli otsA gene, as previously described (29). Complementation of the growth defect of the yeast tps1 tps2 mutant on fructose was a first indication that ripTPS encodes a functional TPS.
FIG 3 .
Complementation of the growth defect of the S. cerevisiae tps1 tps2 strain on fructose by R. solanacearum ripTPS. RipTPS with or without an HA tag was expressed in the tps1 tps2 yeast mutant, and the growth of the strain was compared to that of strains carrying an empty vector or expressing E. coli OtsA with an HA tag or one of the three catalytic mutants of RipTPS (Y154V, W163S, and D208G) with an HA tag. Amounts of 5 µl of liquid cultures were spotted as a series of 10-times dilutions on 2% galactose or 2% fructose SD-LWU plates. The control yeast strain (WT) was grown on plates with LWU. Pictures of a representative experiment at day 2 are shown.
RipTPS catalyzes Tre6P synthesis.
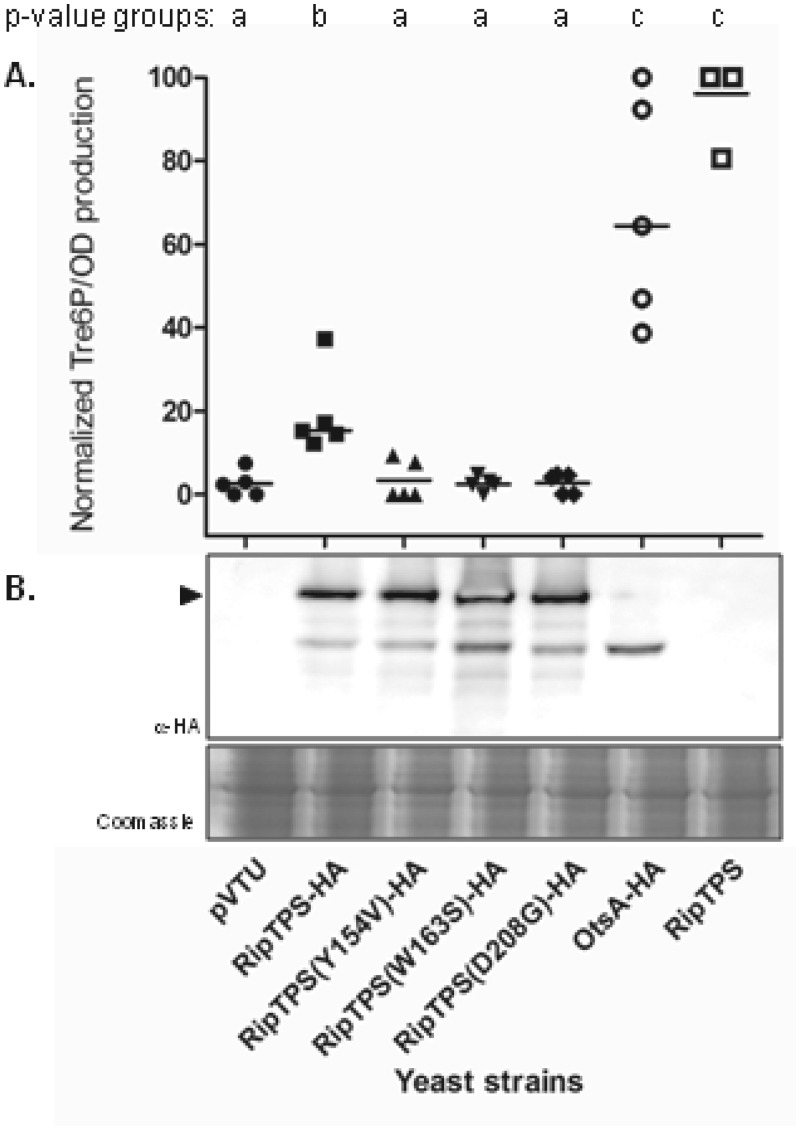
We then used the double tps1 tps2 mutant to test for Tre6P accumulation upon the expression of ripTPS. In this double mutant, the overaccumulation of Tre6P is easier to monitor in the absence of the TPS gene-encoded TPP activity. Tre6P production was quantified in the tps1 tps2 mutant yeast strain complemented by ripTPS after being triggered by a glucose pulse (31). Within 5 min after the pulse, Tre6P synthesis reached a maximum in the RipTPS-expressing strains, whereas Tre6P production was below the detection threshold in the tps1 tps2 mutant carrying the empty vector (Fig. 4, pVTU). Native and hemagglutinin (HA)-tagged versions of RipTPS and native E. coli OtsA were also generated and used to complement the tps1 tps2 mutant strain. As for OtsA, the RipTPS-dependent production of Tre6P was confirmed, although its catalytic activity was decreased due to the addition of the HA tag.
FIG 4 .
Production of trehalose-6-phosphate in yeast tps1 tps2 mutant expressing R. solanacearum ripTPS or one of its catalytic mutants. (A) RipTPS with an HA tag and its catalytic mutants were expressed in the tps1 tps2 yeast, and the levels of Tre6P production were compared to the levels in strains carrying an empty vector or expressing E. coli OtsA with an HA tag or RipTPS without a tag. All samples were analyzed five times, generating biologically independent replicates. The P value groupings indicate which strains cannot be distinguished (at a P value of <5%) using a nonparametric Wilcoxon matched-pairs signed-rank test, i.e., group a: the results for the empty vector pVTU and the three catalytic mutants cannot be distinguished and are all significantly different from the results for the other strains. (B) Western blot analysis of the expression of the HA-tagged TPS variants in the yeast protein extracts. A corresponding Coomassie-stained gel was used as a loading control. The black arrowhead indicates RipTPS (67 kDa).
Based on the crystal structure of E. coli OtsA (26) and on the model generated for Magnaporthe grisea TPS1 (32), conserved residues in the glucose-6-phosphate binding domain were identified and shown to be essential for activity. Corresponding nucleotide changes were introduced into the full-length ripTPS gene to generate 3 amino acid substitutions at these key positions (Fig. 1B; also Fig. S1 in the supplemental material). As shown in Fig. 4, the tps1 tps2 mutant strains carrying the resulting ripTPSY154V (ripTPS with a substitution of valine for tyrosine at position 154), ripTPSW163S, or ripTPSD208G allele were all unable to synthesize Tre6P (the production of Tre6P was indistinguishable from that in the strain carrying the pVTU empty vector), showing the importance of these residues for TPS activity. Western blot analysis confirmed that the wild-type RipTPS and variant alleles were produced in equal amounts in yeast (Fig. 4B). A positive HA band at 58 kDa, as shown in Fig. 4B, suggests that, in yeast, there could be cleavage of the N-terminal extension of RipTPS-HA. As expected, none of the three mutated ripTPS alleles restored yeast growth on fructose when introduced into the tps1 tps2 mutant (Fig. 3).
RipTPS elicits an HR-like response on Nicotiana tabacum independent of TPS activity.
In order to determine the contribution of ripTPS in R. solanacearum pathogenicity, a disruption mutant was created by inserting an interposon into the ripTPS coding sequence. The capacity of the corresponding mutant strain to cause disease was evaluated on several host plants, and no difference in symptom development was detected compared to that of the wild-type strain (see Fig. S2 in the supplemental material). Further inoculation tests on a panel of Arabidopsis ecotypes and competitive index experiments (13) on tomato and bean did not show any difference either (data not shown). We then wanted to evaluate the role of TPS activity during infection by R. solanacearum by testing available transgenic Arabidopsis plants expressing E. coli otsA (33) or ripTPS, generated by ourselves. Both otsA- and ripTPS-expressing plants had stunted growth and very low fertility (33; also our unpublished data), making the test of the contribution of TPS to disease difficult to evaluate.
We then investigated whether the expression of ripTPS could elicit defense responses in other plants. We used an Agrobacterium-mediated transient expression assay after infiltration of leaf mesophyll tissues using a vector carrying the full-length ripTPS coding sequence under the control of the 35S promoter. On Nicotiana tabacum, a strong HR-like necrotic response could be observed 48 h postinfiltration (Fig. 5B, a). This response could be specific to N. tabacum, since no reaction was observed after leaf infiltration of Nicotiana benthamiana or Nicotiana glutinosa (data not shown). The kinetics of this necrotic response on N. tabacum was comparable to the HR response elicited by the ripAA (avrA) avirulence gene product under the same experimental conditions (Fig. 5B, g) (18). Altogether, these data suggest that RipTPS is specifically recognized by the N. tabacum immune system.
FIG 5 .
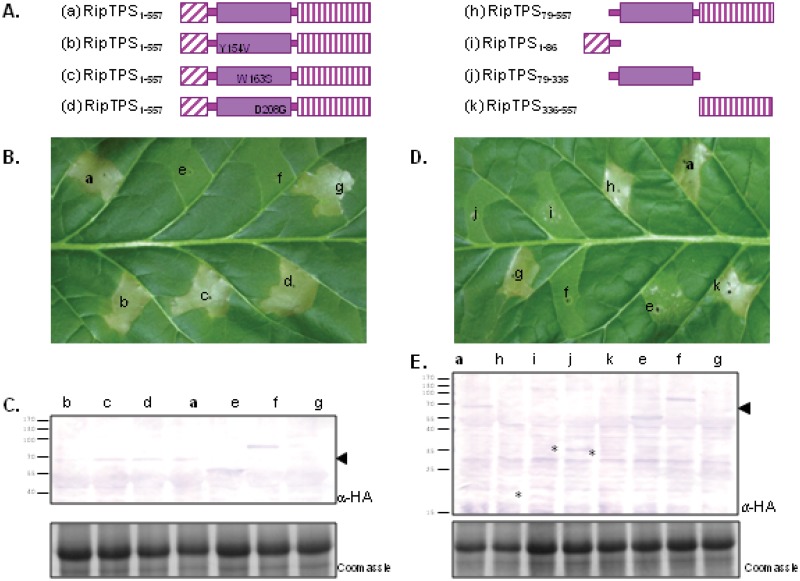
Agrobacterium-mediated transient expression of RipTPS or the C-terminal domain of RipTPS in N. tabacum leaves elicits an HR-like necrotic response. (A) Schematics of the structures of the RipTPS proteins expressed, including WT RipTPS (a) and the three catalytic mutants of RipTPS-HA (b, c, and d). Non-RipTPS proteins used in the experiments are not depicted; these were OtsA-HA (e), GUS-HA (negative control) (f), and AvrA (positive HR control) (g). (B and D) N. tabacum leaf pictures taken 3 days after infiltration of Agrobacterium strains, allowing the expression of the different recombinant proteins. (C and E) Western blotting with anti-HA antibody and Coomassie staining of the protein extracts from leaves infiltrated like those shown in panels B and D, respectively. The black arrowheads indicate RipTPS (67 kDa). The asterisks indicate protein that is visible but at low abundance.
Agrobacterium-mediated transient expression assays were also performed with strains expressing RipTPS carrying single point mutations in the essential catalytic sites described above. Agrobacterium tumefaciens strain GV3101::pMP90RK containing the pAM-PAT plasmids in which the ripTPSY154V, ripTPSW163S, and ripTPSD208G alleles were cloned induced an HR-like response similar to that induced by the strain carrying the wild-type ripTPS gene (Fig. 5B, b, c, and d). This demonstrates that the enzymatic activity of RipTPS is not required for the elicitation of the necrotic response on N. tabacum.
The C-terminal half of RipTPS is sufficient to trigger the HR-like response.
Since plants contain TPS enzymes (34), which are related to prokaryotic TPS (Fig. 2), the finding that RipTPS elicited an HR-like response on a tobacco species was intriguing. We therefore performed Agrobacterium-mediated transient expression assays using the full-length E. coli otsA as performed for ripTPS. Although otsA is closely related to ripTPS (Fig. 2; also Fig. S1 in the supplemental material), we can see from the results shown in Fig. 5 (Fig. 5B, e) that Agrobacterium-mediated expression of otsA did not induce a necrotic response on N. tabacum.
The three functional domains of RipTPS (the T3SS translocation domain, the glucose-6-phosphate binding domain, and the UDP-glucose binding domain) (Fig. 5A) were cloned in a plant expression vector and used for transient expression assays. A necrotic response is observed only when the RipTPS C-terminal domain is present in the agroinfiltrated protein (Fig. 5D, a, h, and k). We can thus hypothesize that the N. tabacum immune system recognizes the C-terminal domain of RipTPS, containing the UDP-glucose binding domain. It is notable that, although the detection of the expressed proteins again proved difficult, the detection of agroinfiltrated recombinant proteins that failed to trigger necrosis was possible (Fig. 5E, see the small dots next to the HA-positive bands from infiltrations i and j). The expression of the different versions of recombinant RipTPS was confirmed by agroinfiltration in N. benthamiana leaves (see Fig. S3 in the supplemental material).
DISCUSSION
Trehalose is a nonreducing disaccharide found in many organisms, including eubacteria, fungi, insects, and plants. Although different biosynthetic routes exist in bacteria, a general pathway involving the successive action of TPS and TPP enzymes is conserved in fungi, plants, and many bacteria (35). Phylogenetic analyses have shown that there is a close evolutionary relationship between the TPS and TPP modules in these diverse organisms; in bacteria, the corresponding otsA and otsB genes are clustered and are generally transcribed from a single promoter (35). ripTPS, while being conserved in taxonomically distant R. solanacearum strains (4), is never associated with a TPP gene module. Instead, the neighboring gene to ripTPS is ripAV, which encodes a T3E of unknown function (4). The presence of an N-terminal T3SS secretion domain is also unique among bacterial TPS enzymes. For RipTPS, this N-terminal extension is a functional T3SS signal, and its presence in TPS enzymes of other plant-pathogenic bacteria makes them good candidates for also being secreted effectors. This unusual modular architecture of a T3SS-dependent TPS enzyme suggests that this effector evolved from a shuffling process that Stavrinides and colleagues have called “terminal reassortment” (36). This evolutionary mechanism has been described for several bacterial effector proteins in which the termini of the effector reassort with other genetic information to create new chimeric proteins with added secretion function.
There are increasing numbers of reports in the literature indicating that trehalose metabolism emerges in the establishment of virulence traits in distantly related microbial species (37). Either in prokaryotes or in several fungal species, trehalose-associated mechanisms have been shown to contribute to cell morphogenesis, cell wall integrity, regulation of metabolism, and evasion of the host immune response (32, 37, 38). For example, a recent study has shown that the opportunistic pathogen Pseudomonas aeruginosa has repurposed a specific trehalose biosynthesis pathway as a virulence factor to increase its replication rate in the intercellular leaf environment (39). However, there is not just one way to tamper with this signaling in host plants, as several distinct trehalose-associated mechanisms operate to promote pathogenesis in species-specific manners (37).
This work provides evidence that RipTPS displays trehalose-6-phosphate synthase activity and is translocated into plant cells by the T3SS. Although Tre6P is present in trace abundance in plant cells, its content is tightly regulated and not merely restricted to trehalose synthesis (34). Several studies in recent years have indeed unraveled the role of Tre6P as a key signal molecule that regulates carbon assimilation and the sugar status in plants (34, 40–43). It is therefore tempting to speculate that the pathogen directly induces Tre6P synthesis during the course of infection to subvert plant cell metabolism through modulation of the endogenous Tre6P content rather than to stimulate trehalose biosynthesis. Tre6P has many physiological effects and probable targets in plants, and the regulatory mechanisms are not yet completely understood. However, Tre6P in plants was shown to inhibit Snf1-related kinase 1 (SnRK1) activity (40), and it also indirectly increases the expression of the abscisic acid-insensitive-4 (ABI4) transcription factor (35). SnRK1 is the plant ortholog of SNF1 from yeast, and both are important regulators of sugar metabolism (44, 45). Interestingly, Arabidopsis SnRK1 was shown to be targeted by the Xanthomonas campestris T3E AvrBsT (46) and the SnRK1 homolog AKIN1 from N. benthamiana was shown to be targeted by two geminivirus proteins (47), suggesting that downregulation of SnRK1 activity could be a strategy common to different plant pathogens. The fact that ripTPS is dispensable for R. solanacearum pathogenicity, as well as the multiple effects induced by Tre6P in plant cells, hampers elucidation of the biological impact of RipTPS on plant physiology that is relevant during infection. Manipulation of plant Tre6P levels could promote either plant defense suppression or, alternatively, a metabolic priming that is beneficial for the pathogen, as recently evidenced with other effectors (48–50). Even though we have not pinpointed it experimentally, we cannot exclude a contribution by RipTPS to the pathogenicity. Indeed, the absence of an apparent role could actually be yet another example of a functional overlap between unrelated effectors. Furthermore, since the R. solanacearum infection process involves multiple distinct steps, from root entry to xylem vessel colonization, a future challenge will be to address whether RipTPS is required at certain specific stages of this process.
Transient expression assays in tobacco revealed that RipTPS elicits strong necrosis in N. tabacum but not in two other tobacco species tested (N. glutinosa and N. benthamiana). This host specificity is suggestive of a hypersensitive response (HR) phenomenon. The observation that an R. solanacearum ripP1 ripAA (popP1 avrA) double mutant is HR negative on N. tabacum (18) also suggests that this probable HR-eliciting activity of RipTPS is suppressed by additional T3E(s) in the wild-type GMI1000 strain. Our results showed that this phenotype is independent of Tre6P synthesis. Moreover, we were able to demonstrate that this HR-like eliciting activity is associated with the RipTPS C-terminal domain. Amino acid comparison of this domain with E. coli OtsA (which does not elicit the plant reaction) reveals that they display 42% identity (62% similarity), with the most divergent region corresponding to the 40-aa C-terminal end, which is only 15% identical between OtsA and RipTPS (see Fig. S1 in the supplemental material). Despite a probable common tridimensional folding of these TPS enzymes, the recognition of RipTPS by a plant immunity component might therefore be associated with some of these sequence differences. Further studies aimed at characterizing the various ripTPS alleles from diverse strains from the R. solanacearum species complex may identify ripTPS variants that are unable to elicit the HR-like response on N. tabacum. This could enable the identification of the sequence determinants actually triggering the plant response.
Many T3SS-translocated effectors mimic eukaryotic proteins in structure and function. Most often, for T3E enzymes where the molecular function was elucidated, the enzymatic activity of these T3E proteins consists of posttranslational modifications of host proteins (e.g., phosphorylation, acetylation, ubiquitination, sumoylation, etc.) or the direct degradation of host components (through hydrolase or protease activities, for example) (9, 19, 51–53). RipTPS appears to be a so-far unique example of a T3E directing the synthesis of an endogenous signal molecule in individual host cells.
Despite considerable progress during the last decade, the role of Tre6P as a regulatory factor in plants is far from being fully understood. The finding that a bacterial pathogen could induce the production of Tre6P in plant cells further highlights the importance of this metabolite in the molecular physiology of plants.
MATERIALS AND METHODS
Bacterial strains, yeast strains, plasmids, growth conditions, and plant material.
The bacterial and yeast strains and plasmids used for this study are described in Table S1 in the supplemental material. Escherichia coli cells were grown in Luria-Bertani medium at 37°C with 100 µg/ml ampicillin or 15 µg/ml gentamycin. Agrobacterium tumefaciens strain GV3101::pMP90RK cells were grown in YEB medium (5 g/liter beef extract, 1 g/liter yeast extract, 5 g/liter peptone, 5 g/liter sucrose, 2 mM MgSO4, pH 7.2) at 28°C with 100 µg/ml rifampin, 50 µg/ml kanamycin, 15 µg/ml gentamicin, and 75 µg/ml carbenicillin. Agar was added at 1.5% (wt/vol) for solid medium. R. solanacearum strains GMI1000, GMI1694 (hrcV, a T3SS mutant derivative), and GRS432 (ripTPS mutant) were grown in complete medium B or minimal medium (MM) at 28°C with or without 40 µg/ml spectinomycin (54). For liquid cultures, spectinomycin was reduced by half. The S. cerevisiae tps1 tps2 mutant strain YSH652 (MATα leu2-3,112 ura3-1 trp1-1 his3-11,15 ade2-1 can1-100 tps1::TRP1 tps2::LEU2 GAL SUC) and the control strain W303-1A (MATa leu2-3,112 ura3-1 trp1-1 his3-11,15 ade2-1 can1-100 GAL SUC) were grown at 30°C in SD-LWU and SD medium (both from Clontech), respectively, supplemented with galactose unless otherwise stated. The plants used in this study were Nicotiana tabacum cv. Bottom special, Nicotiana benthamiana, eggplant (Solanum melongena cv. Zebrina), tomato (Solanum lycopersicum cv. Marmande), and geranium (Geranium sanguineum var. Maverick Ecarlate), grown in the greenhouse, and Arabidopsis thaliana grown in a growth chamber at 22°C with a 9-h day length.
DNA manipulations and genetic constructs.
Constructs were created using the Gateway technology as recommended by the manufacturer (Life Technologies, Carlsbad, CA). ripTPS was cloned from the genomic DNA of R. solanacearum GMI1000 in pDONR207, creating pMP12 (with a stop codon) and pMP15 (without a stop codon). Amplifications were performed in two steps, using the cloning primers (731GW-ATG and 731GW-STOP or 731GW-NOSTOP) in the first PCR and then universal primers (attB1 and attB2) in the second PCR, with Platinum Pfx (Life technologies), Pfu Ultra (StrataGen, Kirkland, WA), or Phusion (NEB, Ipswich, MA). The primers used for the amplifications are provided in Table S1 in the supplemental material. The three mutant alleles of ripTPS were generated using the QuikChange XL site-directed mutagenesis kit (Agilent) on the pMP15 plasmid, creating plasmids pMP90 (Y154V), pMP91 (W163S), and pMP92 (D208G), respectively. Expression plasmids (pMP72, pMP93, pMP94, and pMP95) were created by LR Gateway cloning using pAM-PAT-P35S-GW-3xHA (L. D. Noël and J. E. Parker, unpublished data) and used to transform Agrobacterium tumefaciens GV3101::pMP90RK. The different constructs derived from RipTPS (RipTPS79–557, RipTPS1–86, RipTPS79–335, and RipTPS336–557) were generated by PCR (using primer pairs oMP1-oMP3, 731GW-ATG–oMP4, oMP1-oAuA7, and oAuA5-oMP3, respectively) (Table S1) and cloned in the pDONR207 vector (pMP71, pMP35, pACC758, and pMP87) and then in pAM-PAT-P35S-GW-3xHA (pMP73, pACC678, pACC766, and pMP89), and the resulting plasmids were used to transform A. tumefaciens. The expression levels of GUS (courtesy of L. Deslandes), OtsA (pMP74), and AvrA (pMP99) (18) were used as controls for the A. tumefaciens assay. OtsA from E. coli was amplified using primers otsA-fwd and otsA-rev (Table S1).
Construction of an ripTPS mutant strain.
ripTPS was disrupted by the insertion of an Ω cassette conferring resistance to spectinomycin as previously described (55). A 2.2-kb DNA fragment comprising the ripTPS coding sequence was cloned after PCR amplification using primers GRS432 fwd and GRS432 Rev. The Ω cassette was inserted into the unique EcoRI restriction site within the coding sequence, resulting in plasmid pMP2. This construct was linearized by NcoI and introduced into R. solanacearum GMI1000 by natural transformation. The mutant strain GRS432 generated after selection for a double recombination event using spectinomycin resistance and was further checked by PCR.
Adenylate cyclase assay.
A DNA fragment comprising 509 bp upstream from the ripTPS start codon and the region encoding the first 83 amino acids was PCR amplified (primers BglII731 and HindIII731) (see Table S1 in the supplemental material) and cloned into the pSC266 vector (6) HindIII-BglII restriction sites to create a translational fusion with the cyaA′ gene. The resulting plasmid, named pGG7, was introduced into the GMI1000 and GMI1694 strains by natural transformation to select for a single recombination event, leading to genomic integration. Natural transformation of R. solanacearum strains was performed after growth in minimal medium supplemented with 2% glycerol as described previously (54).
The adenylate cyclase assay was performed as previously described (6). Briefly, strain GMI1000 and derivatives carrying plasmid pGG7 were infiltrated into Nicotiana tabacum leaves at an optical density (OD) of 1. Plants were sampled 7 h later with a cork borer. Leaf disks were transferred into Eppendorf tubes and immediately frozen in liquid nitrogen. For cyclic AMP (cAMP) extraction, samples in Eppendorf tubes were kept frozen while grinding by shaking with 2-mm tungsten beads in a Qiagen MM300 mixer mill for two runs of 2 min at 30 Hz. Samples were stored at −80°C before cAMP quantification. Protein concentration was assessed with the Bio-Rad protein assay kit. Cyclic AMP levels were monitored with a cAMP enzyme immunoassay kit (Biotrak; Amersham Pharmacia Biotech) according to the manufacturer’s instructions.
Complementation of the tps1 tps2 yeast mutant.
To express ripTPS and its derivatives in yeast, the Gateway-compatible yeast episomic vector pVTU102-GW containing the yeast alcohol dehydrogenase (ADH1) gene promoter (56) was used. The full-length ripTPS gene was cloned, resulting in the yeast expression plasmid named pMP31, which was used to transform the tps1 tps2 yeast mutant strain (31). Similarly, the catalytic mutants of ripTPS were cloned in pVTU102-GW (pMP96, pMP97, and pMP98). To create the HA-tagged versions of these vectors, a PCR fragment obtained from pMP72 amplification with primers TPSHA-fwd and TPSHA-rev (see Table S1 in the supplemental material) was inserted into the XhoI-HindIII site. The resulting plasmids (pMP104, pMP105, pMP106, and pMP107) were used to transform the tps1 tps2 mutant yeast strain. As a positive control, the full-length otsA gene from E. coli was cloned in the pVTU102-GW vector with an HA tag in the SalI-HindIII site (pMP108), using primers otsAHA-fwd and otsAHA-rev (Table S1).
The yeast strains were grown on SD-LWU containing 2% galactose and 40 mg/liter adenine. For the fructose growth assay, 10-fold dilution series from exponential growing cells were spotted onto plates containing either fructose or galactose.
To measure the production of Tre6P, the extraction of yeast metabolites following a glucose pulse was carried out as described previously (31). The measured production of Tre6P (mM Tre6P/OD unit) was normalized by attributing a value of 100 to the sample with maximal value. All the samples were analyzed five times, generating independent biologically replicates. To analyze the differences between strains, we used the nonparametric Wilcoxon matched-pairs signed-rank test with a one-tailed P value. This was performed using the GraphPad Prism 5.0 software package.
Pathogenicity assays.
Arabidopsis (Col-0) plantlets were grown for 4 to 5 weeks on Jiffy plugs (Jiffy Products International BV, Netherlands). Geranium (Geranium sanguineum var. Maverick Ecarlate), tomato (cv. Marmande), and eggplant (cv. Zebrina) were grown in greenhouse facilities in potting mixture. For all plant inoculations, the roots were soaked with a bacterial suspension at 107 CFU/ml of GMI1000 or GRS432 (the ripTPS mutant). Solanaceae were then placed at 28°C (light) and 27°C (night) with a 12-h photoperiod. For Arabidopsis, the temperatures were 27°C (light) and 26°C (night). The progression of the disease was reported by scoring the plants on a scale from 1 to 4 based on 25% to 100% of leaves being wilted; to perform the survival analysis (see Fig. S2 in the supplemental material), all scores equal to or higher than 2 (50% wilted leaves) were set as 1 (or dead) when the lower scores were 0 (or alive). The experiments were repeated at least 3 times independently. The statistical analysis was performed as described previously (21).
Agrobacterium-mediated expression of proteins in plant cells.
A. tumefaciens strains carrying plasmid-borne ripTPS constructs were grown overnight in YEB medium. Cells were pelleted at 4,000 rpm, resuspended in infiltration medium (10 mM MgCl2, 10 mM morpholine ethanesulfonic acid, and 150 µM acetosyringone), and incubated for 2 h at room temperature. Resuspended cells were infiltrated into leaves of 4-week-old Nicotiana spp. plants at an optical density at 600 nm (OD600) of 0.6 with a 1-ml needleless syringe. The infiltrated plants were incubated in growth chambers for a 16-h day at 20°C. After infiltration (48 to 72 h), leaf disks taken in infiltrated areas were ground with a mixer mill (MM400; Retsch, Haan, Germany) and homogenized in Laemmli buffer in order to be analyzed by Western blotting to control for the expression levels. Rat anti-HA antibody (1:5,000) (number 11867423001; Roche) was incubated overnight in Tris-buffered saline (TBS)--0.1% Tween with 1% bovine serum albumin (BSA), and then goat anti-rat antibody coupled to alkaline phosphatase (1:10,000) (A8438; Sigma) was added after three washes of 10 min each in TBS-Tween, and the mixture was incubated for 1 h. Incubation was followed by two washes in TBS-Tween and then nitroblue tetrazolium (NBT)-BCIP (5-bromo-4-chloro-3-indolylphosphate)-based staining. The expression of the RipTPS variants was followed in N. benthamiana in parallel to the expression in N. tabacum because the low expression level in N. tabacum was limiting for Western blot analysis. After SDS-PAGE electrophoresis, Coomassie staining was performed to check for equal protein loading (57).
SUPPLEMENTAL MATERIAL
Protein sequence alignment of RipTPS with other TPS sequences. Amino acids that constitute the glucose-6-phosphate binding domain are underlined with a pink arrow, whereas those involved in the conformation of the binding site are blue. The green arrows indicate the residues required for interaction with UDP-glucose. The tyrosine, tryptophan, and aspartate amino acids from the glucose-6-phosphate binding domain that were mutated in RipTPS are also indicated. The sequence alignment was performed using jalview 2.0 software (59). The accession numbers of the aligned proteins are as follows: OtsA_Rs_GMI1000 (NP_522666), OtsA_Rs_CMR15 (YP_006061033), OtsA_Rs_Psi07 (YP_003750101), OtsA_Rs_CFBP2957 (YP_003748420), OtsA_E.coli_K12 (YP_490157), Candidate RipTPS_X.oryzae (WP_019303360), Candidate_RipTPS_A.citrulli (YP_970430), RipTPS_Rs_GMI1000 (NP_522292), RipTPS_Rs_CMR15 (CMR15v4_mp10701), RipTPS_Rs_Psi07 (YP_003749608), and RipTPS_Rs_CFBP2957 (RCFBP_mp20172). Download
R. solanacearum ripTPS
mutant is not affected in its pathogenicity. The R. solanacearum ripTPS mutant GRS432 cannot be distinguished from the wild-type strain GMI1000 on Arabidopsis, tomato, eggplant, or geranium. All four plants were tested at least three times; the results of one representative experiment are shown in a Kaplan-Meier survival analysis representation. This graphical representation indicates the percent survival of the plants inoculated with GMI1000 or GRS432. For this statistical analysis, the phenotyping data (progressive disease index [DI] scale ranging from 0 to 4, or 0% to 100% of plants wilted) is converted to binary data: wilted (DI > 2) or not wilted. The log-rank test is a hypothesis test, with Ho being the fact that both survival curves are identical. The P values for each test indicate that the hypothesis of similarity of curves cannot be rejected. Statistical tests were performed as described previously (21). Download
Agrobacterium-mediated transient expression of ripTPS constructs in N. benthamiana. Expression of RipTPS-HA (a), RipTPS variants labeled h (57 kDa), i (15 kDa), j (34 kDa), and k (30 kDa) in Fig. 5A, and controls OtsAEcoli-HA (e), GUS-HA (f), and AvrA (g). Results of Western blotting with anti-HA antibody and Coomassie staining of the protein extracts from Nicotiana benthamiana are shown. The black arrow indicates RipTPS (67 kDa). The asterisks indicate the same proteins as in Fig. 5. Download
Primers used in this work.
ACKNOWLEDGMENTS
We thank Audrey Anglade (LIPM), Sebastien Cunnac (LIPM), Aurelie Vax (LISBP), and Marie-Odile Loret (LISBP) for their help in several experiments. We also thank Jean-Luc Pariente (LIPM), Claudette Icher (LIPM), and Céline Remblière (LIPM) for the production of the plant material.
This work was supported by the French Laboratory of Excellence project TULIP (ANR-10-LABX-41 and ANR-11-IDEX-0002-02) and by a grant from the Ministère de l’Enseignement Supérieur et de la Recherche to M.P. This work was supported in part by a grant from ANR (ANR-07-Blan-0205-01) and by a grant from Région Midi-Pyrénées (grant no. 07005583) to J.M.F.
Footnotes
Citation Poueymiro M, Cazalé AC, François JM, Parrou JL, Peeters N, Genin S. 2014. A Ralstonia solanacearum type III effector directs the production of the plant signal metabolite trehalose-6-phosphate. mBio 5(6):e02065-14. doi:10.1128/mBio.02065-14.
REFERENCES
- 1. Genin S, Denny TP. 2012. Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50:67–89. 10.1146/annurev-phyto-081211-173000. [DOI] [PubMed] [Google Scholar]
- 2. Peeters N, Guidot A, Vailleau F, Valls M. 2013. Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol. Plant Pathol. 14:651–662. 10.1111/mpp.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Genin S. 2010. Molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum. New Phytol. 187:920–928. 10.1111/j.1469-8137.2010.03397.x. [DOI] [PubMed] [Google Scholar]
- 4. Peeters N, Carrère S, Anisimova M, Plener L, Cazalé AC, Genin S. 2013. Repertoire, unified nomenclature and evolution of the type III effector gene set in the Ralstonia solanacearum species complex. BMC Genomics 14:859. 10.1186/1471-2164-14-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mukaihara T, Tamura N, Iwabuchi M. 2010. Genome-wide identification of a large repertoire of Ralstonia solanacearum type III effector proteins by a new functional screen. Mol. Plant Microbe Interact. 23:251–262. 10.1094/MPMI-23-3-0251. [DOI] [PubMed] [Google Scholar]
- 6. Cunnac S, Occhialini A, Barberis P, Boucher C, Genin S. 2004. Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol. Microbiol. 53:115–128. 10.1111/j.1365-2958.2004.04118.x. [DOI] [PubMed] [Google Scholar]
- 7. Deslandes L, Genin S. 2014. Opening the Ralstonia solanacearum type III effector tool box: insights into host cell subversion mechanisms. Curr. Opin. Plant Biol. 20:110–117. [DOI] [PubMed] [Google Scholar]
- 8. Cunnac S, Boucher C, Genin S. 2004. Characterization of the cis-acting regulatory element controlling HrpB-mediated activation of the type III secretion system and effector genes in Ralstonia solanacearum. J. Bacteriol. 186:2309–2318. 10.1128/JB.186.8.2309-2318.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Angot A, Peeters N, Lechner E, Vailleau F, Baud C, Gentzbittel L, Sartorel E, Genschik P, Boucher C, Genin S. 2006. Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. U. S. A. 103:14620–14625. 10.1073/pnas.0509393103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jacobs JM, Milling A, Mitra RM, Hogan CS, Ailloud F, Prior P, Allen C. 2013. Ralstonia solanacearum requires PopS, an ancient AvrE-family effector, for virulence and to overcome salicylic acid-mediated defenses during tomato pathogenesis. mBio 4(6):e00875-13. 10.1128/mBio.00875-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen L, Shirota M, Zhang Y, Kiba A, Hikichi Y, Ohnishi K. 2014. Involvement of HLK effectors in Ralstonia solanacearum disease development in tomato. J. Gen. Plant Pathol. 80:79–84. 10.1007/s10327-013-0490-2. [DOI] [Google Scholar]
- 12. Solé M, Popa C, Mith O, Sohn KH, Jones JD, Deslandes L, Valls M. 2012. The awr gene family encodes a novel class of Ralstonia solanacearum type III effectors displaying virulence and avirulence activities. Mol. Plant Microbe Interact. 25:941–953. 10.1094/MPMI-12-11-0321. [DOI] [PubMed] [Google Scholar]
- 13. Macho AP, Guidot A, Barberis P, Beuzón CR, Genin S. 2010. A competitive index assay identifies several Ralstonia solanacearum type III effector mutant strains with reduced fitness in host plants. Mol. Plant Microbe Interact. 23:1197–1205. 10.1094/MPMI-23-9-1197. [DOI] [PubMed] [Google Scholar]
- 14. Jones JD, Dangl JL. 2006. The plant immune system. Nature 444:323–329. 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 15. Lavie M, Shillington E, Eguiluz C, Grimsley N, Boucher C. 2002. PopP1, a new member of the YopJ/AvrRxv family of type III effector proteins, acts as a host-specificity factor and modulates aggressiveness of Ralstonia solanacearum. Mol. Plant Microbe Interact. 15:1058–1068. 10.1094/MPMI.2002.15.10.1058. [DOI] [PubMed] [Google Scholar]
- 16. Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. 2003. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. U. S. A. 100:8024–8029. 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nahar K, Matsumoto I, Taguchi F, Inagaki Y, Yamamoto M, Toyoda K, Shiraishi T, Ichinose Y, Mukaihara T. 2014. Ralstonia solanacearum type III secretion system effector Rip36 induces a hypersensitive response in the nonhost wild eggplant Solanum torvum. Mol. Plant Pathol. 15:297–303. 10.1111/mpp.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Poueymiro M, Cunnac S, Barberis P, Deslandes L, Peeters N, Cazale-Noel AC, Boucher C, Genin S. 2009. Two type III secretion system effectors from Ralstonia solanacearum GMI1000 determine host-range specificity on tobacco. Mol. Plant Microbe Interact. 22:538–550. 10.1094/MPMI-22-5-0538. [DOI] [PubMed] [Google Scholar]
- 19. Tasset C, Bernoux M, Jauneau A, Pouzet C, Brière C, Kieffer-Jacquinod S, Rivas S, Marco Y, Deslandes L. 2010. Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R-mediated immunity in Arabidopsis. PLoS Pathog. 6:e1001202. 10.1371/journal.ppat.1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kajava AV, Anisimova M, Peeters N. 2008. Origin and evolution of GALA-LRR, a new member of the CC-LRR subfamily: from plants to bacteria? PLoS One 3:e1694. 10.1371/journal.pone.0001694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Remigi P, Anisimova M, Guidot A, Genin S, Peeters N. 2011. Functional diversification of the GALA type III effector family contributes to Ralstonia solanacearum adaptation on different plant hosts. New Phytol. 192:976–987. 10.1111/j.1469-8137.2011.03854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li L, Atef A, Piatek A, Ali Z, Piatek M, Aouida M, Sharakuu A, Mahjoub A, Wang G, Khan S, Fedoroff NV, Zhu J-K, Mahfouz MM. 2013. Characterization and DNA-binding specificities of Ralstonia TAL-like effectors. Mol. Plant 6:1318–1330. 10.1093/mp/sst006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. De Lange O, Schreiber T, Schandry N, Radeck J, Braun KH, Koszinowski J, Heuer H, Strauss A, Lahaye T. 2013. Breaking the DNA-binding code of Ralstonia solanacearum TAL effectors provides new possibilities to generate plant resistance genes against bacterial wilt disease. New Phytol. 199:773–786. 10.1111/nph.12324. [DOI] [PubMed] [Google Scholar]
- 24. Lunn JE, Delorge I, Figueroa CM, Van Dijck P, Stitt M. 2014. Trehalose metabolism in plants. Plants J. 79:544–567. 10.1111/tpj.12509. [DOI] [PubMed] [Google Scholar]
- 25. Gibson RP, Tarling CA, Roberts S, Withers SG, Davies GJ. 2004. The donor subsite of trehalose-6-phosphate synthase: binary complexes with UDP-glucose and UDP-2-deoxy-2-fluoro-glucose at 2 A resolution. J. Biol. Chem. 279:1950–1955. 10.1074/jbc.M307643200. [DOI] [PubMed] [Google Scholar]
- 26. Gibson RP, Turkenburg JP, Charnock SJ, Lloyd R, Davies GJ. 2002. Insights into trehalose synthesis provided by the structure of the retaining glucosyltransferase OtsA. Chem. Biol 9:1337–1346. [DOI] [PubMed] [Google Scholar]
- 27. Sory MP, Cornelis GR. 1994. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol. Microbiol. 14:583–594. 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 28. Remenant B, de Cambiaire JC, Cellier G, Jacobs JM, Mangenot S, Barbe V, Lajus A, Vallenet D, Medigue C, Fegan M, Allen C, Prior P. 2011. Ralstonia syzygii, the blood disease bacterium and some Asian R. solanacearum strains form a single genomic species despite divergent lifestyles. PLoS One 6:e24356. 10.1371/journal.pone.0024356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bonini BM, Van Vaeck C, Larsson C, Gustafsson L, Ma P, Winderickx J, Van Dijck P, Thevelein JM. 2000. Expression of Escherichia coli otsA in a Saccharomyces cerevisiae tps1 mutant restores trehalose 6-phosphate levels and partly restores growth and fermentation with glucose and control of glucose influx into glycolysis. Biochem. J. 350:261–268. 10.1042/0264-6021:3500261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Walther T, Mtimet N, Alkim C, Vax A, Loret MO, Ullah A, Gancedo C, Smits GJ, François JM. 2013. Metabolic phenotypes of Saccharomyces cerevisiae mutants with altered trehalose 6-phosphate dynamics. Biochem. J. 454:227–237. 10.1042/BJ20130587. [DOI] [PubMed] [Google Scholar]
- 31. Loret MO, Pedersen L, François J. 2007. Revised procedures for yeast metabolites extraction: application to a glucose pulse to carbon-limited yeast cultures, which reveals a transient activation of the purine salvage pathway. Yeast 24:47–60. 10.1002/yea.1435. [DOI] [PubMed] [Google Scholar]
- 32. Wilson RA, Jenkinson JM, Gibson RP, Littlechild JA, Wang ZY, Talbot NJ. 2007. Tps1 regulates the pentose phosphate pathway, nitrogen metabolism and fungal virulence. EMBO J. 26:3673–3685. 10.1038/sj.emboj.7601795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schluepmann H, Pellny T, van Dijken A, Smeekens S, Paul M. 2003. Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 100:6849–6854. 10.1073/pnas.1132018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Paul MJ, Primavesi LF, Jhurreea D, Zhang Y. 2008. Trehalose metabolism and signaling. Annu. Rev. Plant Biol. 59:417–441. 10.1146/annurev.arplant.59.032607.092945. [DOI] [PubMed] [Google Scholar]
- 35. Avonce N, Mendoza-Vargas A, Morett E, Iturriaga G. 2006. Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 6:109. 10.1186/1471-2148-6-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stavrinides J, Ma W, Guttman DS. 2006. Terminal reassortment drives the quantum evolution of type III effectors in bacterial pathogens. PLoS Pathog. 2:e104. 10.1371/journal.ppat.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tournu H, Fiori A, Van Dijck P. 2013. Relevance of trehalose in pathogenicity: some general rules, yet many exceptions. PLoS Pathog. 9:e1003447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Puttikamonkul S, Willger SD, Grahl N, Perfect JR, Movahed N, Bothner B, Park S, Paderu P, Perlin DS, Cramer RA., Jr 2010. Trehalose 6-phosphate phosphatase is required for cell wall integrity and fungal virulence but not trehalose biosynthesis in the human fungal pathogen Aspergillus fumigatus. Mol. Microbiol. 77:891–911. 10.1111/j.1365-2958.2010.07254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Djonović S, Urbach JM, Drenkard E, Bush J, Feinbaum R, Ausubel JL, Traficante D, Risech M, Kocks C, Fischbach MA, Priebe GP, Ausubel FM. 2013. Trehalose biosynthesis promotes Pseudomonas aeruginosa pathogenicity in plants. PLoS Pathog. 9:e1003217. 10.1371/journal.ppat.1003217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Y, Primavesi LF, Jhurreea D, Andralojc PJ, Mitchell RA, Powers SJ, Schluepmann H, Delatte T, Wingler A, Paul MJ. 2009. Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol. 149:1860–1871. 10.1104/pp.108.133934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wahl V, Ponnu J, Schlereth A, Arrivault S, Langenecker T, Franke A, Feil R, Lunn JE, Stitt M, Schmid M. 2013. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 339:704–707. 10.1126/science.1230406. [DOI] [PubMed] [Google Scholar]
- 42. Ponnu J, Wahl V, Schmid M. 2011. Trehalose-6-phosphate: connecting plant metabolism and development. Front. Plant Sci. 2:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schluepmann H, Berke L, Sanchez-Perez GF. 2012. Metabolism control over growth: a case for trehalose-6-phosphate in plants. J. Exp. Bot. 63:3379–3390. 10.1093/jxb/err311. [DOI] [PubMed] [Google Scholar]
- 44. Hardie DG. 2007. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 8:774–785. 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 45. Halford NG, Hey SJ. 2009. Snf1-related protein kinases (SnRKs) act within an intricate network that links metabolic and stress signalling in plants. Biochem. J. 419:247–259. 10.1042/BJ20082408. [DOI] [PubMed] [Google Scholar]
- 46. Szczesny R, Büttner D, Escolar L, Schulze S, Seiferth A, Bonas U. 2010. Suppression of the AvrBs1-specific hypersensitive response by the YopJ effector homolog AvrBsT from Xanthomonas depends on a SNF1-related kinase. New Phytol. 187:1058–1074. 10.1111/j.1469-8137.2010.03346.x. [DOI] [PubMed] [Google Scholar]
- 47. Hao L, Wang H, Sunter G, Bisaro DM. 2003. Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell 15:1034–1048. 10.1105/tpc.009530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, Chermak D, Antony G, White FF, Somerville SC, Mudgett MB, Frommer WB. 2010. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468:527–532. 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Djamei A, Schipper K, Rabe F, Ghosh A, Vincon V, Kahnt J, Osorio S, Tohge T, Fernie AR, Feussner I, Feussner K, Meinicke P, Stierhof YD, Schwarz H, Macek B, Mann M, Kahmann R. 2011. Metabolic priming by a secreted fungal effector. Nature 478:395–398. 10.1038/nature10454. [DOI] [PubMed] [Google Scholar]
- 50. Yadav UP, Ivakov A, Feil R, Duan GY, Walther D, Giavalisco P, Piques M, Carillo P, Hubberten HM, Stitt M, Lunn JE. 2014. The sucrose-trehalose 6-phosphate (Tre6P) nexus: specificity and mechanisms of sucrose signalling by Tre6P. J. Exp. Bot. 65:1051–1068. 10.1093/jxb/ert457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang Y, Li J, Hou S, Wang X, Li Y, Ren D, Chen S, Tang X, Zhou JM. 2010. A Pseudomonas syringae ADP-ribosyltransferase inhibits Arabidopsis mitogen-activated protein kinase kinases. Plant Cell 22:2033–2044. 10.1105/tpc.110.075697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhao B, Dahlbeck D, Krasileva KV, Fong RW, Staskawicz BJ. 2011. Computational and biochemical analysis of the Xanthomonas effector AvrBs2 and its role in the modulation of Xanthomonas type three effector delivery. PLoS Pathog. 7:e1002408. 10.1371/journal.ppat.1002408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dean P. 2011. Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol. Rev. 35:1100–1125. 10.1111/j.1574-6976.2011.00271.x. [DOI] [PubMed] [Google Scholar]
- 54. Plener L, Manfredi P, Valls M, Genin S. 2010. PrhG, a transcriptional regulator responding to growth conditions, is involved in the control of the type III secretion system regulon in Ralstonia solanacearum. J. Bacteriol. 192:1011–1019. 10.1128/JB.01189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Plener L, Boistard P, González A, Boucher C, Genin S. 2012. Metabolic adaptation of Ralstonia solanacearum during plant infection: a methionine biosynthesis case study. PLoS One 7:e36877. 10.1371/journal.pone.0036877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pazhouhandeh M, Dieterle M, Marrocco K, Lechner E, Berry B, Brault V, Hemmer O, Kretsch T, Richards KE, Genschik P, Ziegler-Graff V. 2006. F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc. Natl. Acad. Sci. U. S. A. 103:1994–1999. 10.1073/pnas.0510784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Neuhoff V, Stamm R, Eibl H. 1985. Clear background and highly sensitive protein staining with Coomassie blue dyes in polyacrylamide gels: a systematic analysis. Electrophoresis 6:427–448. 10.1002/elps.1150060905. [DOI] [Google Scholar]
- 58. Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard J-F, Guindon S, Lefort V, Lescot M, Claverie J-M, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469. 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview, version 2. a multiple sequence alignment Editor and analysis workbench. Bioinformatics 25:1189–1191. 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein sequence alignment of RipTPS with other TPS sequences. Amino acids that constitute the glucose-6-phosphate binding domain are underlined with a pink arrow, whereas those involved in the conformation of the binding site are blue. The green arrows indicate the residues required for interaction with UDP-glucose. The tyrosine, tryptophan, and aspartate amino acids from the glucose-6-phosphate binding domain that were mutated in RipTPS are also indicated. The sequence alignment was performed using jalview 2.0 software (59). The accession numbers of the aligned proteins are as follows: OtsA_Rs_GMI1000 (NP_522666), OtsA_Rs_CMR15 (YP_006061033), OtsA_Rs_Psi07 (YP_003750101), OtsA_Rs_CFBP2957 (YP_003748420), OtsA_E.coli_K12 (YP_490157), Candidate RipTPS_X.oryzae (WP_019303360), Candidate_RipTPS_A.citrulli (YP_970430), RipTPS_Rs_GMI1000 (NP_522292), RipTPS_Rs_CMR15 (CMR15v4_mp10701), RipTPS_Rs_Psi07 (YP_003749608), and RipTPS_Rs_CFBP2957 (RCFBP_mp20172). Download
R. solanacearum ripTPS
mutant is not affected in its pathogenicity. The R. solanacearum ripTPS mutant GRS432 cannot be distinguished from the wild-type strain GMI1000 on Arabidopsis, tomato, eggplant, or geranium. All four plants were tested at least three times; the results of one representative experiment are shown in a Kaplan-Meier survival analysis representation. This graphical representation indicates the percent survival of the plants inoculated with GMI1000 or GRS432. For this statistical analysis, the phenotyping data (progressive disease index [DI] scale ranging from 0 to 4, or 0% to 100% of plants wilted) is converted to binary data: wilted (DI > 2) or not wilted. The log-rank test is a hypothesis test, with Ho being the fact that both survival curves are identical. The P values for each test indicate that the hypothesis of similarity of curves cannot be rejected. Statistical tests were performed as described previously (21). Download
Agrobacterium-mediated transient expression of ripTPS constructs in N. benthamiana. Expression of RipTPS-HA (a), RipTPS variants labeled h (57 kDa), i (15 kDa), j (34 kDa), and k (30 kDa) in Fig. 5A, and controls OtsAEcoli-HA (e), GUS-HA (f), and AvrA (g). Results of Western blotting with anti-HA antibody and Coomassie staining of the protein extracts from Nicotiana benthamiana are shown. The black arrow indicates RipTPS (67 kDa). The asterisks indicate the same proteins as in Fig. 5. Download
Primers used in this work.