Abstract
Six coryneforms isolated from blood and dialysate fluid were phenotypically similar to Brevibacterium casei, but 16S rRNA gene sequencing and DNA-DNA hybridization indicate that they belong to a new species for which the name Brevibacterium sanguinis is proposed.
Brevibacteria are opportunistic pathogens, and several species have been isolated from clinical samples (6). Brevibacterium casei is reported as the most frequent human isolate (5), but other species, such as B. epidermidis, B. otitidis, B. mcbrellneri, and B. paucivorans, are also encountered (2, 12, 14, 17). A new species, B. lutescens, was recently described (18), but the species name was later corrected to B. luteolum (4). On the basis of genetic studies, six strains phenotypically resembling B. casei were found in this study to belong to a new Brevibacterium species for which the name Brevibacterium sanguinis is proposed.
Origin of the strains and clinical features.
(i) Strains CF51, CF63, CF79, and CF91 were isolated from blood cultures of a patient with human immunodeficiency virus (HIV) during a 22-month period in November 1997, June 1998, March 1999, and August 1999, respectively. The patient had ambulatory treatment for his HIV infection, and the organisms were isolated during recurrent febrile episodes. Pulsed-field gel electrophoresis (3) performed on the four isolates resulted in identical profiles different from that obtained with a similar strain from another patient. Therefore, the four isolates were considered a single strain and only CF63 was selected for further studies. (ii) Strain CF52 was isolated on two consecutive days from several blood bottles from a young female neurological patient with a Hickman catheter. (iii) Strain CF105 (GH443) was isolated by G. Haase from the blood of a 33-month-old boy with acute myeloid leukemia. (iv) Strain CF127 was isolated from one blood bottle from a febrile hospitalized child. Only one blood culture was performed. (v) Strain CF110 was isolated from blood, but no other information was available. (vi) Strain DSM 9657 was identified by G. Funke and A. Carlotti as B. casei, isolated four times from peritoneal dialysate, and later deposited into the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) (5).
Cellular fatty acids (CFA) of the strains, determined as described previously (16), were of the branched type, mainly anteiso 17:0 and anteiso 15:0, accounting for more than 75% of the total CFAs. Peptidoglycan analysis of strain CF63 was carried out at the DSMZ by N. Weiss according to the method of Schleifer and Kandler (15) and showed an A1γ type, with the presence of meso-diaminopimelic acid as the diamino acid.
The 16S rRNA gene (rDNA) sequence determinations were conducted as described previously (18). The 16S rDNA sequence of strain CF63 displayed the highest similarity (98.8%) to the sequence of an unidentified bacterium (strain GH443) that had been submitted to GenBank in 1993 by A. Podbielski under accession number X76703. Its similarity to the sequence of B. casei DSM 20657T was only 97.7% (Table 1). When a new sequence determination of GH443 was performed at our laboratory along with that of CF63, a similarity of 99.9% was obtained for the two strains. The 16S rDNA sequence homologies of CF63 to the four other study strains were as follows: for CF52, 99.4%; for CF110, 99.8%; for CF127, 99.5%; and for DSM 9657, 99.9%.
TABLE 1.
16S rDNA sequence similarity between B. sanguinis (CF63T) and other Brevibacterium species
| Species | Accession no. | 16S rRNA similarity to CF63T (%) |
|---|---|---|
| Brevibacterium avium | Y17962 | 97.2 |
| Brevibacterium luteolum | AJ488509 | 92.7 |
| Brevibacterium otitidis | X93573 | 92.3 |
| Brevibacterium mcbrellneri | X93594 | 93.3 |
| Brevibacterium paucivorans | AJ231463 | 92.3 |
| Brevibacterium casei | AJ251418 | 97.7 |
| Brevibacterium epidermidis | X76565 | 96.7 |
| Brevibacterium iodinum | X83813 | 97.1 |
| Brevibacterium linens | X77451 | 96.7 |
DNA-DNA hybridization was carried out at DSMZ by P. Schumann as described previously (18). The DNA homologies of strains CF63 and DSM 9657 to B. casei DSM 20657T were 60.9 and 50.8%, respectively. When the study strains were compared to strain CF63, DNA homologies were 88.2% for CF52, 84.8% for CF105 (GH443), 98.5% for CF110, 87.4% for CF127, and 92.1% for DSM 9657. This suggests that the six strains belong to a single species distinct from B. casei. The DNA G+C content of strain CF63 was 69.9 mol%, as determined at DSMZ.
Most biochemical tests were carried out as outlined previously (6, 9, 17, 18). API 32 GN, API 50 CH, and API Biotype 100 strips (Biomerieux, Marcy l'Etoile, France) were used with AUX medium, incubated at 37°C, and read after 2 and 4 days. API ZYM strips (Biomerieux) were used according to the manufacturer's instructions. API CORYNE strips (Biomerieux) were incubated at 37°C and read after 1 and 2 days. Susceptibility to thallium acetate was tested, since this compound is known to inhibit several bacterial species and has been used as a selective agent (8). For this purpose paper disks were impregnated with 10 μl of a 0.5% (wt/vol) thallium acetate (Sigma-Aldrich, St. Louis, Mo.) solution and put onto blood agar. Assimilation-alkalinization of organic compounds was performed on Simmons agar base, replacing citrate by 0.2% (wt/vol) of the substrate according to the method of Martin et al. (11).
The six strains were nonmotile coryneform rods. They grew aerobically on blood agar at 37°C, and the colonies were grayish white and somewhat sticky. Catalase gave positive test results, and oxidase gave negative test results. The metabolism was oxidative, there was proteolytic but no saccharolytic activity, and production of methanethiol was positive. The code generated by the API CORYNE system was 6112004, which was similar to the code generated by B. casei. By conventional methods the phenotypic profile of the six strains was also closely related to that of B. casei (5); however, some differences allowed the study strains to be distinguished from the latter species. When paper disks containing 50 μg of thallium acetate were used on blood agar, B. casei was resistant, growing at the edge of the disk, whereas the new strains exhibited an inhibition zone of more than 25 mm. In the API Biotype 100 system, quinate was assimilated by the six strains but not by B. casei. Assimilation-alkalinization of quinate was also achieved by using Simmons agar base containing 0.2% (wt/vol) quinate (Sigma-Aldrich). On Simmons base containing 0.2% (wt/vol) tyramine hydrochloride (Sigma-Aldrich), similarly, this substrate was utilized by all the study strains but one (CF127) whereas none of the B. casei strains grew. As with other amines like putrescine or cadaverine, acidification, and not alkalinization, of the medium was observed when tyramine was assimilated. The main phenotypic characteristics that differentiate the new strains from Brevibacterium species encountered in humans are reported in Table 2.
TABLE 2.
Characteristics that differentiate B. sanguinis from other Brevibacterium species encountered in humans
| Test or characteristic | Value fora:
|
||||||
|---|---|---|---|---|---|---|---|
| B. sanguinis (n = 6) | B. casei (n = 11) | B. epidermidis (n = 3) | B. otitidis (n = 3) | B. luteolum (n = 4) | B. mcbrellneri (n = 2) | B. paucivorans (n = 11) | |
| Colonies | White-grayish, smooth or sticky | White-grayish, smooth | White-yellow, smooth | Yellowish, smooth | Yellowish, smooth | Grayish, dry, friable | Grayish, smooth or sticky |
| Growth at 20°C | + | + | + | − | + | − | − |
| Growth on NaCl (10%) | + | + | + | − | + | − | − |
| Thallium acetate | S | R | S | R | S | R | S |
| Hydrolysis of casein | + | + | + | + | + | + | − |
| Hydrolysis of gelatin | + | + | + | + | + | +/(+) | (+)w |
| Nitrate | −/+w | −/+ | +/− | − | − | − | − |
| Acid from 2,3-butylene glycol | − | − | − | − | − | + | − |
| Acid from phenyl acetate | + | + | − | − | + | + | + |
| Utilization on API 50 CH: | |||||||
| d-Arabinose | + | + | − | − | − | − | − |
| Mannitol | − | − | + | − | − | − | − |
| Gluconate | + | + | + | − | − | − | − |
| Utilization on Simmons base: | |||||||
| γ-Aminobutyrate | + | + | + | − | + | − | − |
| Quinate | + | − | − | − | − | − | − |
| Tyramine | +/− | − | − | − | − | − | − |
| α-Glucosidase | + | + | − | − | − | − | − |
| N-Acetyl-β-d-glucosaminidase (nitrophenyl compound) | +w | − | + | − | + | + | +/− |
| Pyrrolidone arylamidase | v | +/− | − | + | + | − | − |
| Pyrazinamidase | + | + | + | + | + | − | − |
+, positive; −, negative; (+), delayed +; +/−, most strains positive; −/+, most strains negative; superscript w, weak reaction; v, variable; S, susceptible; R, resistant.
Antibiotic susceptibility was tested on Mueller-Hinton blood agar by the paper disks diffusion method using the criteria for Staphylococcus of the NCCLS (13). All the strains were resistant to penicillin but susceptible to cephalothin, cefotaxime, gentamicin, erythromycin, ciprofloxacin, and vancomycin.
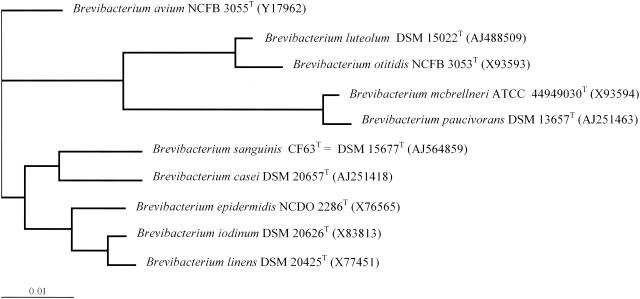
Phenotypic, taxonomic, and genetic properties suggest that the six strains belong to the genus Brevibacterium but constitute a new species related to B. casei for which the name B. sanguinis is proposed (Fig. 1).
FIG. 1.
Unrooted tree showing the phylogenetic position of B. sanguinis (CF63)T within the genus Brevibacterium. The bar represents one nucleotide substitution per 100 nucleotides.
All strains were isolated from sterile body sites, five from blood and one from dialysate fluid. Although not all blood isolates were fully documented, at least two of them were clinically relevant. The isolation of the same organism from the blood of an HIV patient four times within a 22-month period highlights the frequent recurrence of infections by coryneforms in compromised hosts. Recently, Microbacterium paraoxydans was isolated twice from a hematologic patient at an interval of several weeks (9). There are several reports of recurrent infections caused by Brevibacterium species, especially B. casei (1, 7, 10). Although an intravascular catheter was present in the patients whose cases are reported in these studies, its role as source of the organism was not proved in all cases. Our HIV patient had ambulatory treatment, and no intravascular catheter was used. Cultures of nose, mouth, and skin samples gave negative test results, and no persistent focus of the organism could be detected.
B. sanguinis is very similar to B. casei when tested by conventional methods or commercial identification sets. Their CFA compositions are similar and do not differentiate the two species. Therefore, it is likely that some strains previously reported as B. casei were misidentified. This is also suggested by the results seen with strain DSM 9657, which was deposited as B. casei but proved in this study to belong to the new species. A simple test, such as determination of susceptibility to thallium acetate, may help to differentiate the two species in the routine laboratory.
Description of B. sanguinis sp. nov.
Cells of B. sanguinis (san′-gui-nis, meaning blood, because most strains were isolated from blood) are gram-positive coryneform rods growing aerobically at 25 and 37°C. On blood agar, colonies are grayish white, opaque, somewhat sticky, and reach a diameter of 2 mm after 48 h at 37°C. Catalase gives positive test results, and oxidase gives negative test results. Gelatin, casein, tyrosine, and xanthine are hydrolyzed, but esculin is not. Methanethiol is produced. Strains are very susceptible to thallium acetate. Urease gives negative test results, pyrazinamidase gives positive test results, and nitrate reduction assays produce weak or negative results. Glucose and other carbohydrates are not acidified, but on API 50 CH and ID 32 GN strips glucose, d-arabinose, maltose, glycerol, N-acetylglucosamine, l-fucose, myo-inositol, sucrose, turanose, trehalose, and gluconate are assimilated. With API ZYM system tests, alkaline phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, acid phosphatase, phosphoamidase, and α-glucosidase give positive results. N-Acetyl-β-d-glucosaminidase gives negative results by this method, but results are slightly positive when the nitrophenyl conjugate (Rosco, Taastrup, Denmark) is used. Pyrrolidone arylamidase gives variable results. Quinate and γ-aminobutyrate are assimilated and alkalinized on Simmons mineral base. Tyramine is assimilated and acidified on Simmons mineral base by most strains. Acid is produced from phenyl acetate. CFAs are of the branched type, with anteiso C15:0 and anteiso C17:0 being the major components. The diamino acid of the peptidoglycan is meso-diaminopimelic acid.
The organisms are isolated from human clinical samples; the type strain is CF63T (= DSM 15677T = CCUG 47857T). The G+C content of the DNA is 69.9 mol%. Pyrrolidone arylamidase gives weakly positive results, and tyramine is utilized. The strain was isolated from the blood of an HIV patient.
Nucleotide sequence accession numbers.
The 16S rDNA sequences of the following strains were deposited in the EMBL (European Molecular Biology Laboratory) Nucleotide Sequence Database under the following accession numbers: for CF63, AJ564859; for CF52, AJ628351; and for DSM 9657, AJ628352. Two other strains with the following accession numbers have been deposited in the DSMZ and the Culture Collection, University of Göteborg (CCUG): CF105 (= GH443) = DSM 15679 = CCUG 47859 and CF52 = DSM 15678 = CCUG 47858.
Acknowledgments
We are grateful to P. Goffinet and J. Verhaegen for providing us with clinical data for some strains.
REFERENCES
- 1.Brazzola, P., R. Zbinden, C. Rudin, U. B. Schaad, and U. Heininger. 2000. Brevibacterium casei sepsis in an 18-year-old female with AIDS. J. Clin. Microbiol. 38:3513-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins, M. D., J. A. E. Farrow, M. Goodfellow, and D. E. Minnikin. 1983. Brevibacterium casei sp. nov. and Brevibacterium epidermidis sp. nov. Syst. Appl. Microbiol. 4:388-395. [DOI] [PubMed] [Google Scholar]
- 3.Correia, A., J. F. Martin, and J. M. Castro. 1994. Pulsed-field gel electrophoresis analysis of the genome of amino acid producing corynebacteria: chromosome sizes and diversity of restriction patterns. Microbiology 140:2841-2847. [DOI] [PubMed] [Google Scholar]
- 4.Euzéby, J. 2004. Notification list. Int. J. Syst. Bacteriol. 54:3-6. [Google Scholar]
- 5.Funke, G., and A. Carlotti. 1994. Differentiation of Brevibacterium spp. encountered in clinical specimens. J. Clin. Microbiol. 32:1729-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Funke, G., A. von Graevenitz, J. E. Clarridge III, and K. A. Bernard. 1997. Clinical microbiology of coryneform bacteria. Clin. Microbiol. Rev. 10:125-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaukoranta-Tolvanen, S. S., A. Sivonen, A. A. Kostiala, P. Hormila, and M. Vaara. 1995. Bacteremia caused by Brevibacterium species in an immunocompromised patient. Eur. J. Clin. Microbiol. Infect. Dis. 14:801-804. [DOI] [PubMed] [Google Scholar]
- 8.Knisely, R. F. 1966. Selective medium for Bacillus anthracis. J. Bacteriol. 92:784-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laffineur, K., V. Avesani, G. Cornu, J. Charlier, M. Janssens, G. Wauters, and M. Delmée. 2003. Bacteremia due to a novel Microbacterium species in a patient with leukemia and description of Microbacterium paraoxydans sp. nov. J. Clin. Microbiol. 41:2242-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lina, B., A. Carlotti, V. Lesaint, Y. Devaux, J. Freney, and J. Fleurette. 1994. Persistent bacteremia due to Brevibacterium species in an immunocompromised patient. Clin. Infect. Dis. 18:487-488. [DOI] [PubMed] [Google Scholar]
- 11.Martin, R., P. S. Riley, D. G. Hollis, R. E. Weaver, and M. I. Krichevsky. 1981. Characterization of some groups of gram-negative nonfermentative bacteria by the carbon source alkalinization technique. J. Clin. Microbiol. 14:39-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McBride, M. E., K. M. Ellner, H. S. Black, J. E. Clarridge, and J. E. Wolf. 1993. A new Brevibacterium sp. isolated from infected genital hair of patients with white piedra. J. Med. Microbiol. 39:255-261. [DOI] [PubMed] [Google Scholar]
- 13.NCCLS. 2002. Zone diameter interpretative standards and equivalent minimal inhibitory concentration (MIC) breakpoints for Staphylococcus spp. NCCLS document M100-S12 (M2-A7 disk diffusion). National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 14.Pascual, C., M. D. Collins, G. Funke, and D. G. Pitcher. 1996. Phenotypic and genotypic characterisation of two Brevibacterium strains from the human ear: description of Brevibacterium otitidis sp. nov. Med. Microbiol. Lett. 5:113-123. [Google Scholar]
- 15.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wauters, G., A. Driessen, E. Ageron, M. Janssens, and P. A. D. Grimont. 1996. Propionic acid-producing strains previously designated as Corynebacterium xerosis, C. minutissimum, C. striatum, and CDC group I2 and group F2 coryneforms belong to the species Corynebacterium amycolatum. Int. J. Syst. Bacteriol. 46:653-657. [Google Scholar]
- 17.Wauters, G., J. Charlier, M. Janssens, and M. Delmée. 2001. Description of Brevibacterium paucivorans sp. nov. isolated from human clinical specimens Int. J. Syst. Evol. Microbiol. 51:1703-1707. [DOI] [PubMed] [Google Scholar]
- 18.Wauters, G., V. Avesani, K. Laffineur, J. Charlier, M. Janssens, B. Van Bosterhaut, and M. Delmée. 2003. Brevibacterium lutescens sp. nov., from human and environmental samples. Int. J. Syst. E vol. Microbiol. 53:1321-1325. [DOI] [PubMed] [Google Scholar]