Abstract
Background
Group 5 allergens are small proteins that consist of two domains. They belong to the most potent respiratory allergens.
Objective
To determine the binding sites and to study allergic patients' IgE recognition of the group 5 allergen (Phl p 5) from timothy grass pollen using human monoclonal IgE antibodies that have been isolated from grass pollen allergic patients.
Methods
Using recombinant isoallergens, fragments, mutants and synthetic peptides of Phl p 5, as well as peptide-specific antibodies, the interaction of recombinant human monoclonal IgE and Phl p 5 was studied using direct binding and blocking assays. Cross-reactivity of monoclonal IgE with group 5 allergens in several grasses was studied and inhibition experiments with patients' polyclonal IgE were performed.
Results
Monoclonal human IgE showed extensive cross-reactivity with group 5 allergens in several grasses. Despite its small size of 29 kDa, four independent epitope clusters on isoallergen Phl p 5.0101, two in each domain, were recognized by human IgE. Isoallergen Phl p 5.0201 carried two of these epitopes. Inhibition studies with allergic patients' polyclonal IgE suggest the presence of additional IgE epitopes on Phl p 5.
Conclusions & Clinical Relevance
Our results reveal the presence of a large number of independent IgE epitopes on the Phl p 5 allergen explaining the high allergenic activity of this protein and its ability to induce severe allergic symptoms. High-density IgE recognition may be a general feature of many potent allergens and form a basis for the development of improved diagnostic and therapeutic procedures in allergic disease.
Keywords: allergenicity, conformational epitope, epitope mapping, group 5 grass pollen allergen, human monoclonal antibody, recombinant allergen fragment, recombinant antibody technology
Introduction
The interaction between IgE and allergen is a critical molecular interaction that initiates a cellular cascade of events that result in allergic inflammation 1. For instance, cross-linking of IgE on mast cells and basophils initiates cellular degranulation and the release of inflammatory mediators, proteases and pro-inflammatory cytokines whereas IgE-facilitated allergen presentation contributes to T cell activation.
While most common allergens are well characterized, for instance in terms of sequence, structure and often function, the allergen-specific IgE component of allergic disease is poorly defined 2. This lack of knowledge in part stems from the low concentration of IgE in biological fluids and the rarity of IgE-producing cells. In fact, B cells producing allergen-specific IgE have not yet been isolated and characterized from allergic patients. The only access to the molecular structures of human allergen-specific IgE has been via combinatorial cloning approaches using blood-derived cells of the B cell lineage 3. Indeed, only a handful of allergen-specific human IgE have been isolated by these means and characterized so far (reviewed by Gadermaier et al. 4), and only two structures of complexes of such human IgE and allergen have been determined to date 5,6.
Due to the relative lack of allergen-specific human IgE, many studies of antibody interactions with clinically relevant allergens had to rely on relatively artificial systems as they have utilized mouse monoclonal antibodies and recombinant chimeric human IgE created from such mouse antibodies. In such a series of investigations factors like IgE concentration, affinity and clonality were shown to be important for the biological outcome of allergen-IgE interaction 7,8. Similarly, it has been demonstrated that the IgE concentration and number of epitopes on the grass pollen allergen Phl p 1 determined the degree of effector cell degranulation 9. Overall, despite their shortcomings such monoclonal antibodies have provided crucial insight into the molecular mechanisms that govern key processes in the allergic reaction.
However, human allergen-specific IgE may differ in important respects from allergen-specific human IgG or mouse antibodies 10,11, in particular regarding their mode of allergen recognition 5,6. Human monoclonal IgE may thus represent a better mirror of human allergy-causing humoral immune responses and is therefore a preferred option for such studies 4. Unfortunately, human monoclonal IgE representing heavy and light chain combinations as present in human IgE-producing B cells of allergic donors are not currently available to address matters of IgE-allergen recognition 4. However, allergen-specific antibody fragments derived from the IgE-encoding transcriptome using combinatorial library technology are, as outline above, available 4, and these reagents have been used successfully for assessment of human IgE-allergen interactions, as summarized by Gadermaier et al. 4.
The largest available set of human monoclonal IgE 3,12–14 that targets a single allergen is that specific for the grass pollen allergen Phl p 5 15. Phl p 5 and related group 5 pollen allergens are highly potent and allergenic and were reported to be involved in triggering severe asthma attacks 16,17. Using a set of 10 human recombinant IgE obtained from grass pollen allergic patients, we now define the presence of at least four non-overlapping epitopes detected by these human IgE with potential additional binding sites for polyclonal IgE on this allergen. The presence of such an array of epitopes detected by IgE on a relatively small allergen (29 kDa) like Phl p 5 provides an explanation for the high allergenic potency of this protein.
Materials and methods
Recombinant antibody fragments
Single-chain fragment variable (scFv) and Fab specific for Phl p 5 have been selected by phage display technology. These proteins are summarized in Table S1. These proteins and variants thereof, produced as described in Supplementary Methods, were used in these investigations.
Recombinant scFv–Fc protein
ScFv fused to the Fc fragment of human IgE and IgG2 were produced in HEK293 cells as described in Supplementary Methods. Such fusion proteins that differ from conventional antibodies primarily by the presence of a short, flexible linker between the heavy and light chain variable domains and the absence of the first constant domain of the heavy chain and the constant domain of the light chain, have been used successfully in the past in a variety of applications 18,19.
Recombinant allergens, allergen fragments and synthetic peptides
Recombinant Phl p 5.0101 and Phl p 6.0101 were obtained from Biomay AG (Vienna, Austria) while Phl p 5.0201 was kindly donated by Dr. Jonas Lidholm (Thermo Fisher Scientific AB, Uppsala, Sweden). The C-terminal fragment of Phl p 5.0201, and mutants thereof made as described in the past 20, was kindly donated by Dr. Kirsten Gehlhar (Ruhr-Universität, Bochum, Germany). The C-terminal fragment (residues 181-312) of Phl p 5.0101, mutants thereof, as well as Phl p 5-derived peptides were made as described in Supplementary Methods.
Immunological analysis of allergen binding properties
Immunological analysis to determine the ability of scFv displayed on phage or scFv–Fc fusion proteins (scFv linked to Fcε or Fcγ2) to bind allergen was performed using standard methodology. These assays, described in detail in Supplementary Methods, assessed (1) the ability of these proteins to directly bind immobilized recombinant allergens and allergen fragments, (2) the affinity of scFv–Phl p 5.0101 interaction, (3) the ability of soluble allergen fragments to block binding of scFv to immobilized allergen, (4) the ability of a given specific scFv to block the binding of other scFv to Phl p 5, (5) the ability of pairs of recombinant scFv–Fc to bind allergen in a sandwich assay format, (6) the ability of anti-sera raised against defined peptides of Phl p 5 to block the binding of scFv to immobilized allergen, (7) the ability of scFv–Fcγ2 to block binding of polyclonal IgE to immobilized Phl p 5 and (8) the ability of serum IgE to bind complexes of allergen and recombinant scFv–Fcγ2 created in solution. Anonymized serum samples were used for this purpose with approval (EK 565/2007) by the ethics committee of the Medical University of Vienna. Furthermore, the reactivity of recombinant scFv–Fcε towards pollen allergen extracts and recombinant Phl p 5 was analyzed using the ImmunoCAP platform (Phadia AB, Uppsala, Sweden).
Results
IgE heavy chains in Phl p 5-specific IgE show broad germline gene usage
Table S1 provides a summary of the human antibody fragments that so far have been selected for binding to Phl p 5.0101 from antibody fragment libraries developed from human IgE transcriptomes. Two additional sets of antibody fragments have been selected for specificity to Phl p 6, both of which cross-react to Phl p 5 13. All of these antibodies but one (clone 5) were derived from an antibody library established from transcripts of a single donor affected by allergic rhinitis. The ten IgE clones have a diverse heavy chain-encoding germline gene origin both in terms of usage of gene subgroups (IGHV1, 3 and 5) and individual genes (IGHV1-18, IGHV1-69, IGHV3-11, IGHV3-21, IGHV3-23, IGHV3-30/IGHV3-30-3, IGHV5-a) and a diverse composition of the third, often specificity-determining 21, complementarity determining region (CDR) of the heavy chain, including a diverse length (16.9 ± 4.2 residues) (Table S1). The binders were all mutated in comparison with the corresponding heavy chain variable gene and carried a frequency of mutations of 5.5 ± 2.8%, a level similar to that of many other immunoglobulin repertoires 22. Altogether, the collection of antibody fragments represents a clonally diverse population of allergen-specific antibodies. We now produced several of these binders (4.2, 4.3, 4.4, 4.12, 4.13, p5-AB5, p5-MA5, clone 5 and p6-12) in scFv format fused to the Fc of human IgG2 and/or human IgE to perform further analysis of their reactivity patterns.
IgE targets both domains of Phl p 5 and preferentially the Phl p 5.0101 isoallergen
Phl p 5 is composed of two major domains 23,24. In the past, we defined three antibodies (4.2, 4.3 and clone 5) that bind to the N-terminal domain of Phl p 5.0101, as exemplified by their binding to an allergen fragment comprising residues 56-165 that showed high allergenic activity and was frequently recognized by grass pollen allergic patients IgE 12,23,25. In addition, the C-terminal part of the allergen has also been identified as an IgE-reactive domain 26. We produced a recombinant form of the C-terminal part of Phl p 5.0101 fused to GST (Glutathione S-transferase) and determined the ability of our monoclonal human antibodies to bind this protein. None of the antibodies that had been shown in the past to bind the N-terminal domain of Phl p 5 recognized its C-terminal fragment (Fig.1a). Clones 4.4, 4.12, 4.13, p5-AB5 and p5-MA5 bound the C-terminal fragment on its own (Fig.1a) showing that both domains contain epitopes bound by the recombinant human allergen-specific IgE fragments.
Figure 1.
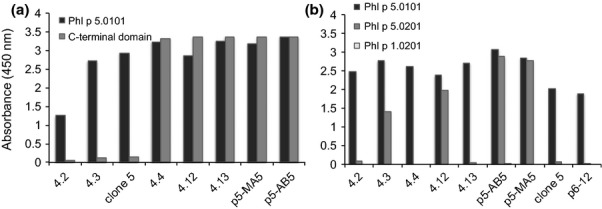
Reactivity of human scFv (x-axis) to the Phl p 5 C-terminal domain (a) and to the two major isoallergens of Phl p 5 (b). The results represent means of duplicate determinations with variations of < 8%.
Two major isoallergens 15,27 and 16 isoforms (variants) of Phl p 5 are recognized in the WHO/IUIS allergen nomenclature (http://www.allergen.org/). Mass spectrometry analysis of purified allergen even suggests that the number of isoforms of this allergen may be even larger 28. The reactivity of most patients' polyclonal IgE against different recombinant isoallergens is often of the same magnitude 29. However, the IgE of some patients may display a distinct difference in isoallergen reactivity 29. The isoallergen reactivity profile of one recombinant human IgE (clone 5) included in this study has been assessed in the past 25, and it was shown to bind only the Phl p 5.0101 isoallergen. We now tested additional recombinant human allergen-specific IgE antibody fragments to determine the degree of discrimination between the isoallergens of Phl p 5. We found that while all nine investigated scFv recognized the 01 isoallergen only four (4.3, 4.12, p5-AB5 and p5-MA5) recognized the 02 isoallergen (Fig.1b). ImmunoCAP analysis confirmed the ability of scFv 4.3 and the inability of scFv 4.2, 4.4 and 4.13, as scFv–Fcε, to bind the 02 isoallergen (data not shown). Furthermore, selection of the library used to isolate all but one of the Phl p 5-specific binders on Phl p 5.0201 did not result in identification of new scFv specific only for Phl p 5.0201 (data not shown). In conclusion, there are differences between isoallergens of Phl p 5 that translate into substantial differences in recognition by some human IgE and Phl p 5.0101 is the preferentially recognized isoallergen by this set of antibodies.
Recombinant human IgE antibody fragments recognize several conformational epitopes on both domains of group 5 pollen allergen
First, the relative location of epitopes recognized by different specific antibody fragments was determined by cross-wise inhibition studies. Clone 5 and 4.3 did not interfere with each other's binding to the Phl p 5 N-terminal domain (Table1), whereas scFv 4.2 bound a third epitope on the N-terminal domain that overlaps with the epitopes of clone 5 and 4.3 12. p6-12 selected on Phl p 6, an allergen with high sequence similarity to the N-terminal domain of Phl p 5 (Fig.2a) bound an epitope that overlaps the epitope recognized by scFv 4.3 (Table1). There are thus several, only in part overlapping, epitopes on the N-terminal domain of Phl p 5. Using scFv–Fcγ2 to block the reactivity of scFv–Fcε it was possible to demonstrate that binders 4.12, p5-AB5 and p5-MA5, known to bind the C-terminal domain of the allergen, represented one group of specific binders targeting an overlapping epitope, while 4.4 and 4.13 represent another group of overlapping binders targeting other epitopes on the C-terminal domain. Although some overlap between the epitope clusters on the C-terminal domain appeared to exist using this assay methodology, several of the antibodies targeting one of the clusters-bound Phl p 5 even in the presence of antibodies targeting the other cluster (Table1). To confirm these findings sandwich assays were carried out in which the ability of an scFv–Fcε to bind allergen caught by another binder (scFv–Fcγ2) was assessed. The signals of such assays are affected by epitope specificity but also the affinity of the antibodies for the antigen. Nevertheless, as demonstrated in Figure S1, these studies confirmed the existence of four largely independent epitopes bound by (1) 4.3, (2) clone 5, (3) 4.4 and 4.13, and (4) 4.12, p5-MA5 and p5-AB5.
Table 1.
Assessment of the ability of different human antibodies to block each other's binding to immobilized Phl p 5
| Inhibitory scFv–Fcγ2 | Test clone (scFv–Fcε) | Inhibitory scFv–Fcε | Test clone (scFv–Fcγ2) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 4.3 | 4.4 | 4.13 | 4.12 | p5-AB5 | p5-MA5 | p6-12 | clone 5 | ||
| 4.3 | ++ | − | − | − | − | − | 4.3 | ++ | − |
| Clone 5 | − | − | − | − | − | − | Clone 5 | nd | nd |
| 4.4 | − | + | + | − | − | − | 4.4 | − | − |
| 4.13 | − | ++ | ++ | + | − | − | 4.13 | − | − |
| 4.12 | − | − | − | + | − | + | 4.12 | − | − |
| p5-AB5 | − | − | − | ++ | + | ++ | p5-AB5 | − | − |
| p5-MA5 | − | ++ | − | ++ | − | ++ | p5-MA5 | − | − |
++: > 80% inhibition; +: 50–80% inhibition; −: < 50% inhibition.
nd, not determined as clone 5 (and p6-12) scFv–Fcε could not be produced.
Figure 2.
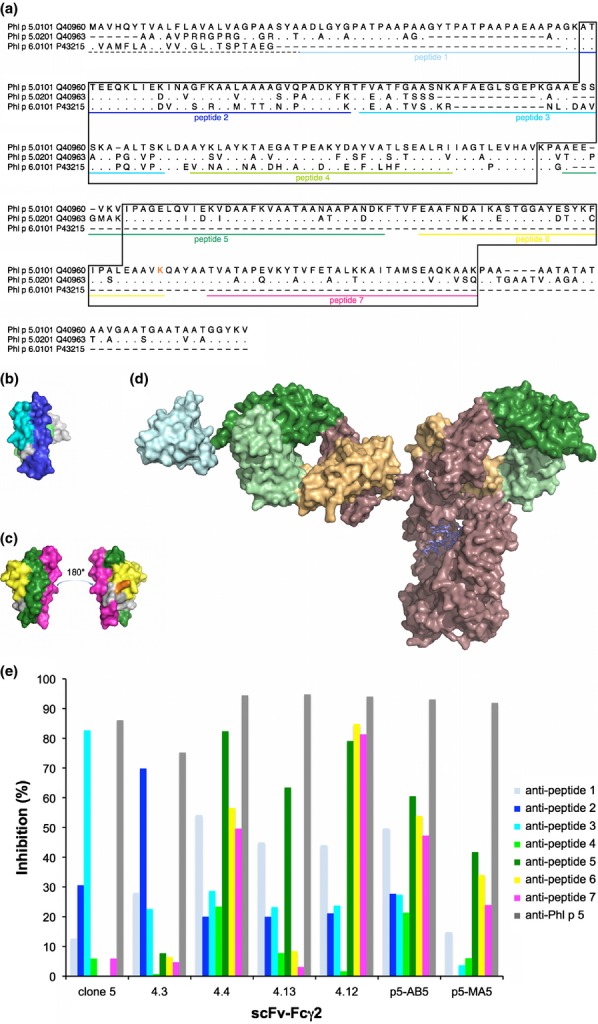
Localization of epitopes on Phl p 5. The sequences (including the signal sequences (dashed line)) of the two isoallergens of Phl p 5 and the related allergen Phl p 6 (GenBank accession numbers are indicated) are aligned (a). Residues identical to those found in Phl p 5.0101 are indicated by dots; missing residues are indicated by dashes. The parts of Phl p 6 and the C-terminal domain of Phl p 5 that are visible in 3D-structures (PDB: 1NLX and 1L3P, respectively) are boxed. Sequences of peptides derived from the Phl p 5.0101 sequence that were used for epitope mapping are underlined in colour. Residue K246 (incorporated in peptide 6) of Phl p 5.0101 is printed in orange. The position of peptides are highlighted in the structures of Phl p 6 (PDB: 1NLX) (b) and the C-terminal domain of Phl p 5.0201 (PDB: 1L3P) (c). A schematic illustration of the size difference between an antibody, in this case an IgG (PDB: 1HZH) and the C-terminal domain of Phl p 5 (PDB: 1L3P; cyan) is shown (d). Antibody variable and constant domains are coloured green and brown with sequences that belong to heavy and light chains shown in dark and pale colour, respectively. (e) Percentages inhibition (y-axis) of binding of scFv–Fcγ2 (x-axis) to Phl p 5 by rabbit anti-peptide and anti-Phl p 5 anti-sera.
To further characterize the epitopes of 4.12, p5-AB5 and p5-MA5 scFv derivatives, we tested scFv–Fcε against a set of mutants of the C-terminal domain of Phl p 5.0201 20. These proteins had been mutated, using an alanine-scanning approach, at 10 surface-exposed lysine residues of the C-terminal domain. 4.12, p5-MA5 and p5-AB5 scFv derivatives recognized the C-terminal domain of Phl p 5.0201 if lysines were retained at residues 214, 238 and 248 (Table2, Figure S2). The importance of residue 214 was confirmed by mutation of residue 246 of the C-terminal domain of Phl p 5.0101 (corresponding to residue 214 of Phl p 5.0201), which eliminated the reactivity of 4.12, p5-MA5 and p5-AB5 scFv (Fig.3). In contrast, 4.4 and 4.13, which could not be tested for reactivity to mutants of isoform Phl p 5.0201 as they did not recognize that isoform, were not sensitive to mutation of any of these three residues (246, 270 and 280, corresponding to residues 214, 238 and 248 of Phl p 5.0201) introduced into the C-terminal domain of Phl p 5.0101 (Fig.3). These studies confirm the existence of at least two different epitopes/epitope clusters detected by human IgE on the C-terminal domain of Phl p 5.
Table 2.
Recognition of recombinant group 5 or 6 allergens and versions of the C-terminal domain of Phl p 5.0201 in a direct binding assay
| Clone | Recombinant protein | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Phl p 1 (negative control) | Phl p 5.0101 | Phl p 5.0201 | Phl p 6.0101 | Phl p 5.0201 C-terminal domain*,† | |||||
| Wild-type | Mutations 1-6 | Mutations 1 + 10 | Mutations 1-6, 10 | Mutations 7-10 | |||||
| 4.3 | −‡ | +++ | ++ | +++ | − | − | − | − | − |
| 4.12 | − | +++ | ++ | − | − | − | − | ++ | − |
| p5-MA5 | − | +++ | +++ | − | ++ | +++ | + | +++ | − |
| p5-AB5 | − | +++ | +++ | − | + | +++ | ++ | +++ | − |
The following mutations in the C-terminal domain of Phl p 5.0201 were investigated: mutation number 1: K149A; 2: K160A; 3: K166A; 4: K190A; 5: K193A; 6: K204A; 7: K214A; 8: K238A; 9: K248A; 10: K283A.
Residues 214, 238 and 248 (modified by mutations 7-9) in Phl p 5.0201 correspond to residues 246, 270 and 280 in Phl p 5.0101 (Figure2A).
Absorbance values: −: A450 = 0–0.5; +: A450 = 0.5–1; ++: A450 = 1–2; +++: A450 = 2–3.
Figure 3.
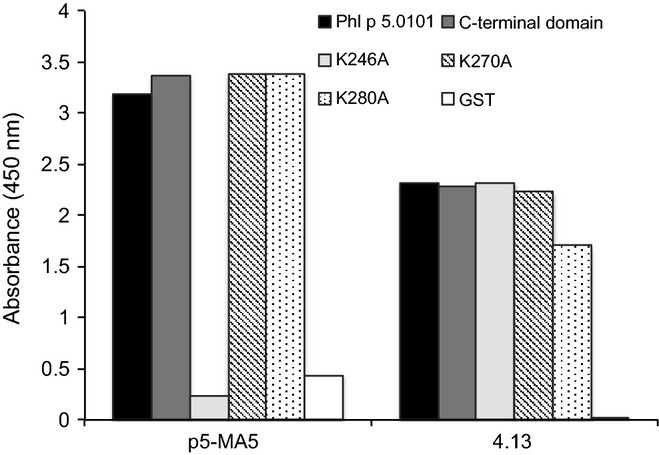
Effect on antibody binding of mutations in the C-terminal domain of Phl p 5. Binding of p5-MA5 (a), and 4.13 (b) to Phl p 5, the C-terminal domain of Phl p 5 and mutants (K246A, K270A or K280A) thereof, and GST. The results represent means of duplicate determinations with variations of < 8%.
We made use of synthetic peptides 30 covering much of the sequence of Phl p 5.0101 (Fig.2a) to further characterize the epitope specificity and nature of epitopes of our human monoclonal antibodies. According to circular dichroism analysis the synthetic peptides were unfolded and hence represented sequential epitopes 30. Interestingly, none of the tested scFv–Fcγ2 (4.3, 4.4, 4.12, 4.13, p5-AB5, p5-MA5 and, clone 5) bound directly to any of these peptides demonstrating that they did not carry sufficient sequence and/or structure to allow binding (data not shown). These binders thus capture the profile of serum IgE that also is unable to recognize these peptides 30. However, rabbit anti-sera 30 developed against some of these seven peptides inhibited the reactivity of IgE-derived scFv derivatives to Phl p 5.0101. In agreement with the non-overlapping nature of their respective epitopes, the binding of 4.3 and clone 5 was inhibited by anti-sera targeting peptides representing different parts of the allergen's N-terminal domain (i.e. residues 59–92 and 98–128, respectively) (Figs2b and e). The finding that antibodies targeting a peptide comprising residues 98–128 block binding of clone 5 confirmed past studies of this binder 25,31. Binders specific for the C-terminal domain were blocked to different degrees by anti-sera specific for three different peptides derived from this domain. In particular, the binding of scFv 4.13 was blocked by anti-sera targeting residues 176-212 while scFv 4.12, p5-AB5 and to a lesser extent, p5-MA5 detected an epitope blocked by anti-sera targeting three peptides (representing residues 176-212, 217-246 (i.e. including residue K246 defined by alanine scanning as described above) and 252–283 of Phl p 5.0101) that together represent the C-terminal domain of Phl p 5 (Figs2c and e). Using the peptide-specific anti-sera, it was thus possible to pinpoint the location of epitopes recognized by IgE-derived scFv on the folded Phl p 5 allergen suggesting that they are conformational in nature. The combined epitope mapping strategies demonstrate that Phl p 5 contains multiple conformational epitopes recognized by monoclonal human IgE antibodies.
Allergic subjects are not only likely to encounter different group 5 allergens from timothy but also to encounter such allergens from other grasses. Indeed, IgE antibodies from allergic patients are known to cross-react to allergens found in several different grasses 32 but the cross-reacting behaviour of human monoclonal IgE antibodies targeting the different epitopes of these allergens is not well established. Past studies of the Fab of clone 5 indicated that it reacted strongly not only to components of timothy grass (Phleum pratense) pollen but also to pollen extracts of other grasses 25,33. We now assessed the binding of seven of our IgE-derived human antibody fragments, to pollen extracts of several grass species. Each of the tested Phl p 5-specific binders cross-reacted with extracts of pollen derived from six different grasses although the precise levels differed (Fig.4). ScFv 4.4. and 4.13, both of which recognize the same epitope cluster in the C-terminal domain of the allergen, showed lower reactivity to barley pollen extract.
Figure 4.
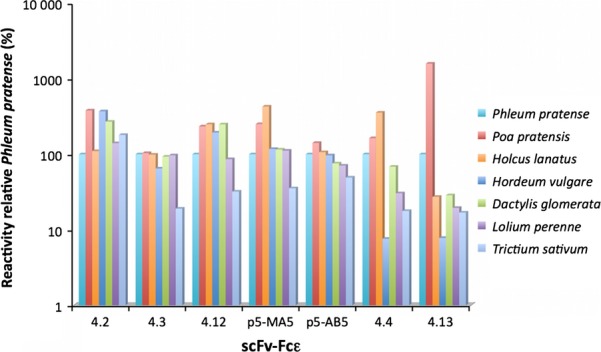
Relative binding of seven human scFv–Fcε (x-axis) to pollen extracts from different grasses as percentage of reactivity to timothy grass pollen extract (y-axis).
High-density recognition of Phl p 5 by allergic patients IgE
To obtain further data regarding the density of IgE binding to the Phl p 5 allergen, we used a mix of four Phl p 5-specific scFv–Fcγ2 derivatives (4.3, 4.13, p5-AB5 and clone 5) to determine the extent of inhibition of binding of allergic patients' polyclonal IgE to the allergen. The scFv-bound Phl p 5 with affinities in the range 3.9 × 106–5.3 × 109 m−1 (Table S1). Despite the fact that these scFv–Fcγ2 derivatives were present in excess in relation to the amount of immobilized allergen, bound to four different epitopes on Phl p 5 and each of them was more than threefold as big as the Phl p 5 allergen itself, although substantial, a yet only partial inhibition of allergic patients' serum IgE binding was achieved (52–76% inhibition; mean inhibition: 65%) by their presence (Table3). A further 2.5-fold increase in scFv–Fcγ2 inhibitory concentration only slightly increased the blocking capacity by a few percent (data not shown). By contrast, an 87% mean inhibition (range 80–92%) of patients IgE binding was obtained with polyclonal Phl p 5-specific IgG antibodies (Table3). Thus, it appears that allergic patients contain IgE antibodies that can bind to Phl p 5 even in the presence of the four monoclonal IgE antibodies suggesting that high-density recognition of Phl p 5 by allergic patients' IgE is achieved.
Table 3.
Inhibition of allergic patients' IgE binding to Phl p 5 by allergen-specific recombinant antibodies or antisera
| Patient sample | Remaining binding (%) in the presence of antibody preparation | |||
|---|---|---|---|---|
| Phl p 5-specific scFv–CHγ2* | Bet v 1–specific IgG | Phl p 5-specific rabbit antiserum | Bet v 1-specific rabbit antiserum | |
| 1 | 48 | 116 | 12 | 106 |
| 2 | 41 | 103 | 14 | 102 |
| 3 | 30 | 112 | 11 | 110 |
| 4 | 24 | 193 | 8 | 101 |
| 5 | 36 | 116 | 20 | 116 |
| 6 | 28 | 105 | 13 | 105 |
| Average ± SD | 35 ± 9 | 108 ± 9 | 13 ± 4 | 107 ± 6 |
4.3, 4.13, p5-AB5 and clone 5.
To further investigate this aspect, allergen-antibody complexes were established by incubation of Phl p 5.0101 with one (clone 5) or four (4.3, 4.13, p5-AB5 and clone 5) scFv–Fcγ2. Such complexes immobilized onto a surface were demonstrated to still expose a substantial amount of serum IgE-binding epitopes (Figure S3). Indeed, the presence of four instead of one scFv–Fcγ2 detecting different epitopes only partly (≤38%) reduced the IgE-binding capacity of allergen immobilized this way. Altogether, two different assay systems demonstrated that epitopes beyond the four epitopes defined by our set of recombinant scFv are present in the relatively small allergen Phl p 5.
Discussion
Group 5 allergens such as the timothy grass pollen allergen, Phl p 5, are present in a variety of grass species and have been implicated in the induction of severe asthma attacks 16. They have been recognized as highly allergenic proteins that are targeted by IgE early during the development of grass pollen allergy 34 and that account for high percentage of grass pollen-specific IgE 15,17,25. Recombinant Phl p 5 is a key molecule used for diagnosis of grass pollen allergy 35, and it is incorporated as a component of new vaccines developed for specific immunotherapy of grass pollen allergy 36. In this investigation, we have made use of the largest available panel of human monoclonal allergen-specific IgE described to date to analyze the molecular interaction of these antibodies and the Phl p 5 allergen. This set of binders was derived by combinatorial library technology and relies on the important role of the heavy chain variable domain in determining allergen specificity 7,37,38. It therefore does not capture all aspects of the IgE that existed in vivo. For instance, the affinity of the interactions may be influenced by the molecular format (such as the use of the scFv format or the scFv–Fc fusion protein construct) or the precise light chain variable domain incorporated into the antibody fragment as allowed by the conditions employed during phage display selection. Despite the technological shortcomings, this type of binders likely represents a close match to allergic patients' IgE.
Available IgE specific for Phl p 5 represent a diverse, somatically hypermutated repertoire in terms of immunoglobulin gene usage with an origin in several different heavy chain variable domain subgroups. The fact that all but one (clone 5) of these antibodies were derived from the repertoire of a single allergic individual demonstrates that despite the oligoclonality of IgE responses in general 4,12,39, the group 5 allergen is targeted by a quite complex repertoire of IgE. Such complexity is not always observed in the IgE responses to allergens. The group 2 grass pollen allergen represents a very much different target for IgE as such antibodies tend to be structurally very similar and to primarily recognize one set of epitopes 40,41. This finding is in agreement with the fact that Phl p 5 accounts for a high percentage of the grass pollen-specific polyclonal IgE response in patients, whereas IgE levels specific for group 2 allergens are low 42. Phl p 1 on the other hand is a highly allergenic protein targeted by IgE primarily specific for one of its two domains, the C-terminal module 43. IgE specific for this domain also recognize at least two non-overlapping epitopes within this small folded structure 18. In conclusion, different allergens (even from the same allergen source) may be targeted by IgE populations of very different complexity.
The humoral immune response against Phl p 5 is not only diverse in terms of antibody variable domain sequence but also by the fact that it detects several non-overlapping epitopes distributed on both domains of the allergen. Epitope mapping has in the past identified IgE epitopes, some of which overlap each other, in the N-terminal domain of Phl p 5 12,25,31. The present investigation further dissects the location of these epitopes in the N-terminal domain and defines additional epitopes in the C-terminal domain of the allergen. Some reactivities following blocking (Table1) or unexpected interference in sandwich assay binding capability (Figure S1) are not completely congruent with the epitope allocation; however, four epitopes (recognized by e.g. 4.3, clone 5, 4.13 and p5-AB5, respectively) stand out as independent. Epitope allocations are further supported by differences in blocking by Phl p 5 peptide-specific antisera and/or differential sensitivity to mutation of the allergen. It is thus obvious that despite the small size of group 5 pollen allergen domains in comparison with antibodies (Fig.2d), Phl p 5 offers accommodation for several antibodies at largely independent sites. As even the presence of a mix of recombinant antibodies targeting the four epitopes on Phl p 5 was unable to completely block the reactivity of serum IgE to Phl p 5, it furthermore appears likely that additional epitopes targeted by patients' IgE exist on Phl p 5 for which there are as yet no human monoclonal IgE representatives.
The N-terminal part of mature Phl p 5 (residues 26-58) is not required for binding of any of the herein-described human recombinant IgE as they recognize either the N- 12 or the C-terminal (this study) domain of the allergen. Yet, polyclonal antibodies specific for this protein segment are particularly effective in blocking serum IgE binding to intact allergen 30. That polyclonal antiserum also in part blocked the binding of recombinant human IgE to allergen (Fig.2e). Either this N-terminal segment resides in proximity to (or even partly contribute to) epitopes mainly defined by residues in other domains within the folded structure, resulting in blocking through steric hindrance, or epitopes within this sequence are, due to sequence similarity, also found elsewhere in Phl p 5 (Figure S4). It may also be a target of at least part of the human IgE reactivity outlined above that has not yet been captured into the available set of recombinant human IgE. Altogether, this N-terminal sequence represents a prime candidate for future studies of its contribution to Phl p 5-specific IgE reactivity.
The complexity of the response against an allergen is likely to enhance its potential for initiation of the early events of an allergic reaction through efficient cross-linking of FcεR-bound IgE on mast cells and basophils 7. We envisage that the potential for IgE binding to numerous independent epitopes on an allergen such as Phl p 5 renders it potent in terms of allergenic potential as has been described in studies of antibodies targeting the major timothy grass pollen allergen 9 and a mite allergen 7. The high-density IgE recognition of Phl p 5 described in our study is also in agreement with findings made for the major birch pollen allergen Bet v 1 which likewise can accommodate the binding of several different IgE antibodies on a relatively small surface 44. It becomes evident that any specific immunotherapy strategy targeting Phl p 5 will have to address the many epitopes of group 5 allergens. Development of safe hypoallergen variants of this allergen may thus be challenging, as we need to account for these many epitopes. Nevertheless, the present investigation and past studies 25,31 have identified residues important for binding of some human IgE to Phl p 5. We foresee that these residues or residues located in their proximity, or hypoallergenic variants described by Wald et al. 45, might serve as candidates for a future evidence-based mutagenesis approach 18 to iteratively develop improved hypoallergenic variants of Phl p 5.
Pollen contains multiple isoallergens and isoforms of major allergens such as the group 5 allergens 28 and several grasses produce similar cross-reacting allergens 32. Our results identify that monoclonal IgE with reactivity towards the Phl p 5.0101 isoallergen display broad cross-reactivity to group 5 allergens of a large variety of grass species. These findings and the efficiency of Phl p 5.0101 in an allergen cocktail to absorb grass pollen-specific IgE 11,15 provide evidence that this particular isoallergen is a major candidate representing group 5 allergens to be incorporated into a grass pollen allergy vaccine.
In conclusion, we provide evidence that the presence of a large number of independent IgE epitopes and their tight packing on an allergen is likely an important factor contributing to the high allergenic potency of an allergen.
Acknowledgments
The authors are grateful to Dr. Kirsten Gehlhar (Ruhr-Universität, Bochum, Germany) for her donation of recombinant versions of the C-terminal domain of Phl p 5.0201, to Dr. Jonas Lidholm (Thermofisher Scientific AB, Uppsala, Sweden) for his donation of Phl p 5.0201 and to Dr. Franck Perez (Institut Curie, CNRS UMR144, Paris, France) for his donation of the pFUSE-hIgG-Fc2 vector. This study was supported by grants from the Swedish Research Council (grant numbers: 521-2008-3614 and 521-2011-3282), by Alfred Österlunds stiftelse and in part by grants P23318-B11, F4607 and F4605 of the Austrian Science Fund (FWF). SR was supported by a EU ERASMUS grant.
Conflict of interest
M.L. and M.O. are co-inventors of a patent application in part related to this study. M.O. received recombinant allergen from Thermo Fisher Scientific AB to carry out some of these studies and grant funding from the Swedish research Council and Alfred Österlunds stiftelse. R.V. has received research grant funding from the EU, the Austrian Science Fund (FWF), Biomay AG, Vienna, Austria and Phadia/Thermofisher, Uppsala, Sweden and serves as a consultant for Biomay and Thermofisher. S.F. and N.N. have received grant funding from the Austrian Science Fund. M.F.T. has received grant funding from the Christian Doppler Research Association. The other authors of this investigation have no conflict of interests to declare.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Data S1. Materials and methods.
Table S1. Genes encoding and summary of properties of Phl p 5-specific IgEa.
Figure S1. Sandwich immunoassay detecting the ability of a second antibody (scFv-Fcε) to bind allergen caught on an immobilized antibody (scFv-Fcγ2).
Figure S2. Blocking of the binding of scFv P5b-7 (identical to p5-MA5) (top) and P5b-31 (a sequence variant of 4.12 that uses a light chain different from that found in 4.12) (bottom) (both selected by phage display for binding to Phl p 5.0201 (data not shown)), displayed on phage, to immobilized Phl p 5.0201 by pre-incubation of the displayed antibody fragment with recombinant allergens or recombinant C-terminal domain of Phl p 5.0201 or mutated variants thereof (1: K149A; 2: K160A; 3: K166A; 4: K190A; 5: K193A; 6: K204A; 7: K214A; 8: K238A; 9: K248A; 10: K283A).
Figure S3. Binding of serum IgE from 12 grass pollen allergic donors to complexes of Phl p 5.0101 and one (clone 5; red) or four (clone 5, 4.3, 4.13, and p5-AB5; blue) scFv-Fcγ2 fusion proteins that had been caught on immobilized anti-human IgG.
Figure S4. Sequence similarities (light grey shading) and identities (dark grey shading) between the N-terminal sequence of Phl p 5.0101, and the N-terminal parts of the N- and C-terminal domains of Phl p 5.0101.
References
- Gould HJ, Sutton BJ. IgE in allergy and asthma today. Nat Rev Immunol. 2008;8:205–17. doi: 10.1038/nri2273. [DOI] [PubMed] [Google Scholar]
- Valenta R, Ferreira F, Focke-Tejkl M, et al. From allergen genes to allergy vaccines. Annu Rev Immunol. 2010;28:211–41. doi: 10.1146/annurev-immunol-030409-101218. [DOI] [PubMed] [Google Scholar]
- Steinberger P, Kraft D, Valenta R. Construction of a combinatorial IgE library from an allergic patient. Isolation and characterization of human IgE Fabs with specificity for the major timothy grass pollen allergen, Phl p 5. J Biol Chem. 1996;271:10967–72. doi: 10.1074/jbc.271.18.10967. [DOI] [PubMed] [Google Scholar]
- Gadermaier E, Levin M, Flicker S, Ohlin M. The human IgE repertoire. Int Arch Allergy Immunol. 2014;163:77–91. doi: 10.1159/000355947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemi M, Jylhä S, Laukkanen ML, et al. Molecular interactions between a recombinant IgE antibody and the beta-lactoglobulin allergen. Structure. 2007;15:1413–21. doi: 10.1016/j.str.2007.09.012. [DOI] [PubMed] [Google Scholar]
- Padavattan S, Flicker S, Schirmer T, et al. High-affinity IgE recognition of a conformational epitope of the major respiratory allergen Phl p 2 as revealed by X-ray crystallography. J Immunol. 2009;182:2141–51. doi: 10.4049/jimmunol.0803018. [DOI] [PubMed] [Google Scholar]
- Christensen LH, Holm J, Lund G, Riise E, Lund K. Several distinct properties of the IgE repertoire determine effector cell degranulation in response to allergen challenge. J Allergy Clin Immunol. 2008;122:298–304. doi: 10.1016/j.jaci.2008.05.026. [DOI] [PubMed] [Google Scholar]
- Christensen LH, Riise E, Bang L, Zhang C, Lund K. Isoallergen variations contribute to the overall complexity of effector cell degranulation: effect mediated through differentiated IgE affinity. J Immunol. 2010;184:4966–72. doi: 10.4049/jimmunol.0904038. [DOI] [PubMed] [Google Scholar]
- Gieras A, Focke-Tejkl M, Ball T, et al. Molecular determinants of allergen-induced effector cell degranulation. J Allergy Clin Immunol. 2007;119:384–90. doi: 10.1016/j.jaci.2006.09.034. [DOI] [PubMed] [Google Scholar]
- Aalberse RC, Crameri R. IgE-binding epitopes: a reappraisal. Allergy. 2011;66:1261–74. doi: 10.1111/j.1398-9995.2011.02656.x. [DOI] [PubMed] [Google Scholar]
- Van Milligen FJ, Craig S, Rogers BL, Van Swieten P, Aalberse RC. Differences between specificities of IgE and IgG4 antibodies: studies using recombinant chain 1 and chain 2 of the major cat allergen Felis domesticus (Fel d) I. Clin Exp Allergy. 1995;25:247–51. doi: 10.1111/j.1365-2222.1995.tb01036.x. [DOI] [PubMed] [Google Scholar]
- Andréasson U, Flicker S, Lindstedt M, et al. The human IgE-encoding transcriptome to assess antibody repertoires and repertoire evolution. J Mol Biol. 2006;362:212–27. doi: 10.1016/j.jmb.2006.06.062. [DOI] [PubMed] [Google Scholar]
- Persson H, Karbalaei Sadegh M, Greiff L, Ohlin M. Delineating the specificity of an IgE-encoding transcriptome. J Allergy Clin Immunol. 2007;120:1186–92. doi: 10.1016/j.jaci.2007.06.041. [DOI] [PubMed] [Google Scholar]
- Persson J, Augustsson P, Laurell T, Ohlin M. Acoustic microfluidic chip technology to facilitate automation of phage display selection. FEBS J. 2008;275:5657–66. doi: 10.1111/j.1742-4658.2008.06691.x. [DOI] [PubMed] [Google Scholar]
- Vrtala S, Sperr WR, Reimitzer I, et al. cDNA cloning of a major allergen from timothy grass (Phleum pratense) pollen; characterization of the recombinant Phl pV allergen. J Immunol. 1993;151:4773–8. [PubMed] [Google Scholar]
- Suphioglu C, Singh MB, Taylor P, et al. Mechanism of grass-pollen-induced asthma. Lancet. 1992;339:569–72. doi: 10.1016/0140-6736(92)90864-y. [DOI] [PubMed] [Google Scholar]
- Westritschnig K, Horak F, Swoboda I, et al. Different allergenic activity of grass pollen allergens revealed by skin testing. Eur J Clin Invest. 2008;38:260–7. doi: 10.1111/j.1365-2362.2008.01938.x. [DOI] [PubMed] [Google Scholar]
- Levin M, Rydnert F, Källström E, et al. Phl p 1-specific human monoclonal IgE and design of a hypoallergenic group 1 grass pollen allergen fragment. J Immunol. 2013;191:551–60. doi: 10.4049/jimmunol.1202051. [DOI] [PubMed] [Google Scholar]
- Braren I, Blank S, Seismann H, et al. Generation of human monoclonal allergen-specific IgE and IgG antibodies from synthetic antibody libraries. Clin Chem. 2007;53:837–44. doi: 10.1373/clinchem.2006.078360. [DOI] [PubMed] [Google Scholar]
- Gehlhar K, Rajashankar KR, Hofmann E, et al. Lysine as a critical amino acid for IgE bBinding in Phl p 5b C terminus. Int Arch Allergy Immunol. 2006;140:285–94. doi: 10.1159/000093706. [DOI] [PubMed] [Google Scholar]
- Venet S, Ravn U, Buatois V, et al. Transferring the characteristics of naturally occurring and biased antibody repertoires to human antibody libraries by trapping CDRH3 sequences. PLoS ONE. 2012;7:e43471. doi: 10.1371/journal.pone.0043471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M, Ohlin M. Inconclusive evidence for or against positive antigen selection in the shaping of human IgE repertoires – a call for new approaches. Int Arch Allergy Immunol. 2013;161:122–6. doi: 10.1159/000345421. [DOI] [PubMed] [Google Scholar]
- Maglio O, Saldanha JW, Vrtala S, Spitzauer S, Valenta R, Pastore A. A major IgE epitope-containing grass pollen allergen domain from Phl p 5 folds as a four-helix bundle. Protein Eng. 2002;15:635–42. doi: 10.1093/protein/15.8.635. [DOI] [PubMed] [Google Scholar]
- Rajashankar K, Bufe A, Weber W, Eschenburg S, Lindner B, Betzel C. Structure of the functional domain of the major grass-pollen allergen Phlp 5b. Acta Crystallogr D Biol Crystallogr. 2002;58:1175–81. doi: 10.1107/s0907444902007254. [DOI] [PubMed] [Google Scholar]
- Flicker S, Vrtala S, Steinberger P, et al. A human monoclonal IgE antibody defines a highly allergenic fragment of the major timothy grass pollen allergen, Phl p 5: molecular, immunological, and structural characterization of the epitope-containing domain. J Immunol. 2000;165:3849–59. doi: 10.4049/jimmunol.165.7.3849. [DOI] [PubMed] [Google Scholar]
- Bufe A, Becker WM, Schramm G, Petersen A, Mamat U, Schlaak M. Major allergen Phl p Va (timothy grass) bears at least two different IgE-reactive epitopes. J Allergy Clin Immunol. 1994;94:173–81. doi: 10.1016/0091-6749(94)90037-x. [DOI] [PubMed] [Google Scholar]
- Bufe A, Schramm G, Keown MB, Schlaak M, Becker WM. Major allergen Phl p Vb in timothy grass is a novel pollen RNase. FEBS Lett. 1995;363:6–12. doi: 10.1016/0014-5793(95)00259-c. [DOI] [PubMed] [Google Scholar]
- Chabre H, Gouyon B, Huet A, et al. Molecular variability of group 1 and 5 grass pollen allergens between Pooideae species: implications for immunotherapy. Clin Exp Allergy. 2010;40:505–19. doi: 10.1111/j.1365-2222.2009.03380.x. [DOI] [PubMed] [Google Scholar]
- Andersson K, Lidholm J. Characteristics and immunobiology of grass pollen allergens. Int Arch Allergy Immunol. 2003;130:87–107. doi: 10.1159/000069013. [DOI] [PubMed] [Google Scholar]
- Focke-Tejkl M, Campana R, Reininger R, et al. Dissection of the IgE and T-cell recognition of the major group 5 grass pollen allergen Phl p 5. J Allergy Clin Immunol. 2014;33:836–45. doi: 10.1016/j.jaci.2013.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker J, Diethers A, Etzold S, Seismann H, Michel Y, Plum M, et al. Generation and epitope analysis of human monoclonal antibody isotypes with specificity for the Timothy grass major allergen Phl p 5a. Mol Immunol. 2011;48:1236–44. doi: 10.1016/j.molimm.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Bullimore A, Batten T, Hewings S, Fischer von Weikersthal-Drachenberg KJ, Skinner M. Cross-reactivity in brasses: biochemical attributes define exemplar relevance. World Allergy Organ J. 2012;5:111–9. doi: 10.1097/WOX.0b013e31826a10cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madritsch C, Flicker S, Scheiblhofer S, et al. Recombinant monoclonal human immunoglobulin E to investigate the allergenic activity of major grass pollen allergen Phl p 5. Clin Exp Allergy. 2011;41:270–80. doi: 10.1111/j.1365-2222.2010.03666.x. [DOI] [PubMed] [Google Scholar]
- Hatzler L, Panetta V, Lau S, et al. Molecular spreading and predictive value of preclinical IgE response to Phleum pratense in children with hay fever. J Allergy Clin Immunol. 2012;130:894–901.e5. doi: 10.1016/j.jaci.2012.05.053. [DOI] [PubMed] [Google Scholar]
- Kazemi-Shirazi L, Niederberger V, Linhart B, Lidholm J, Kraft D, Valenta R. Recombinant marker allergens: diagnostic gatekeepers for the treatment of allergy. Int Arch Allergy Immunol. 2002;127:259–68. doi: 10.1159/000057742. [DOI] [PubMed] [Google Scholar]
- Jutel M, Jaeger L, Suck R, Meyer H, Fiebig H, Cromwell O. Allergen-specific immunotherapy with recombinant grass pollen allergens. J Allergy Clin Immunol. 2005;116:608–13. doi: 10.1016/j.jaci.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Laffer S, Steinberger P, Kraft D, Valenta R. Promiscuous use of light chains by human IgE antibodies specific for three major grass pollen allergens. Int Arch Allergy Immunol. 2001;124:29–30. doi: 10.1159/000053660. [DOI] [PubMed] [Google Scholar]
- Gadermaier E, Flicker S, Lupinek C, Steinberger P, Valenta R. Determination of allergen specificity by heavy chains in grass pollen allergen-specific IgE antibodies. J Allergy Clin Immunol. 2013;131:1185–93. doi: 10.1016/j.jaci.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerzel S, Rogosch T, Struecker B, Maier RF, Zemlin M. IgE transcripts in the circulation of allergic children reflect a classical antigen-driven B cell response and not a superantigen-like activation. J Immunol. 2010;185:2253–60. doi: 10.4049/jimmunol.0902942. [DOI] [PubMed] [Google Scholar]
- Flicker S, Steinberger P, Norderhaug L, et al. Conversion of grass pollen allergen-specific human IgE into a protective IgG(1) antibody. Eur J Immunol. 2002;32:2156–62. doi: 10.1002/1521-4141(200208)32:8<2156::AID-IMMU2156>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Persson H, Flicker S, Karbalaei Sadegh M, Valenta R, Greiff L, Ohlin M. A common idiotype in IgE and its relation to recognition of the grass pollen allergen Phl p 2. Mol Immunol. 2008;45:2715–20. doi: 10.1016/j.molimm.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Niederberger V, Stübner P, Spitzauer S, et al. Skin test results but not serology reflect immediate type respiratory sensitivity: a study performed with recombinant allergen molecules. J Invest Dermatol. 2001;117:848–51. doi: 10.1046/j.0022-202x.2001.01470.x. [DOI] [PubMed] [Google Scholar]
- Flicker S, Steinberger P, Ball T, et al. Spatial clustering of the IgE epitopes on the major timothy grass pollen allergen Phl p 1: importance for allergenic activity. J Allergy Clin Immunol. 2006;117:1336–43. doi: 10.1016/j.jaci.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Gieras A, Cejka P, Blatt K, et al. Mapping of conformational IgE epitopes with peptide-specific monoclonal antibodies reveals simultaneous binding of different IgE antibodies to a surface patch on the major birch pollen allergen, Bet v 1. J Immunol. 2011;186:5333–544. doi: 10.4049/jimmunol.1000804. [DOI] [PubMed] [Google Scholar]
- Wald M, Kahlert H, Reese G, et al. Hypoallergenic mutants of the Timothy grass pollen allergen Phl p 5 generated by proline mutations. Int Arch Allergy Immunol. 2012;159:130–42. doi: 10.1159/000336651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Materials and methods.
Table S1. Genes encoding and summary of properties of Phl p 5-specific IgEa.
Figure S1. Sandwich immunoassay detecting the ability of a second antibody (scFv-Fcε) to bind allergen caught on an immobilized antibody (scFv-Fcγ2).
Figure S2. Blocking of the binding of scFv P5b-7 (identical to p5-MA5) (top) and P5b-31 (a sequence variant of 4.12 that uses a light chain different from that found in 4.12) (bottom) (both selected by phage display for binding to Phl p 5.0201 (data not shown)), displayed on phage, to immobilized Phl p 5.0201 by pre-incubation of the displayed antibody fragment with recombinant allergens or recombinant C-terminal domain of Phl p 5.0201 or mutated variants thereof (1: K149A; 2: K160A; 3: K166A; 4: K190A; 5: K193A; 6: K204A; 7: K214A; 8: K238A; 9: K248A; 10: K283A).
Figure S3. Binding of serum IgE from 12 grass pollen allergic donors to complexes of Phl p 5.0101 and one (clone 5; red) or four (clone 5, 4.3, 4.13, and p5-AB5; blue) scFv-Fcγ2 fusion proteins that had been caught on immobilized anti-human IgG.
Figure S4. Sequence similarities (light grey shading) and identities (dark grey shading) between the N-terminal sequence of Phl p 5.0101, and the N-terminal parts of the N- and C-terminal domains of Phl p 5.0101.


