Abstract
A multilocus sequence typing (MLST) scheme was developed to study the genetic relationships and population structure of 72 Clostridium difficile isolates from various hosts, geographic sources, PCR ribotypes, and toxigenic types (determined by PCR targeting tcdA and tcdB genes). MLST was performed by DNA sequence analysis of seven housekeeping genes (aroE, ddl, dutA, tpi, recA, gmk, and sodA). The number of alleles ranged from five (dutA and ddl) to eleven (recA). Allelic profiles allowed the definition of 34 different sequence types (STs). These STs lacked correlation with geographic source but were well correlated to toxigenic type. The dendrogram generated from a matrix of pairwise genetic distances showed that animal isolates did not constitute a distinct lineage from human isolates and that there was no hypervirulent lineage within the population of toxigenic human isolates (isolates recovered from pseudomembranous colitis and antibiotic-associated diarrhea did not cluster in distinct lineages). However, A− B+ variant isolates shared the same ST that appeared as a divergent lineage in the population studied, indicating a single evolutionary origin. The population structure was further examined by analysis of allelic polymorphism. The dendrogram generated from composite sequence-based analysis revealed a homogeneous population associated with three divergent lineages, one of which was restricted to A− B+ variant isolates. C. difficile exhibited a clonal population structure, as revealed by the estimation of linkage disequilibrium (Ia) between loci. The analysis of alleles within clonal complexes estimated that point mutation generated new alleles at a frequency eightfold higher than recombinational exchange, and the congruence of the dendrograms generated from separate housekeeping loci confirmed the mutational evolution of this species.
Clostridium difficile is a frequent cause of antibiotic-associated diarrhea (AAD) and is responsible for virtually all cases of pseudomembranous colitis (PMC) (5, 48). Since it is recognized as a nosocomial enteropathogen (24, 34), many molecular typing methods such as pulsed-field gel electrophoresis (PFGE), PCR ribotyping, random amplified polymorphic DNA (RAPD) analysis, or amplified fragment length polymorphism have been used to investigate nosocomial outbreaks of C. difficile infections (3, 10, 27, 28) or to distinguish relapse from reinfection (4, 26). Conversely, very little data specifically addresses the population genetics and long-term epidemiology of this species. Indeed, several problems remain unclear about the population genetics of this organism. (i) Are there hypervirulent lineages, which spread by clonal diffusion? (ii) Is there human or animal host specificity? (iii) What is the evolutionary history of the toxin A-negative, toxin B-positive (A− B+) variant isolates recently reported in human pathogenic situations (1)? (iv) What are the relative rates of mutations and recombinations in evolutionary dynamics of C. difficile?
Previous studies, although focusing on short-term molecular epidemiology, tentatively answered some of these questions. Several reports have shown that some C. difficile isolates from different countries and without any direct epidemiological link harbored the same PCR ribotype (49, 50). Furthermore, until recently, almost all A− B+ isolates also harbored the same PCR ribotype, whatever their geographical origin (2, 42). Together, these two results suggest a clonal population structure of this species. Likewise, Rupnik et al. (41) developed toxinotyping and found that many isolates harbored the same toxinotype despite very different origins, suggesting a clonal diffusion of these strains. However, these data were obtained from methods based on DNA banding patterns, which generate results difficult to compare between laboratories despite strenuous efforts at standardization. In addition, although PFGE or PCR ribotyping successfully identified clusters of epidemiologically related isolates (7), these methods may be less suitable for global and long-term epidemiological studies and are inadequate for population genetics analysis.
Multilocus sequence typing (MLST) has been recently developed for the study of clonal relationships within bacterial populations and has been successfully used for population genetics and global epidemiological analysis of Neisseria meningitidis (22), Streptococcus pneumoniae (18), Staphylococcus aureus (16), Streptococcus pyogenes (19), Campylobacter jejuni (15), Salmonella enterica (29), and Enterococcus faecium (23). MLST characterizes multilocus genotypes of bacterial isolates by using 400- to 500-bp intragenic sequences of several (generally seven) housekeeping genes (33). Thus, MLST is similar in principle to multilocus enzyme electrophoresis, which has been largely developed in bacterial population genetics (45, 46) but presents a higher sensitivity due to its ability to detect neutral genetic variations. MLST has also been suggested as offering several advantages over other molecular typing methods. First, the data (DNA sequences) are unambiguous and so readily comparable between different laboratories and can be stored in a shared central database to provide a broader resource for epidemiological studies. Second, evolutionary genetics studies can be performed, since MLST describes variations affecting housekeeping genes. In the present study, we describe an MLST analysis of C. difficile based on the nucleotide sequences of seven housekeeping genes. Using this approach, we study the allelic diversity and population structure of a collection of 72 C. difficile isolates from various hosts, geographic sources, and toxigenic types.
MATERIALS AND METHODS
Bacterial isolates.
A total of 72 C. difficile isolates from various hosts and geographic sources and collected over a 12-year period were studied. Of these, 64 isolates were recovered from human stools: 36 from patients with AAD, 11 from patients with PMC, and 11 from patients with asymptomatic carriage (the data for 6 human isolates were unknown). Eight isolates from animal hosts suspected of clostridial intestinal infection were also included. Isolates were identified as C. difficile by Gram stain, colony morphology, and fluorescence, API 20A (BioMerieux, Marcy l'Etoile, France) biochemical profiles and, for some isolates, by sequencing the first 500 bp of 16S ribosomal DNA (rDNA) and of an internal fragment of the tpi gene (14) to confirm their species identification. Toxigenic types were determined by PCR targeting the toxins A (25) and B genes (36). Among the total 72 C. difficile isolates, 52 harbored the tcdA gene (encoding toxin A) and the tcdB gene (encoding toxin B), 8 harbored a deleted variant of tcdA gene and the tcdB gene (A− B+ variants), and 12 lacked the tcdA and the tcdB genes.
PCR ribotyping.
PCR ribotyping was performed according to a procedure described elsewhere (7) to assess the genetic diversity of the isolates included in the present study, particularly their lack of a direct epidemiological link.
MLST.
Seven housekeeping loci were selected for the characterization of C. difficile isolates by MLST (Table 1): aroE (shikimate dehydrogenase), ddl (d-alanine:d-alanine ligase), dutA (dUTP pyrophosphatase), gmk (guanylate kinase), recA (recombinase), sodA (superoxide dismutase), and tpi (triosephosphate isomerase). The choice of these housekeeping genes was based on their use in MLST schemes of other bacterial species and/or on the availability of sequence data from C. difficile (http://www.sanger.ac.uk/) and from other species. Only one copy of each of the seven housekeeping genes was found on the C. difficile 630 genome.
TABLE 1.
Genetic polymorphism of the seven housekeeping genes analyzed by MLST
| Gene | PCR and sequencing primers
|
Size (bp) of analyzed fragments | No. of alleles | No. of polymorphic sites | % Polymorphic sites | No. (%) of nucleotide differences between alleles | Mean % G + C content | dN/dSa | |
|---|---|---|---|---|---|---|---|---|---|
| Orientation | Sequence (5′→3′) | ||||||||
| aroE | Forward | CTAGTAGGTGAAAAACTCTCTCA | 410 | 10 | 32 | 7.8 | 1-23 (0.3-6.7) | 29.71 | 0.0682 |
| Reverse | ACTGGTGTAGCATTTAATATTATATC | ||||||||
| ddl | Forward | CATAAACTTGTTCATTCAGAAGG | 424 | 5 | 3 | 0.7 | 1-3 (0.2-0.7) | 30.32 | 0.1654 |
| Reverse | CTATGAGAAGTAAAGCCAGGAAT | ||||||||
| dutA | Forward | CCTAATTTTGCTCACAAAGGT | 325 | 5 | 14 | 4.3 | 1-11 (0.4-3.4) | 34.78 | 0.1125 |
| Reverse | AAATCCAGTTGAGCCAAACC | ||||||||
| gmk | Forward | TCA GGT GCA GGA AAA GGT AC | 292 | 7 | 11 | 3.7 | 1-8 (0.4-2.8) | 32.12 | 0.1074 |
| Reverse | TCT GTT TCT GTA CCT CTT CCA AC | ||||||||
| recA | Forward | CCA GAT ACA GGT GAA CAG GC | 379 | 11 | 17 | 4.5 | 1-13 (0.3-3.5) | 32.78 | 0.0000 |
| Reverse | TTT AAC ATT TTC TCT TCC TTG TCC | ||||||||
| sodA | Forward | TATSCWTATGATGCWYTWGARCC | 416 | 7 | 16 | 3.8 | 1-14 (0.3-3.4) | 30.98 | 0.0813 |
| Reverse | TARTAAGCATGYTCCCAAACATC | ||||||||
| tpi | Forward | GCAGGAAACTGGAAAATGCATAA | 395 | 7 | 10 | 2.5 | 1-7 (0.3-1.8) | 29.36 | 0.2611 |
| Reverse | CAGATTGGCTCATATGCAACAAC | ||||||||
Ratio of nonsynonymous to synonymous substitutions.
To prepare a DNA sample for PCR amplification, a bacterial colony was taken from blood agar culture and resuspended in 1 ml of distilled water in a microcentrifuge tube. The sample was then boiled for 20 min prior to being centrifuged for 2 min to settle bacterial debris, and 10 μl of supernatant, containing the genomic DNA, was used for subsequent PCR amplification. Internal 400- to 500-bp fragments of the selected genes were amplified with primers (Table 1) designed from sequence alignments of homologuous genes from low %G+C gram-positive bacteria.
PCRs were performed on a GeneAmp System 2400 thermal cycler (Applied Biosystems) in a final volume of 50 μl containing 0.5 μM concentrations of each primer, 200 μM concentrations of each deoxynucleoside triphosphate, and 1.25 U of TaqDNA polymerase (Applied Biosystems) in a 1× amplification buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 2.5 mM MgCl2). The PCR mixtures were heated for 3 min at 95°C and then a touch-down procedure followed, consisting of 30 s at 95°C, annealing for 30 s at temperatures decreasing from 60°C to 50°C during the first 11 cycles (with 1°C decremental steps in cycles 1 to 11), and ending with an extension step at 72°C for 30 s. Forty cycles were performed. PCR products were then purified with a QiaQuick gel extraction kit (Qiagen) and sequenced (200 to 500 ng of DNA) with PCR forward or reverse primers by using an ABI-PRISM BigDye terminator sequencing kit on an ABI-Prism 310 genetic analyzer (Applied Biosystems). Different sequences of a given locus were given allele numbers, and each unique combination of alleles (multilocus allelic profile) was assigned a sequence type (ST). Single point polymorphisms were assessed by sequencing both DNA strands from two separate PCR experiments.
Computer analysis of MLST data.
Clustering of the 72 isolates from the matrix of pairwise similarities between the allelic profiles was performed by using the START program (http://www.mlst.net) by the unweighted pair-group method with arithmetic averages (UPGMA). Nucleotide sequences were aligned by using BioEdit sequence alignment editor (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Average numbers of nucleotide differences between alleles and ratios of nonsynonymous to synonymous substitutions (dN/dS) were calculated to test the degree of selection operating on a locus by using the START program. Gene trees were constructed by using the neighbor-joining method and bootstrapping algorithms contained in the PHYLIP package (http://www.genebee.msu.su/services/phtree_full.html). Nucleotide composite sequences (2,217 bp, derived from seven concatenated gene fragments) were also aligned, and a phylogenetic dendrogram was generated by using the same procedure.
The index of association (Ia) between alleles (47) was used to test for linkage disequilibrium between alleles of the seven loci analyzed (http://www.mlst.net). The observed variance in the distribution of allelic mismatches in all pairwise comparisons of the allelic profiles was compared to that expected in a freely recombining population (linkage equilibrium). The significance of the difference in the observed and expected variances was evaluated by computing the maximum variance in the distribution of allelic mismatches obtained by using 100 randomizations of the data set.
Another estimation of recombination rates was made by using the BURST program (http://www.mlst.net), which defines clonal complexes (groups in which every isolate shares at least five identical alleles with at least one other isolate) and characterizes ancestral genotypes and their corresponding single-locus variants (SLVs; isolates that differ at only one of the seven loci) within these clonal complexes. Comparisons between the sequences of the alleles in each of the SLVs and in the corresponding putative ancestral genotype allowed to us to determine the relative rates of mutations and recombinations in the short-term evolution of the population studied (21).
Nucleotide sequence accession numbers.
Nucleotide sequences of the internal fragment genes analyzed in the present study have been deposited in the GenBank database under accession numbers AY533246 to AY533255 (for aroE), AY533256 to AY533260 (for dutA), AY530799 to AY530802 (for ddl), AY533261 to AY533267 (for gmk), AY533268 to AY533278 (for recA), AY533279 to AY533285 (for sodA), and AY533286 to AY533292 (for tpi).
RESULTS
Allelic variation in C. difficile.
Data reporting the allelic variations of the seven housekeeping genes are summarized in Table 1. The number of individual alleles for each of the seven housekeeping genes ranged from 5 for dutA and ddl to 11 for recA (for ddl; one of these five alleles was a null allele, since several amplification attempts with various primers spanning over the whole gene remained unsuccessful for 16 strains). The number of polymorphic sites on a given locus varied from 3 (for ddl) to 32 (for aroE), and the number of nucleotide differences between alleles of a given locus varied from one to 3 for ddl and from one to 23 for aroE. The variations in the sequences extended over the whole stretch of each of the seven genes investigated (data not shown). Most polymorphisms resulted in synonymous substitutions, the ratios of nonsynonymous to synonymous substitutions (dN/dS) varying from 0 (for recA) to 0.2611 (for tpi). These low ratios indicate a limited contribution of environmental selection to the sequence variations in the seven housekeeping genes analyzed; therefore, these loci are assumed to be suitable for population genetic study.
MLST analysis of C. difficile isolates.
The lack of an epidemiological link between the 72 isolates studied was confirmed by PCR ribotyping; a total of 62 PCR ribotypes were recorded. Among these 72 isolates, MLST generated 34 different STs (Table 2). The majority of these (22 of 34 STs) were represented by single isolates. Among STs shared by several isolates, the most frequently encountered were ST1 (9 isolates), ST2 (8 isolates), ST3 (5 isolates), and ST4 (5 isolates). No correlation was found between genotype and geographic origin: for example, ST1 isolates were from Belgium, Italy, Japan, and the United States and from different French hospitals; ST2 isolates were from the United States, Japan, and France. Conversely, a good correlation was observed between STs and toxigenic types: for example, ST1, ST3, ST4, and ST5 isolates were all A+ B+, whereas ST7 and ST11 isolates were all nontoxigenic, and ST2 isolates were all A− B+ variant isolates. In addition, the same correlation was also observed for four clonal complexes (including 18, 3, 3, and 2 STs, respectively) that were characterized by BURST analysis (http://www.mlst.net; data not shown).
TABLE 2.
Characteristics of the 34 allelic profiles (STs)
| ST | Allelic profiled
|
No. of isolates | Geographic origina | Hostb | No. of toxinogenic isolatesc | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| aroE | dutA | ddl | tpi | recA | gmk | sodA | |||||
| 1 | 2 | 2 | 2 | 1 | 2 | 1 | 2 | 9 | USA, It, Be, RF, NF, CF | Ha, Hc, horse, u | 9 |
| 2 | 4 | 3 | 5 | 3 | 3 | 3 | 4 | 8 | USA, Jp, PF | Ha, Hc, u | 8 (A− B+) |
| 3 | 2 | 2 | 2 | 1 | 1 | 1 | 5 | 5 | Jp, It, RF, CF | Ha, horse | 5 |
| 4 | 5 | 2 | 2 | 1 | 1 | 1 | 2 | 5 | RF, NF | Ha, Hc | 5 |
| 5 | 2 | 2 | 2 | 2 | 1 | 1 | 2 | 4 | GB, Be | Ha | 4 |
| 6 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 3 | USA, NF, RF | Ha | 3 |
| 7 | 6 | 2 | 3 | 1 | 6 | 1 | 1 | 3 | NF, CF | Hc, horse | 0 |
| 8 | 8 | 5 | 5 | 5 | 7 | 6 | 6 | 3 | It, RF, CF | Ha, cow | 2 |
| 9 | 5 | 2 | 4 | 1 | 1 | 2 | 1 | 3 | It, RF | Ha | 3 |
| 10 | 7 | 4 | 5 | 1 | 1 | 1 | 2 | 3 | RF, NF | Ha | 3 |
| 11 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | USA, RF | Hc | 0 |
| 12 | 2 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | It, CF | Dog, horse | 2 |
| 13 | 3 | 2 | 1 | 1 | 1 | 2 | 3 | 1 | USA | u | 1 |
| 14 | 3 | 2 | 3 | 1 | 1 | 4 | 2 | 1 | It | Ha | 0 |
| 15 | 2 | 2 | 2 | 1 | 1 | 2 | 5 | 1 | It | Rabbit | 1 |
| 16 | 2 | 2 | 1 | 2 | 1 | 1 | 5 | 1 | It | Ha | 1 |
| 17 | 5 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | It | Ha | 1 |
| 18 | 6 | 2 | 3 | 1 | 4 | 1 | 1 | 1 | Jp | Ha | 0 |
| 19 | 5 | 2 | 4 | 2 | 1 | 2 | 1 | 1 | Jp | Ha | 1 |
| 20 | 6 | 2 | 3 | 1 | 5 | 1 | 1 | 1 | RF | Hc | 0 |
| 21 | 2 | 2 | 1 | 4 | 1 | 2 | 2 | 1 | RF | Hc | 0 |
| 22 | 2 | 2 | 2 | 1 | 7 | 5 | 6 | 1 | NF | Hc | 1 |
| 23 | 2 | 2 | 2 | 6 | 1 | 1 | 2 | 1 | It | Ha | 1 |
| 24 | 5 | 2 | 4 | 1 | 8 | 2 | 1 | 1 | NF | Ha | 1 |
| 25 | 7 | 4 | 4 | 1 | 1 | 1 | 2 | 1 | NF | Ha | 1 |
| 26 | 4 | 3 | 5 | 7 | 9 | 7 | 5 | 1 | NF | Hc | 0 |
| 27 | 2 | 2 | 2 | 2 | 2 | 1 | 1 | 1 | RF | Ha | 1 |
| 28 | 9 | 2 | 2 | 1 | 1 | 1 | 7 | 1 | RF | Ha | 1 |
| 29 | 8 | 5 | 5 | 5 | 10 | 6 | 6 | 1 | RF | Ha | 1 |
| 30 | 3 | 2 | 2 | 1 | 11 | 2 | 1 | 1 | RF | Ha | 1 |
| 31 | 3 | 1 | 1 | 1 | 3 | 2 | 1 | 1 | RF | Ha | 1 |
| 32 | 2 | 2 | 2 | 4 | 1 | 2 | 2 | 1 | RF | Hc | 0 |
| 33 | 10 | 2 | 2 | 1 | 2 | 1 | 2 | 1 | PF | Ha | 1 |
| 34 | 8 | 5 | 2 | 5 | 7 | 6 | 6 | 1 | PF | Ha | 1 |
Be, Belgium; It, Italia; Jp, Japan; NF, Nancy (France); RF, Rouen (France); CF, Caen (France); PF, Paris; USA; United States of America; GB, Great Britain.
Ha, Human adult; Hc, Human child; u, unknown.
A+ B+ isolates, except for ST2 (A− B+ isolates).
Numbers are identification numbers of alleles.
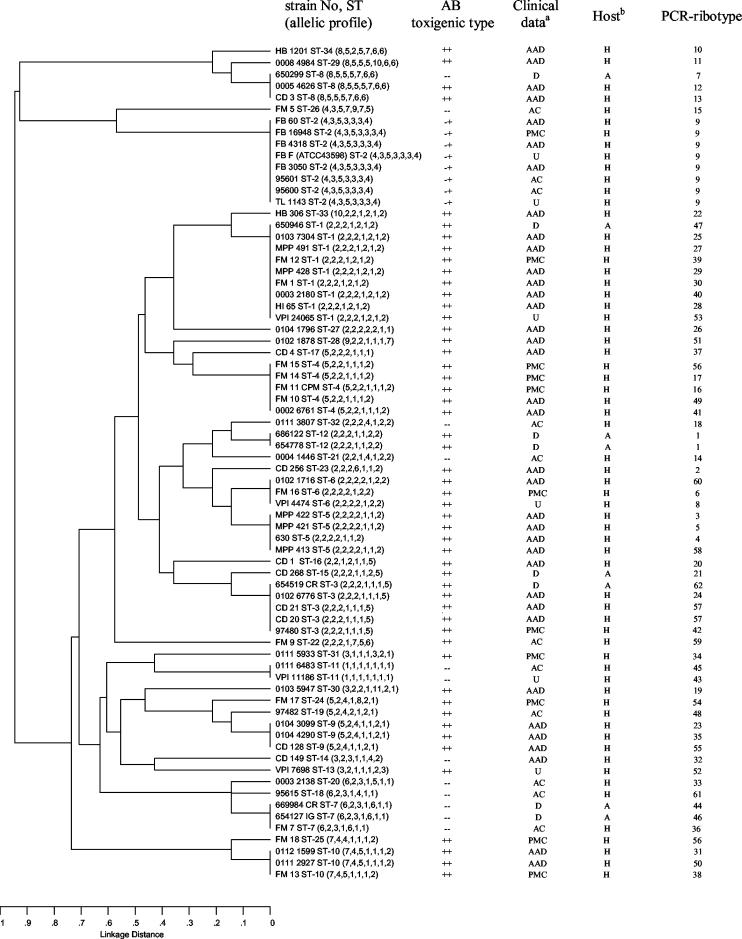
The results of clustering of the allelic profiles by UPGMA are shown in Fig. 1. The dendrogram confirms the correlation between toxigenic type and ST: only ST8 comprised both toxigenic and nontoxigenic isolates, and all A− B+ isolates are included in the ST2. This ST differed from ST26 (the nearest ST) in four of the seven alleles and from all other STs in at least six of the seven alleles. The dendrogram reveals also that animal isolates (belonging to ST1, ST3, ST7, ST8, ST12, and ST15) did not constitute a distinct subpopulation from human isolates. Toxigenic animal isolates clustered with toxigenic human isolates, and nontoxigenic animal isolates clustered with nontoxigenic human isolates, with the exception of ST8 (one nontoxigenic animal with two toxigenic human isolates). Concerning human isolates, PMC isolates belonged to 9 STs which contain only A+ B+ isolates except for ST2 which contains only A− B+ variant strains. They did not cluster in distinct lineages from AAD human isolates, and thus no lineage could be characterized as PMC specific. Clustering analysis also revealed very divergent STs (ST8, ST26, ST29, ST34, and ST2), which differed in at least six of the seven alleles from all other STs. The isolates of ST2 (all A− B+ variant isolates) shared tpi3, gmk3, and sodA4 alleles, which were not found among any of the other isolates and thus could be alternative markers of this lineage.
FIG. 1.
Dendrogram showing cluster analysis (UPGMA) of the 72 C. difficile isolates. Clinical data: D, diarrhea; AC, asymptomatic carriage; U, unknown. Other abbreviations are as defined in the text. Host: H, Human; A, Animal.
Composite sequence-based analysis.
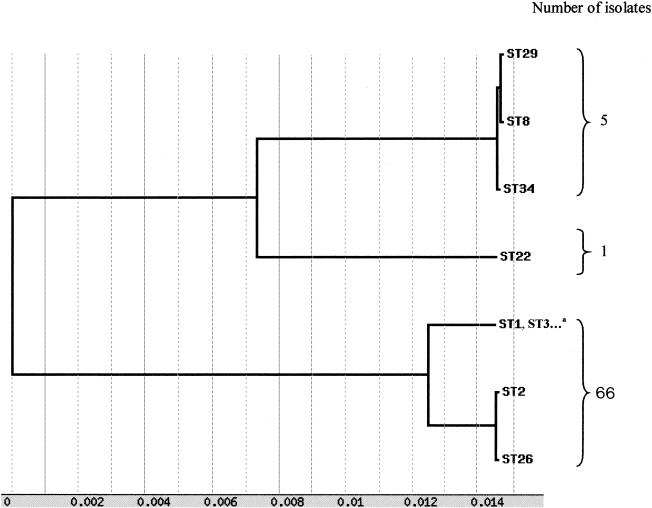
In order to determine the overall divergence of the sequenced gene fragments of the loci studied, the sequences of six loci (without ddl because of lack of sequence data for the null allele) were spliced together to obtain a concatenated composite sequence for each of the isolates. The identity between the 72 composite sequences was found between 96.8 and 100%. A dendrogram created from the matrix of pairwise sequence divergences of composite sequences is shown in Fig. 2. A majority of strains constituted a homogeneous population because of the very high degree of conservation observed in the housekeeping genes studied. Nevertheless, 3 divergent lineages were characterized: the first contained all A− B+ variants (ST2) and a nontoxigenic isolate (ST26); the second contained ST8, ST29, and ST34; and the third was restricted to one isolate (ST22).
FIG. 2.
Dendrogram showing genetic relationships of the 72 C. difficile isolates based on composite sequence of six housekeeping gene fragments. Other STs of this lineage: STs 4 to 7, 9 to 21, 23 to 25, 27, 28, and 30 to 33.
Dendrograms based on allelic variation of each housekeeping gene.
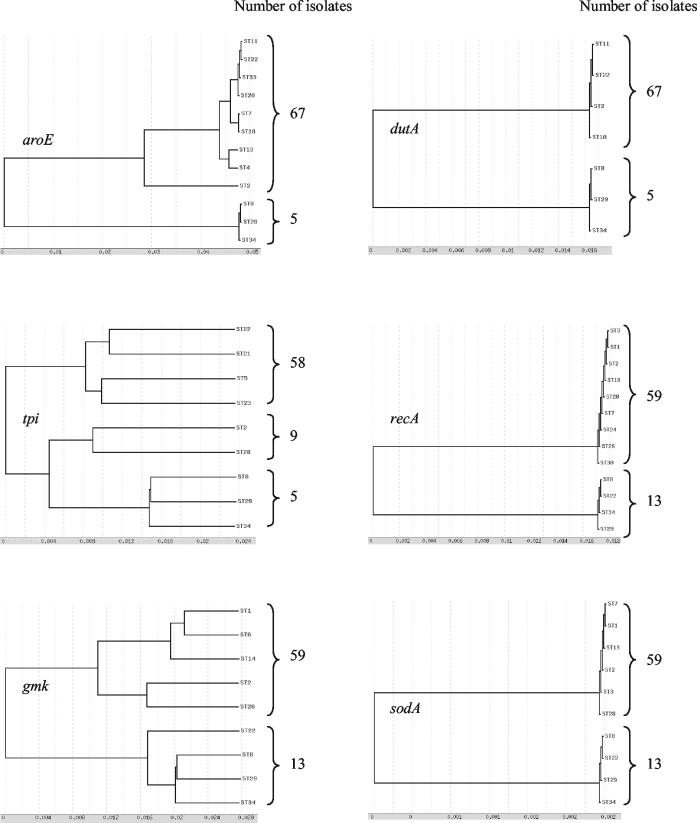
Dendrograms from the alleles of the six separate housekeeping genes were found to be congruent, as shown in Fig. 3 (ddl was not analyzed as upper justified). A clear bifurcation into majority and minority allelic populations was observed for all of the six genes studied. ST8 (three isolates), ST29 (one isolate), and ST34 (one isolate) belonged only to the minority allelic population of these six housekeeping genes. ST2 (variant A− B+ isolates) belonged to the majority allelic population for dutA, recA, and sodA loci and to the minority allelic population for aroE, tpi, and gmk loci.
FIG. 3.
Dendrograms showing genetic relationships of the 72 C. difficile isolates based on allele sequences of individual loci used for MLST. Only one ST is mentioned for each allele, except for STs 8, 29, and 34, which characterize the minority allelic population.
The five most divergent isolates (belonging to STs 8, 29, and 34), clustered in a distantly related lineage on the dendrograms constructed from both the multilocus allelic profiles (Fig. 1) and the composite sequence analysis (Fig. 2). In addition, considering the separate housekeeping genes, they differed in six of the seven alleles from all of the remaining isolates investigated. To confirm that these isolates actually belonged to C. difficile, we sequenced the first 500 bp of their 16S rDNA genes and an alternative species-specific target, the tpi gene encoding triosephosphate isomerase (14). 16S rDNA analysis revealed that these isolates shared 99.4% identity with the C. difficile ATCC 9689 strain and that they shared only 93% identity with C. sordellii and C. bifermentans type strains. On the basis of tpi sequences, these isolates were grouped in a cluster having at least 96.2% homology with the C. difficile type strain sequence, well separated (81% homology) from C. sordellii and C. bifermentans type strain sequences (14). Thus, these isolates diverging from the whole population studied belonged unambiguously to C. difficile.
Estimation of the relative contributions of recombination and mutation to genomic evolution of C. difficile.
A quantitative analysis of the linkage between alleles from the different loci was performed by calculating the index of association (Ia) (47). When all of the 72 isolates were included in the analysis, significant linkage disequilibrium was detected (Ia = 1.78). At the level of STs (one isolate from each ST to avoid bias due to a possible epidemic population structure), significant linkage disequilibrium was also detected (Ia = 0.89). After exclusion of the five genetically most divergent isolates, significant linkage disequilibrium between alleles of the remaining 67 isolates or the 31 STs was also detected (Ias = 1.53 and 0.50 for isolates and STs, respectively).
Another estimation of the relative contributions of recombination and mutation to clonal divergence was made by using the method and the criteria described by Feil et al. (21). It was estimated that point mutation generated new alleles at a frequency more than eightfold higher than recombinational exchange. These analyses, together with the congruence of trees constructed from individual loci, indicate that mutation events appeared much more frequently than recombination events in the population studied.
DISCUSSION
The aim of the present study was to provide a molecular approach that should be suitable for population genetics and global epidemiology analysis of C. difficile. Molecular methods such as PFGE (27) and PCR ribotyping (10) have been demonstrated accurate for short-term epidemiology of C. difficile but are inadequate for long-term epidemiology or population genetics of this organism. Hence, we chose to develop an MLST scheme, since MLST indexes neutral variations accumulating relatively slowly within the nucleotide sequences of housekeeping genes, which are thus suitable for phylogenetic analysis.
We determined the degree of allelic variations in seven housekeeping genes of 72 C. difficile isolates from various hosts and geographic sources. The degree of isolate differentiation by MLST (34 STs among 72 isolates) appears to be adequate for use in a population genetics setting. The allelic polymorphism (number of distinct STs among the isolates and number of polymorphic sites per locus) was comparable to that of S. aureus (16), a species whose population structure is very clonal (20), but lower than those of N. meningitidis and S. pneumoniae, two naturally recombinant species (21, 22). Of note, PCR ribotyping generated a higher number of genotypes than MLST. However, since our study mainly aimed at population genetics, we needed to analyze epidemiologically unrelated isolates, which therefore exhibited numerous different PCR ribotypes, whereas MLST genotypes reflect more ancestral genetic relationships.
This MLST analysis allowed us to study the possible correlations between STs and geographic source, host, toxigenic type, and pathogenic potential. We could not detect any correlation between ST and geographic origin. This lack of correlation was also seen for C. difficile with other typing methods such as toxinotyping (41) and PCR ribotyping (50) and for S. aureus with MLST (17). Thus, at least some of the STs should be regarded as stable subpopulations of C. difficile that are spread worldwide, a typical case for organisms with a clonal population structure.
C. difficile has been isolated from the feces of numerous animal species such as horses, cattle, cats, dogs, camels, donkeys, or hamsters (40). A correlation between a primary antibiotic treatment and the animal disease was demonstrated, as for the human disease (6, 40). However, few studies were interested in genetic relationships of human and animal C. difficile isolates (37, 50). Restriction endonuclease analysis found no correlation between C. difficile isolates recovered from pets and human isolates (37), but the isolates studied were collected from different locations. Nevertheless, based on the intestinal carriage rate of C. difficile in cats and dogs (40), it was speculated that domestic animals could constitute an important reservoir of C. difficile isolates and a potential source for human acquisition (37). Since the present MLST study did not characterize any host specificity, we can also hypothesize that domestic animals could constitute a source for human community infections.
It is still unclear why certain toxigenic strains can be recovered from asymptomatic patients or induce moderate to severe diarrhea or even PMC. Several studies have hypothesized that C. difficile infection is more a host-related phenomenon rather than being related to the characteristics of the organism (35, 37). Nevertheless, previous studies characterized various levels of intrinsic virulence in C. difficile, classifying toxigenic strains as highly or less virulent in the hamster model (12). These different virulence potentials of strains, however, could not be confirmed by other researchers (44), who were unable to discriminate in the hamster model between toxigenic strains with various levels of postulated virulence in humans. In our MLST analysis, isolates recovered from PMCs did not constitute a subpopulation distinct from AAD or asymptomatic carriage associated isolates. Therefore, if hypervirulent lineages do exist, the genetic determinants of virulence are probably not only vertically transmitted, but also horizontally exchanged between strains.
Toxin A variants (A− B+) strains have been recently reported (1, 8, 9). Two types of A− B+ variant strains are well characterized: the main one, similar to the reference strain of serogroup F (13), has been reported from multiple countries (25, 32), but the other one is restricted to only one isolate (8, 31). The clinical significance of these A− B+ variant strains remains unclear. Although strains from serogroup F were initially reported to be nonpathogenic and mainly isolated from asymptomatic children (12), cases of PMC or diarrhea associated with A− B+ variant strains have been recently reported (1, 25, 30). The majority of the A− B+ variant strains share the same genetic changes in the toxin A gene (1.7-kb deletion in the repetitive 3′-end domain and a nonsense mutation at position 47) (41), despite the further description of several other toxin A variants (43). They also share the same DNA profile using PCR ribotyping (50) and PFGE (38), although a few different genotypes were recently described (43). In the present study, A− B+ variant isolates exhibited the same PCR ribotype and clustered in a very homogeneous MLST phylogenetic lineage, although they were isolated from epidemiologically unrelated patients. A null allele was encountered for ddl in all of these A− B+ isolates, which were also found highly divergent for the six other housekeeping genes, confirming the originality of this lineage. Taken together, all of these results support the hypothesis of a low genetic diversity of A− B+ variant strains and of the international spread of this phylogenetic lineage. However, the origin and evolutionary history of this lineage requires further investigation, since the isolates we analyzed did not allow us to identify any ST relating this A− B+ lineage to the other isolates analyzed.
Several previous observations led us to presume that the population structure of C. difficile was very clonal: (i) isolates from diverse geographic origins could be included in the same toxinotype (41) or PCR ribotype (50), (ii) PCR ribotyping and toxinotyping display a strong correlation (42), and (iii) effective in vitro gene transfer is difficult to perform (39). In the present work, we also observed a lack of correlation between STs and geographic sources and a strong correlation between multilocus genotypes (STs) and toxigenic types. In addition, the index of association (Ia) between alleles of the different loci revealed linkage disequilibrium between alleles, confirming the presumed clonal population structure of C. difficile.
In bacteria with a clonal population structure, mutation plays a more important role than recombination in the genomic evolution. We undertook two approaches to estimate the relative impact of recombination and mutation on clonal divergence in C. difficile: (i) comparison of allele sequences within clonal complexes and (ii) visual inspection of individual MLST gene trees. The analysis of clonal complexes showed that a single allele is 8- to 10-fold more likely to change by point mutation than by recombination. This result is in agreement with that of S. aureus, another very clonal pathogen (20), but contrasts with those of N. meningitidis and S. pneumoniae, in which an allele changes between 5- and 10-fold more frequently by recombination than by mutation (21). However, (i) it is not obvious that the microevolutionary events occurring within clonal complexes reflect the relative importance of recombination and mutation over longer evolutionary time scales (20), and (ii) the accuracy of the method depends on the proportion of alleles that are effectively represented in the data set and then on the number of isolates analyzed. Therefore, we also examined the congruence of individual gene trees, since this approach requires a smaller number of isolates (chosen to be distantly related on the basis of source, date of isolation and MLST data) to examine the effect of recombination over the long-term. The strong congruence between single-locus phylogenetic trees suggests a consistent phylogenetic signal between the loci and gives an additional argument for a mutational evolution of this species. However, we detected a probable recombinational event between the FM9 strain (ST22) and another uncharacterized strain: ST22 clustered with the majority population on aroE, dutA, tpi, and recA gene trees and with the divergent minority population on gmk and sodA gene trees and was located in an intermediate position between majority and minority populations by the composite sequence approach. This indicates that recombinational events occurred in some regions of gmk and sodA genes. Previous data also suggest the occurrence of recombination in C. difficile, such as strains sharing the same PCR ribotype despite very different toxinotypes (42), or the suggestion that the cytotoxin gene of serotype F C. difficile strains should be a functional hybrid between the C. difficile toxin B gene and the C. sordellii lethal toxin gene (11). Finally, although homologous recombination may occasionally contribute to the evolution of C. difficile, there is evidence that the long-term evolution is mainly based on mutation events.
Finally, the MLST scheme proposed for C. difficile allowed us to establish the lack of host specificity and the lack of hypervirulent lineage within toxigenic strains and gives strong arguments for a clonal population structure and mutational evolution of this pathogen. Since MLST provides unambiguous and exportable sequence data, a central database could be proposed to collect allelic variation data from worldwide sources, which would be available for further population genetics and global epidemiology analysis of C. difficile.
Acknowledgments
We thank Anne Collignon (Chatenay-Malabry, France), Karine Maillard (Caen, France), Frederic Barbut (Paris, France), and Francine Mory (Nancy, France) for supplying strains.
REFERENCES
- 1.Alfa, M. J., A. Kabani, D. Lyerly, S. Moncrief, L. M. Neville, A. Al-Barrak, G. K. Harding, B. Dyck, K. Olekson, and J. M. Embil. 2000. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile responsible for a nosocomial outbreak of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 38:2706-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbut, F., V. Lalande, B. Burghoffer, H. V. Thien, E. Grimprel, and J. C. Petit. 2002. Prevalence and genetic characterization of toxin A variant strains of Clostridium difficile among adults and children with diarrhea in France. J. Clin. Microbiol. 40:2079-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbut, F., N. Mario, M. Delmee, J. Gozian, and J. C. Petit. 1993. Genomic fingerprinting of Clostridium difficile isolates by using a random amplified polymorphic DNA (RAPD) assay. FEMS Microbiol. Lett. 114:161-166. [DOI] [PubMed] [Google Scholar]
- 4.Barbut, F., A. Richard, K. Hamadi, V. Chomette, B. Burghoffer, and J. C. Petit. 2000. Epidemiology of recurrences or reinfections of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 38:2386-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartlett, J. G., N. Moon, T. W. Chang, N. Taylor, and A. B. Onderdonk. 1978. Role of Clostridium difficile in antibiotic-associated pseudomembranous colitis. Gastroenterology 75:778-782. [PubMed] [Google Scholar]
- 6.Baverud, V. 2002. Clostridium difficile infections in animals with special reference to the horse: a review. Vet. Q. 24:203-219. [DOI] [PubMed] [Google Scholar]
- 7.Bidet, P., V. Lalande, B. Salauze, B. Burghoffer, V. Avesani, M. Delmee, A. Rossier, F. Barbut, and J. C. Petit. 2000. Comparison of PCR-ribotyping, arbitrarily primed PCR, and pulsed-field gel electrophoresis for typing Clostridium difficile. J. Clin. Microbiol. 38:2484-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borriello, S. P., B. W. Wren, S. Hyde, S. V. Seddon, P. Sibbons, M. M. Krishna, S. Tabaqchali, S. Manek, and A. B. Price. 1992. Molecular, immunological, and biological characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile. Infect. Immun. 60:4192-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brazier, J. S., S. L. Stubbs, and B. I. Duerden. 1999. Prevalence of toxin A negative/B positive Clostridium difficile strains. J. Hosp. Infect. 42:248-249. [PubMed] [Google Scholar]
- 10.Cartwright, C. P., F. Stock, S. E. Beekmann, E. C. Williams, and V. J. Gill. 1995. PCR amplification of rRNA intergenic spacer regions as a method for epidemiologic typing of Clostridium difficile. J. Clin. Microbiol. 33:184-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaves-Olarte, E., P. Low, E. Freer, T. Norlin, M. Weidmann, C. von Eichel-Streiber, and M. Thelestam. 1999. A novel cytotoxin from Clostridium difficile serogroup F is a functional hybrid between two other large clostridial cytotoxins. J. Biol. Chem. 274:11046-11052. [DOI] [PubMed] [Google Scholar]
- 12.Delmee, M., and V. Avesani. 1990. Virulence of ten serogroups of Clostridium difficile in hamsters. J. Med. Microbiol. 33:85-90. [DOI] [PubMed] [Google Scholar]
- 13.Delmee, M., M. Homel, and G. Wauters. 1985. Serogrouping of Clostridium difficile strains by slide agglutination. J. Clin. Microbiol. 21:323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhalluin, A., L. Lemee, M. Pestel-Caron, F. Mory, G. Leluan, J. F. Lemeland, and J. L. Pons. 2003. Genotypic differentiation of twelve Clostridium species by polymorphism analysis of the triosephosphate isomerase (tpi) gene. Syst. Appl. Microbiol. 26:90-96. [DOI] [PubMed] [Google Scholar]
- 15.Dingle, K. E., F. M. Colles, R. Ure, J. A. Wagenaar, B. Duim, F. J. Bolton, A. J. Fox, D. R. Wareing, and M. C. Maiden. 2002. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg. Infect. Dis. 8:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 19.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69:2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feil, E. J., M. C. Enright, and B. G. Spratt. 2000. Estimating the relative contributions of mutation and recombination to clonal diversification: a comparison between Neisseria meningitidis and Streptococcus pneumoniae. Res. Microbiol. 151:465-469. [DOI] [PubMed] [Google Scholar]
- 22.Feil, E. J., M. C. Maiden, M. Achtman, and B. G. Spratt. 1999. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol. Biol. Evol. 16:1496-1502. [DOI] [PubMed] [Google Scholar]
- 23.Homan, W. L., D. Tribe, S. Poznanski, M. Li, G. Hogg, E. Spalburg, J. D. Van Embden, and R. J. Willems. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40:1963-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, S., C. R. Clabots, F. V. Linn, M. M. Olson, L. R. Peterson, and D. N. Gerding. 1990. Nosocomial Clostridium difficile colonization and disease. Lancet 336:97-100. [DOI] [PubMed] [Google Scholar]
- 25.Kato, H., N. Kato, K. Watanabe, N. Iwai, H. Nakamura, T. Yamamoto, K. Suzuki, S. M. Kim, Y. Chong, and E. B. Wasito. 1998. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J. Clin. Microbiol. 36:2178-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato, H., N. Kato, K. Watanabe, K. Ueno, Y. Sakata, and K. Fujita. 1996. Relapses or reinfections: analysis of a case of Clostridium difficile-associated colitis by two typing systems. Curr. Microbiol. 33:220-223. [DOI] [PubMed] [Google Scholar]
- 27.Kato, H., N. Kato, K. Watanabe, K. Ueno, H. Ushijima, S. Hashira, and T. Abe. 1994. Application of typing by pulsed-field gel electrophoresis to the study of Clostridium difficile in a neonatal intensive care unit. J. Clin. Microbiol. 32:2067-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klaassen, C. H., H. A. van Haren, and A. M. Horrevorts. 2002. Molecular fingerprinting of Clostridium difficile isolates: pulsed-field gel electrophoresis versus amplified fragment length polymorphism. J. Clin. Microbiol. 40:101-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kotetishvili, M., O. C. Stine, A. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2002. Multilocus sequence typing for characterization of clinical and environmental Salmonella strains. J. Clin. Microbiol. 40:1626-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Limaye, A. P., D. K. Turgeon, B. T. Cookson, and T. R. Fritsche. 2000. Pseudomembranous colitis caused by a toxin A− B+ strain of Clostridium difficile. J. Clin. Microbiol. 38:1696-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyerly, D. M., L. A. Barroso, T. D. Wilkins, C. Depitre, and G. Corthier. 1992. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile. Infect. Immun. 60:4633-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lyerly, D. M., L. M. Neville, D. T. Evans, J. Fill, S. Allen, W. Greene, R. Sautter, P. Hnatuck, D. J. Torpey, and R. Schwalbe. 1998. Multicenter evaluation of the Clostridium difficile TOX A/B TEST. J. Clin. Microbiol. 36:184-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McFarland, L. V., M. E. Mulligan, R. Y. Kwok, and W. E. Stamm. 1989. Nosocomial acquisition of Clostridium difficile infection. N. Engl. J. Med. 320:204-210. [DOI] [PubMed] [Google Scholar]
- 35.McFarland, L. V., C. M. Surawicz, and W. E. Stamm. 1990. Risk factors for Clostridium difficile carriage and C. difficile-associated diarrhea in a cohort of hospitalized patients. J. Infect. Dis. 162:678-684. [DOI] [PubMed] [Google Scholar]
- 36.Norwood, D. A., Jr., and J. A. Sands. 1997. Physical map of the Clostridium difficile chromosome. Gene 201:159-168. [DOI] [PubMed] [Google Scholar]
- 37.O'Neill, G., J. E. Adams, R. A. Bowman, and T. V. Riley. 1993. A molecular characterization of Clostridium difficile isolates from humans, animals and their environments. Epidemiol. Infect. 111:257-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pituch, H., N. van den Braak, W. van Leeuwen, A. van Belkum, G. Martirosian, P. Obuch-Woszczatynski, M. Luczak, and F. Meisel-Mikolajczyk. 2001. Clonal dissemination of a toxin-A-negative/toxin-B-positive Clostridium difficile strain from patients with antibiotic-associated diarrhea in Poland. Clin. Microbiol. Infect. 7:442-446. [DOI] [PubMed] [Google Scholar]
- 39.Purdy, D., T. A. O'Keeffe, M. Elmore, M. Herbert, A. McLeod, M. Bokori-Brown, A. Ostrowski, and N. P. Minton. 2002. Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol. Microbiol. 46:439-452. [DOI] [PubMed] [Google Scholar]
- 40.Riley, T. V., J. E. Adams, G. L. O'Neill, and R. A. Bowman. 1991. Gastrointestinal carriage of Clostridium difficile in cats and dogs attending veterinary clinics. Epidemiol. Infect. 107:659-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rupnik, M., V. Avesani, M. Janc, C. von Eichel-Streiber, and M. Delmee. 1998. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J. Clin. Microbiol. 36:2240-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rupnik, M., J. S. Brazier, B. I. Duerden, M. Grabnar, and S. L. Stubbs. 2001. Comparison of toxinotyping and PCR ribotyping of Clostridium difficile strains and description of novel toxinotypes. Microbiology 147:439-447. [DOI] [PubMed] [Google Scholar]
- 43.Rupnik, M., N. Kato, M. Grabnar, and H. Kato. 2003. New types of toxin A-negative, toxin B-positive strains among Clostridium difficile isolates from Asia. J. Clin. Microbiol. 41:1118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambol, S. P., J. K. Tang, M. M. Merrigan, S. Johnson, and D. N. Gerding. 2001. Infection of hamsters with epidemiologically important strains of Clostridium difficile. J. Infect. Dis. 183:1760-1766. [DOI] [PubMed] [Google Scholar]
- 45.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selander, R. K., T. K. Korhonen, V. Vaisanen-Rhen, P. H. Williams, P. E. Pattison, and D. A. Caugant. 1986. Genetic relationships and clonal structure of strains of Escherichia coli causing neonatal septicemia and meningitis. Infect. Immun. 52:213-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, J. M., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spencer, R. C. 1998. Clinical impact and associated costs of Clostridium difficile-associated disease. J. Antimicrob. Chemother. 41(Suppl. C):5-12. [DOI] [PubMed] [Google Scholar]
- 49.Spigaglia, P., R. Cardines, S. Rossi, M. G. Menozzi, and P. Mastrantonio. 2001. Molecular typing and long-term comparison of Clostridium difficile strains by pulsed-field gel electrophoresis and PCR-ribotyping. J. Med. Microbiol. 50:407-414. [DOI] [PubMed] [Google Scholar]
- 50.Stubbs, S. L., J. S. Brazier, G. L. O'Neill, and B. I. Duerden. 1999. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J. Clin. Microbiol. 37:461-463. [DOI] [PMC free article] [PubMed] [Google Scholar]