Summary
Genomic function is dictated by a combination of DNA sequence and the molecular mechanisms controlling access to genetic information. Access to DNA can be determined by the interpretation of covalent modifications that influence the packaging of DNA into chromatin, including DNA methylation and histone modifications. These modifications are believed to be forms of “epigenetic codes” that exist in discernable combinations that reflect cellular phenotype. Although DNA methylation is known to play important roles in gene regulation and genomic function, its contribution to the encoding of epigenetic information is just beginning to emerge. Here we discuss paradigms associated with the various components of DNA methylation/demethylation and recent advances in the understanding of its dynamic regulation in the genome, integrating these mechanisms into a framework to explain how DNA methylation could contribute to epigenetic codes.
Keywords: epigenetics, chromatin, DNA methylation, DNA demethylation
Introduction
Epigenetic mechanisms impart layers of information beyond what is hard-wired into the DNA sequence, allowing for differential expression of common genetic information (Box 1) [1]. At the chromatin level, epigenetic regulation involves the packaging of DNA and its regulated accessibility. This occurs mostly through differential covalent modification of DNA and histones in conjunction with the interpretation of those various modifications. In mammals, epigenetic modifications to DNA primarily involve methylation of cytosine at carbon five, while modifications to histones include the addition of an array of chemical groups to histone tails. In combination, these modifications in the context of chromatin have been described as “epigenetic codes” that underpin gene expression programs and cellular phenotypes [2].
There has been much study and speculation about how epigenetic marks are developmentally regulated in a cell type- and tissue-specific manner, as well as how such processes are dysregulated in human diseases. One emerging theme has been that, as a cell differentiates, epigenetic states are modified and then set, such that cellular phenotype can be maintained, often for a very long period of time throughout life. X-inactivation, silencing of repetitive elements, and genomic imprinting are good examples of how epigenetic states can be stably maintained through mechanisms involving DNA methylation. However, there are also notable exceptions. Early zygotic development, germ cell reprogramming, and cellular reprogramming (induced pluripotency) represent key examples of how epigenetic states can be dynamically regulated. In most somatic cells, the general notion is that different cell types are locked into distinct epigenetic states, coordinated by changes in DNA methylation and histone modifications. More recent evidence from biochemical studies of both DNA methylation and histone modifications indicates that epigenetic states may be more dynamic than previously thought, even in fully differentiated postmitotic cells, such as neurons [3–7]. DNA methylation and histone methylation, previously believed to be stable and long-lasting covalent modifications, are now known to be subject to enzymatic modification followed by removal, meaning they are reversible. Thus, the regulatory mechanisms of epigenetic modifications during normal cellular function and differentiation have now become the subject of changing paradigms.
Here we focus our discussion on the various epigenetic codes associated with DNA methylation, which exemplifies the changing paradigms of how epigenetic marks are written, read, and erased, hence dynamically encoding regulatory information in the genome. The integration of DNA methylation and its dynamic regulation into existing concepts of epigenetic encoding has occurred rapidly over the last several years. Given the widespread relevance of DNA methylation in defining cellular phenotypes, the integration of DNA methylation into a defined framework for epigenetic codes is essential for a full understanding of genomic function.
Sequence-based features direct the deposition of 5-methylcytosine
The role of 5-methylcytosine (5mC) as a component in the epigenetic regulation of mammalian systems was originally put forth by Holliday and Pugh [8] and Arthur Riggs [9] in 1975. They proposed that systems analogous to those known to exist in bacteria, namely sequence-specific DNA methyltransferases that can preferentially act on hemi-methylated palindromic sequences, might also act in mammalian systems to regulate gene expression (Fig. 1). They posited that the palindromic nature of methylated sequences provided a sequence-based template for the heritability of DNA methylation. Indeed, one of the best-understood mechanisms associated with heritable epigenetic marks to date is the faithful propagation of DNA methylation through the cell cycle at symmetrical CpG dinucleotides [10, 11,12]. In mammalian cells, as unmodified cytosines are incorporated into symmetrical CpG sites during replication, the maintenance DNA methyltransferase, DNMT1, selectively recognizes the hemi-methylated CpG dyad, and transfers a methyl group to the unmethylated cytosine [13,14]. Of the same protein family are the de novo DNMT3 proteins (DNMT3a, DNMT3b, and DNMT3L). DNMT3a and DNMT3b are believed to play roles in the initial establishment of DNA methylation states that are subsequently maintained by DNMT1 as DNA is replicated through the cell cycle. DNMT3L, which lacks a conserved catalytic methyltransferase domain, facilitates de novo methylation by DNMT3a/b through recognition of unmethylated histone H3 lysine 4 residues (H3K4me0) [15]. The known preference of DNA methyltransferases for CpG dinucleotides represents the first example of how epigenetic information can be encoded upon the genome in a sequence-specific manner. The palindromic nature of the CpG dyad allows hemimethylated CpGs to be sensed and used as templates for the deposition of methyl groups to the opposing cytosine.
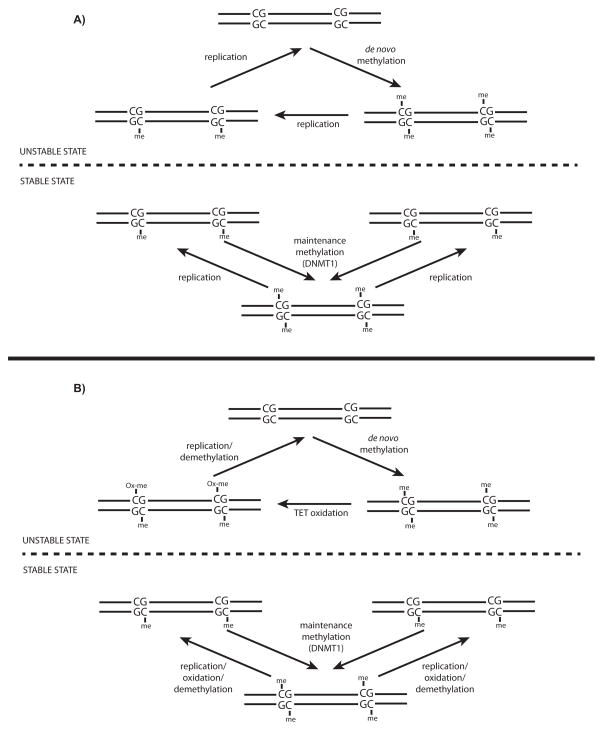
Figure 1. CpG-templated encoding of DNA methylation.
A: An updated schematic of Holliday and Pugh’s [8] original model, similarly proposed by Riggs in the same year [9], for the involvement of symmetrical CpG dinucleotides in the templating of DNA methylation in mammalian genomes. B: An updated and modified version of Holliday and Pugh’s model, incorporating more recent observations involving the destabilization of 5mC by TET family dioxygenases, leading to either active or passive DNA demethylation. Note that the original description of “STABLE” reflects the ability of DNMT1 to heritably maintain a fully methylated state at CpG dinucleotides, while the “UNSTABLE” state reflects the reversibility of DNA methylation.
The primary function of 5mC is largely transcriptional repression of both genes and repetitive elements, although this could depend on genomic context. DNA methylation also plays key roles in epigenetic phenomena, such as imprinting and X-inactivation. 5mC plays a repressive role at CpG-rich transcription start sites and many repetitive elements. The repressive effects of 5mC are generally thought to occur by either preventing the binding of methylation-sensitive transcriptional activators or through the direct affinity of some transcriptional repressors for 5mC [16]. We still do not fully understand the role of DNA methylation at transcribed regions of the genome, regions with reduced CpG density, and subsets of repetitive elements that coincide with enhancer elements [17,18]. However, its presence may facilitate gene expression and influence enhancer activity.
Local CpG density, as well as proteins whose binding is either sensitive to or specific for 5mC, can also influence patterns of 5mC across the genome. In mammalian genomes, the distribution of CpG dinucleotides is non-random. Most genomic sequence is depleted of CpGs relative to other dinucleotide sequences, except for within so-called “CpG islands” (CGIs) (Box 1) [19]. Often, unmethylated CpGs can be recognized by specific protein-binding domains, such as CXXC (Cys–X–X–Cys) domains [20], preventing access to the DNA methylation machinery and allowing for maintenance of the unmethylated state. In this way CXXC domain-containing proteins serve as “epigenetic readers” of information related to DNA methylation. CGIs are frequently associated with transcription start sites (TSSs), and differential methylation at CGIs, in particular, is known to function essentially as an epigenetic switch: the unmethylated state being “ON” and the methylated state “OFF.”
One mechanism by which the non-random distribution of CpG nucleotides is thought to have formed through evolution is via the mutable capacity of 5mC. Sporadic deamination of 5mC in the germline would lead to C-T transition mutations in a way that would track with 5mC. As a result, regions that are recognized and bound by CXXC domains are more likely to be maintained in an unmethylated state and less likely to allow germline C-T transitions that would reduce the overall CpG density across the genome. Methyl-CpG-specific binding domains (MBDs) of proteins exhibit affinity for 5mC and can facilitate 5mCpG-associated transcriptional repression. This may be one mechanism by which 5mC can be protected from sporadic deamination in order to balance the mutagenic capacity of C-T transitions, albeit that direct roles for MBD proteins in such mechanisms remain to be tested. Nonetheless, proteins with affinity for 5mC or whose binding is sensitive to the presence of 5mC play important adaptor-like roles in dictating the outcomes of differentially encoded DNA methylation states. As we discuss below, however, the realization that 5mC may also be actively modified and removed from DNA could also have interesting implications for the mechanisms by which the regulatory landscape of mammalian genomes is shaped. Whatever the case, the actual distributions of methylated/unmethylated CpGs represent discernable patterns throughout the genome that reflect regulatory function, and may thus help further define epigenetic codes. This phenomenon also highlights the fact that, although the term “epigenetics” refers to heritable gene expression changes that do not the result from changes in the underlying DNA itself, such changes are still influenced by DNA sequence.
Non-CpG methylation contributes to the epigenetic regulation of transcription
Most DNA methylation in mammalian genomes occurs at CpG dinucleotides. However, non-CpG methylation is present at appreciable levels in ES cells as well as mouse oocytes [21,22,23], and is particularly high in the mouse and human brain, accounting for over 25% of methylated cytosines genome-wide [5,24,25]. Non-CpG methylation occurs in all dinucleotide contexts (CpA, CpT, CpC), but preferentially occurs at CpAs. Deposition of non-CpG methylation appears to be dependent on DNMT3a-DNMT3L [5,23]. In support of a functional epigenetic role for non-CpG methylation, its deposition in both ES cells and brain is developmentally regulated and linked to gene expression levels [5,21,22]. However, the correlation between non-CpG methylation and gene expression appears to be opposite in these distinct contexts. In ES cells, non-CpG methylation is positively correlated with gene expression [21], whereas in brain there is a negative correlation between such methylation and gene expression [5]. As a result, the integration of DNA methylation into new definitions of epigenetic codes will also require careful consideration of non-CpG methylation.
GC skew can antagonize DNA methylation
Another sequence-based feature associated with differential regulation of DNA methylation is “GC skew” [26,27]. GC skew reflects a general property of many CGIs and TSSs: a strand-specific bias of guanine to cytosine [26]. In mammalian cells, this bias coincides with the presence of RNA:DNA hybrids, or so called “R-loops,” which are capable of protecting CGIs from methylation by the DNTM3B variant, DNMT3B1 (Box 1) [26]. Although GC skew itself is a static sequence-based component of the genome, there is evidence that active enzymatic processes could be involved in R-loop formation in a cell type-specific manner. A genome-wide study of human embryonic stem cells profiling an oxidized form of 5mC, 5-hydroxymethylcytosine (5hmC, see below and Fig. 2), found that regions enriched for 5hmC display GC skew [27]. Resolution of 5hmC at base resolution also identified G-bias surrounding 5hmC bases in human ES cells [28]. It is possible that local GC skew facilitates the formation of R-loops, which can influence the activities of DNA methylation and/or demethylation to program DNA methylation states. The coupling of DNA demethylation machinery to R-loop formation could serve as a potential mechanism to solidify a particular methylation state refractory to de novo methylation in a cell type-specific manner. Whether R-loops have such effects on DNA methylation in cis, trans, or both is for future work to determine.
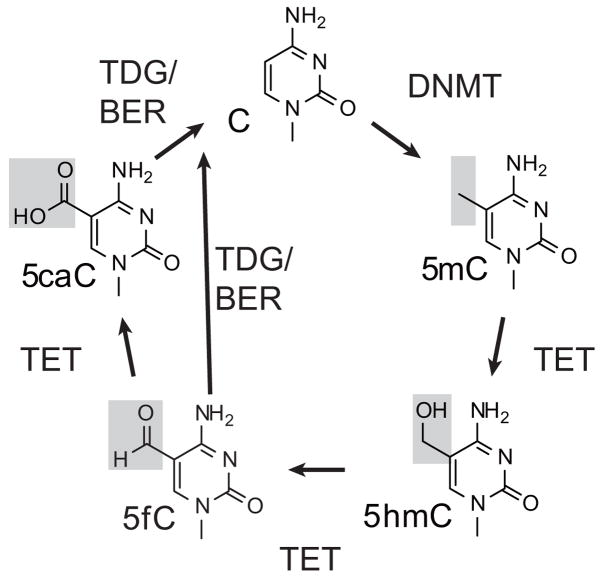
Figure 2. DNA methylation and demethylation.
A summary of DNMT-mediated methylation of cytosine, stepwise TET-mediated 5mC oxidation to 5hmC/5fC/5caC, and coupling to TDG glycosylase activity and base excision repair (BER) to complete the removal of 5mC for reversion to unmodified cytosine.
Oxidized derivatives of 5-methylcytosine and DNA demethylation
5-hydroxymehtylcytosine (5hmC), an oxidized form of 5mC, was rediscovered in 2009 [29,30]. Concurrently, the enzymes responsible for the oxidation of 5mC to 5hmC were identified as the TET family dioxygenases [30–32], named after the ten-eleven translocation (t(10;11)(q22;23)) that results in fusions with the MLL1 gene in rare cases of lymphocytic and acute myeloid leukemias [33,34]. TET proteins later proved to be capable of iterative oxidation of 5mC to 5hmC, then 5-formylcytosine (5fC) and 5-carboxycytosine (5caC) (Fig. 2) [35,36]. The latter oxidation products, 5fC and 5caC, are recognized with high specificity by the DNA glycosylase TDG (thymidine DNA glycosylase) and excised from DNA, which when coupled with base excision repair pathways results in the reversion of 5mC to unmethylated cytosine [35,37,38]. Notably, alternative pathways for the demethylation of 5mC have also been proposed [4,39] and are being actively investigated [4,39,40].
Whatever the exact mechanisms associated with the reversion of 5mC to cytosine, the fact that 5mC can be iteratively oxidized has led now to a total of five types of modified cytosine (C, 5mC, 5hmC, 5fC, and 5caC). The repercussions of this on genomic function are certainly far from clear, and this rapidly changing area of inquiry has been the subject of numerous recent reviews [41–49]. To date, the process of 5mC oxidation has led to two opposing concepts of the functions of 5hmC, 5fC, and 5caC. First, 5mC oxidation may serve simply as an intermediate to DNA demethylation, allowing for reversion of 5mC to unmodified C, as described above. However, it is also possible that distinct “epigenetic readers” could specifically recognize 5hmC, 5fC, and/or 5caC. Thus, rather than serving simply as intermediates to DNA demethylation, 5hmC, 5fC, and 5caC could themselves encode epigenetic information distinct from 5mC.
The existence of multiple DNA methylation states that can be dynamically regulated raises particularly challenging questions about exactly how epigenetic information is encoded. How do these dynamic activities interact with those directing histone modifications to regulate access to DNA? Similarly, how do various transcription factors function in terms of variably regulated DNA methylation states? Because DNA methylation is intricately linked to transcription, how do these newly discovered enzymatic processes interface with the transcriptional machinery? Although insights into a few of these issues are now emerging, to fully answer these questions we will likely need additional molecular tools for dissecting these processes. For instance, the ability to selectively target DNA methylation, 5mC oxidation, and/or excision of 5fC/5caC to defined genomic loci would help determine the role of these processes in encoding regulatory information for both chromatin and transcription.
As mentioned above, beyond their potential role in active DNA demethylation, oxidized 5mC may also serve as distinct epigenetic marks sensed by oxidized 5mC-specific binding proteins. A recent study used a proteomics-based approach to identify 5hmC-, 5fC-, and 5caC-specific binding proteins in ES cells, neural progenitors, and adult mouse brain [50]. These candidates included the UHRF1-related protein, UHRF2, which could stimulate TET dioxygenase activity in vitro. Multiple MBD family proteins, including MeCP2 (methyl-CpG-binding protein 2) and MBD4 (methyl-CpG-binding domain protein 4), were also found to bind 5hmC. The affinity of MeCP2 for 5hmC in brain was corroborated by an independent group [51]. Similar experiments sought to identify 5fC- and 5caC-binding proteins, as well, although it was unclear whether the candidate binding proteins identified simply had affinity for the formyl or carboxyl group, rather than 5fC/5caC. Nonetheless, in addition to the adaptor-like roles MBD proteins play in carrying out regulatory functions based on the presence of 5mC, there may be other layers of adaptor function at work, depending on the presence of 5hmC, 5fC, or 5caC. Such regulatory functions may be complex if MBD proteins, such as MeCP2, have variable affinity for multiple DNA methylation states, as previously reported [51,52].
Genomic views of DNA methylation states reveal three distinct DNA methylation states: FMRs, LMRs, and UMRs
Through advances in epigenomic analyses, there are ever more studies of DNA methylation throughout mammalian genomes across a diverse array of cell and tissue types using a variety of methods. These studies have given us unprecedented details about new aspects of the roles that DNA methylation plays in genomic function, leading to new paradigms of how epigenetic information is encoded through the methylation of DNA. Previous dogma held that the genome existed in essentially a bimodal state, most CpGs being heavily methylated (~70–80%), while a small fraction of highly concentrated CpGs within CGIs are largely devoid of 5mC (<10%). In particular, the use of bisulfite-based methods for the detection of DNA methylation at each individual cytosine throughout the genome has yielded new insights into the distinct states DNA methylation can take, depending on genomic context, cell type, or diseases.
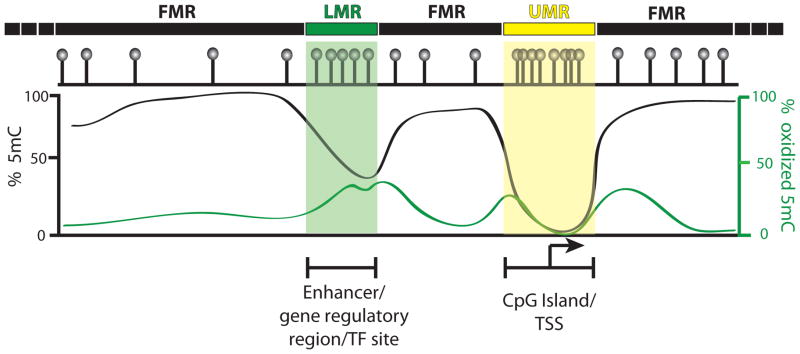
A number of independent groups have now achieved the application of whole-genome bisulfite sequencing (WGBS) in human and mouse ES cells, as well as ES cells differentiated to distinct lineages [53–55]. Moreover, WGBS datasets from many somatic tissues, cultured cell lines, and various cancer cells have also been produced and analyzed [5,56–61]. Together with the integration of RNA-Seq (Box 1) expression datasets and panels of histone modification ChIP-Seq (Box 1) experiments, these studies have established a number of general features associated with the encoding of DNA methylation, with the most detailed information being derived largely from ES cells. In combination, these efforts have confirmed the long-held notion that most of the genome is highly methylated (~80–90% of CpGs with >50% methylation, referred to as fully methylated regions (FMRs), Fig. 3). However, as was originally noticed in mouse ES cells [54], within the remaining portion of the genome were found two distinct methylation states, one that was essentially unmethylated (<10% methylation, referred to as unmethylated regions (UMRs), Fig. 3), and another that was an intermediate state (10–50% methylation, referred to as low-methylated regions (LMRs), Fig. 3). UMRs, as expected, occur largely at unmethylated CGIs corresponding to TSSs. Meanwhile, LMRs occur primarily distal to transcription start sites, have a lower CpG content than UMRs, and coincide with promoter distal gene regulatory elements enriched for transcription factor binding sites [54]. Because WGBS experiments are derived from large numbers of cells, whether the intermediate methylation levels at LMRs represent mosaic patterns of methylation within individual cells or whether the average methylation levels actually reflect subpopulations with distinct methylation states at LMRs remains to be determined. Nonetheless, this study highlighted three key methylation states in mouse ES cells: FMRs, LMRs, and UMRs (Fig. 3). At least two of these states, LMRs and UMRs, were distinct in CpG content that was inversely correlated with methylation, shifting the earlier paradigms, in which DNA methylation existed in a bimodal state.
Figure 3. Genome-wide patterns of DNA methylation and 5mC oxidation.
A simplified schematic summarizing the three DNA methylation states observed in mammalian genomes along with their relation to 5mC oxidation. Fully methylated regions (FMRs) constitute the majority of the genome and generally harbor low levels of oxidized 5mC. Unmethylated regions (UMRs) make up a small fraction of the genome and generally occur at CpG-dense TSSs and are essentially devoid of both 5mC and oxidized 5mC. Low-methylated regions (LMRs) contain intermediate levels of methylation, occur distal to TSSs/UMRs, and are enriched for gene regulatory elements and TF binding sites. LMRs also tend to be highly enriched for oxidized 5mC.
Subsequently, the existence of LMRs and UMRs was found to be conserved in human ES cells [53,55]. Together these studies revealed that most of the DNA methylation dynamics during ES cell differentiation occurred largely within the UMR and LMR fractions of the genome and were indicative of cell type-specific DNA methylation states. Interestingly, however, among the regions exhibiting DNA methylation dynamics during differentiation, most actually occurred at LMRs. Since LMRs correspond to promoter distal gene regulatory elements, such as enhancers, these observations make good sense, as histone modifications at enhancers reflect cell type specificity more than modifications at promoters and CGIs [62]. A pair of subsequent studies in which extensive WGBS in the developing mammalian brain [5] and 30 diverse human cell and tissue types was performed [61] also found that the majority of differentially modified loci occurred within a relatively small portion of the genome corresponding to promoter distal gene regulatory elements. Together, these results support the emerging view that most CpGs throughout the genome exist in a default methylation state that is likely established during early embryonic development as DNA methylation is reset. The processes associated with subsequent cellular differentiation and active/passive DNA methylation dynamics may then serve to modify DNA methylation states, in particular at promoter distal gene regulatory elements. In this way patterns of cell type-specific DNA methylation emerge, reflective of cellular phenotype. Such developmental tuning of DNA methylation via active DNA methylation/demethylation pathways may be an important factor in defining cell type-specific epigenetic codes.
Combined, these findings point to distinct patterns of DNA methylation defining a component of the functional epigenomic gene regulatory landscape that contributes to ES cell differentiation. An important feature distinguishing LMRs from UMRs in ES cells is also reduced CpG content, again highlighting the key role that this variable plays in the encoding of DNA methylation. These studies prompt a number of questions regarding how the dynamic regulation of DNA methylation may be linked to the coinciding histone modifications, as well as the underlying sequence contexts (i.e. high CpG content at CGIs/UMRs vs. low CpG content at LMRs/enhancers and other regulatory regions), in order to encode epigenetic information. Moreover, the fact that LMRs are also enriched for the binding sites of various transcription factors, and can even be induced by the presence of certain transcription factors [54], raises new questions about the interaction between transcription factors and dynamic DNA methylation states.
Genome-wide analyses link 5mC oxidation to transcription and programming of DNA methylation at gene regulatory elements
With the discovery of 5hmC came the realization that, using conventional bisulfite-based methods, 5hmC could not be distinguished from 5mC [63,64], and 5fC and 5caC could not be distinguished from unmodified cytosine [35,65]. This raised technical issues about how to distinguish each of these base modifications from 5mC and C specifically in genomic DNA. In the four short years since 5mC oxidation was discovered, there has been a flurry of new technological developments that allow for the distinction of these various cytosine modifications in genomic DNA (Summarized in Table 1, [66–83]). Application of these methodologies has now given us detailed genome-wide views of 5hmC, and more recently 5fC and 5caC, providing fresh insight into how 5mC oxidation impacts epigenetic encoding through the regulation of DNA methylation states [65,73].
Table 1.
Methods for detecting DNA methylation and its oxidation
| Affinity purification | Base-resolution | |
|---|---|---|
| 5mC | DNA Immunoprecipitation (DIP/MeDIP)[72], MBD-Seq[75–76], TET-assisted methyl-cytoisne selective labeleing (biotin, TAmC-Seq)[74] | Bisulfite sequencing [80], Single-molecule Real-Time Sequencing (SMRT) [76], Oxidative Bisulfite Sequencing (OxBS-Seq)[79] |
| 5hmC | DIP (5hmC-DIP)[64,73,78], JBP1[68], 5hmC selective labeling (biotin, 5hmC-Seal) [71] Glucosylation/Periodate oxidation/Biotinylation (GLIB) [66] |
TET Assisted Bisulfite Sequencing (TAB-Seq)[25], SMRT[77] |
| 5fC | DIP (5fC-DIP)[70], 5fC-Seal (biotin)[62] | 5fC Chemical Assisted Bisulfite Sequencing (fCAB-Seq)[70] |
| 5caC | DIP (caC-DIP)[70] | 5caC Chemical Assisted Bisulfite Sequencing [65] |
One feature that has emerged from the initial studies mapping 5hmC genome-wide is that 5mC oxidation is generally associated with genes and gene regulatory elements (general features summarized in Fig. 3). Nonetheless, its presence at promoters, transcribed gene bodies, gene distal regulatory elements, and even repetitive elements does have unique characteristics. At promoters, 5hmC is most enriched at those with low CpG content and is essentially absent from CGIs and high-CpG TSSs [65,73], as expected since these regions also lack 5mC, the substrate for TET oxidation. The distribution of 5hmC at and around promoters also appears to be dependent on the level of transcription, where lowly transcribed genes tend to be marked with 5hmC directly at the TSS, and more highly expressed genes tend to have a bimodal distribution surrounding the TSS. Examining 5hmC distributions at promoter regions offers a unique perspective from which to consider how, in addition to CpG content and distribution, other factors like gene expression level may also influence the encoding of 5mC oxidation. Intriguingly, LMRs are also highly enriched for 5hmC and TET1. These findings may implicate DNA demethylation processes in the formation and perhaps regulation of LMRs, and more generally, gene distal regulatory elements.
5mC oxidation is highly enriched in gene regulatory elements
Among all known functional genomic regulatory elements, 5hmC appears to be most highly enriched at gene distal enhancers. However, 5hmC is also present at distinct regulatory regions, such as CTCF-binding sites [28,65,73]. Notably, at these types of gene regulatory elements, there is a distinct pattern of both 5mC and 5hmC: 5mC is generally depleted directly at sites of transcription factor-DNA interaction, and 5hmC is enriched surrounding that site of interaction [28]. These features associated with the distribution and patterns of 5mC and 5hmC at such gene regulatory elements hint at unique properties for the encoding of these marks. It would be interesting to determine whether, like unmethylated CGIs, methylation patterns associated with the functional genomic elements can be used for their identification or prediction.
Genome-wide mapping of 5fC and 5caC has also been achieved now using affinity-based approaches [65,70,73]. These studies identified some unique characteristics associated with the differential oxidation of 5mC across the genome and present interesting new paradigms for the encoding of epigenetic information. In general, 5hmC oxidation to 5fC appears to follow the expected pattern of correlating very well with 5hmC as detected by both affinity enrichment and base-resolution methods. At steady state, 5caC can only be detected at appreciable amounts at heavily methylated (5mC) major satellite repeats, despite the fact that such repeats are depleted of 5hmC. What is perhaps more telling are the gains in 5fC and 5caC that occur when TDG is removed or depleted from mouse ES cells [65,73], revealing locations where TDG was actively excising these more oxidized states. In TDG-deficient ES cells, 5fC and 5caC accumulates strongly within the LMR fraction of the genome. Accordingly, there is a strong gain in 5fC and 5caC at enhancers and other diverse types of regulatory elements that is widespread across the genome [65,73].
The fact that the enzymatic output of TET-dependent 5mC oxidation and TDG-dependent 5fC and 5caC excision are highly enriched with the LMR fraction of the genome may point to a functional role for active DNA demethylation in modulating these elements. Rather than a simple passive end-point caused by transcription factor binding, the active oxidation and demethylation of cytosines could be involved in regulating the function of gene regulatory elements.
The links between histone modifications and DNA methylation states
A full understanding of the mechanisms by which DNA methylation is involved in encoding epigenetic mechanisms will require a more complete picture of the interactions between modified histones and DNA of various methylation states. There has been a fair amount of progress in this area, leading to several core concepts about how such interactions are likely to occur more broadly [20,84,85]. Here we will highlight a few examples of the new concepts by which DNA methylation and histone modification dynamics may be linked to explore the role these processes play in encoding epigenetic information in the genome.
DNA methylation states can often be coupled to histone modifications through interactions between methyl-sensitive or methyl-specific binding proteins, which serve as “epigenetic readers” of DNA methylation and are associated with other chromatin-modifying complexes that enzymatically target histone tails. In general, MBD proteins have been biochemically purified along with a number of different histone deacetylase (HDAC) repressor complexes. The recruitment of HDACs by MBD proteins is believed to facilitate a repressive state in chromatin via deacetylation of histone tails [86,87].
Among the most highly correlated of the histone modifications and DNA methylation is that between histone H3-lysine-4-trimethyl (H3K4me3) and an unmethylated state at CGIs and TSSs. This combination of marks can involve MLL family CXXC domains that facilitate targeting of MLL histone H3 methyltransferase complexes to unmethylated DNA [88–91]. Similarly, histone demethylase enzymes, such as Jumonji domain-containing histone demethylase 1a JHDM1a (also known as KDM2a or CXXC8), can also be recruited to unmethylated CpG-dense regions of the genome for demethylation of histone H3 lysine 36 (H3K36me) by way of a CXXC domain [92,93]. Interestingly, TET family proteins also contain CXXC domains and are frequently associated with unmethylated promoters, indicating links between CXXC binding to unmethylated CpGs and the underlying patterns of H3K4me3. CXXC containing histone methyltransferases/demethylases and TET family proteins are good examples of how DNA methylation and histone modifications can be coupled through modular proteins, which contain both enzymatic activity toward chromatin as well as domains capable of recognizing unmethylated DNA. Alternatively, such coupling can also be achieved via accessory proteins. An example of this type of mechanism involves CXXC finger protein 1 (Cfp1) and Set1, a H3K4 methyltransferase. Set1 lacks a CXXC domain, but directly interacts with Cfp1; Cfp1 directly binds unmethylated CGIs, allowing for targeting of Set1 and deposition of H3K4me3 [94,95].
Recently, using affinity purification, multiple groups have shown that OGT (O-linked β-D-N-acetylglucosamine (O-GlcNAc) transferase) physically interacts with TET1, TET2, and TET3 [96–99]. OGT connects to chromatin via its ability to transfer GlcNAc sugars to Ser and Thr residues of histones, including histone H2B and other chromatin-modifying enzymes [100,101]. TET proteins are able to recruit OGT to CpG-rich TSSs, where it subsequently glycosylates HCF-1 (host cell factor 1), a component of the SET/COMPASS H3K4 methyltransferase complex, regulating its activity on H3K4 [96,97]. In this way CXXC-mediated binding of TET to CpG-rich promoters influences levels of H3K4me3. However, TET (Tet1 and Tet2) binding does not necessarily correlate well with 5hmC levels, and in fact the strongest TET-binding sites tend to be unmethylated CpG-rich sequences [76,96]. This may imply that TET proteins could provide a protective function counteracting aberrant methylation at such regions, that there could be underappreciated non-enzymatic roles for TET binding, or that TET enzymatic activity could be targeted to regulatory elements interacting with the unmethylated promoter to fine-tune transcription of a given gene.
5hmC may also negatively influence the maintenance of DNA methylation by DNMT1. In vitro, hemi-hydroxymethylated DNA serves as a poor template for DNMT1-mediated methyltransferase activity and is also poorly recognized by UHRF1, DNMT1’s obligate binding partner, which recognizes hemimethylated CpGs [13,14,102]. Thus, 5hmC may serve as a modification that facilitates passive DNA demethylation by blocking DNMT1 activity (Fig. 1B). Interestingly, UHRF1 is also a tandem-tudor domain-containing protein capable of coordinately recognizing histone H3-K9-trimethyl (H3K9me3), linking maintenance of 5mC to H3K9me3. Since 5hmC is poorly recognized by UHRF1, this may point to an additional role for 5hmC in antagonizing the presence of H3K9me3 at selective genomic loci. On the other hand, recent findings that 5hmC may be selectively recognized by the closely related tandem-tudor domain protein, UHRF2 [50], could have implications for determining links between 5hmC and defined histone modifications in the future.
Outside of promoters, the links between DNA methylation states and histone modifications have been limited primarily to correlational analyses using epigenomic datasets. However, given the strong enrichment of 5mC oxidation and reduced levels of 5mC in LMRs corresponding to gene distal functional regulatory elements, it is worth noting the specific histone modifications that may link 5mC oxidation and histone modification. Among the distinct types of gene regulatory elements enriched for 5hmC, enhancers harbor the highest levels [28]. As such, there is a strong positive correlation between 5hmC levels and H3K4me1. Similarly, there is a relatively good correlation between 5hmC and histone acetylation, including H3K27ac. These correlations provide a genomic basis for further examination of the links between 5mC oxidation and chromatin-modifying complexes mediating the underlying histone modification status.
A framework for the integration of dynamic DNA methylation into epigenetic codes
Through the genome-wide profiling of dynamic cytosine modifications, the principles that influence epigenetic encoding can be built into a framework within which the associated codes are characterized. Specifically, this framework includes static marks, epigenetic readers, and active processes (Fig. 4).
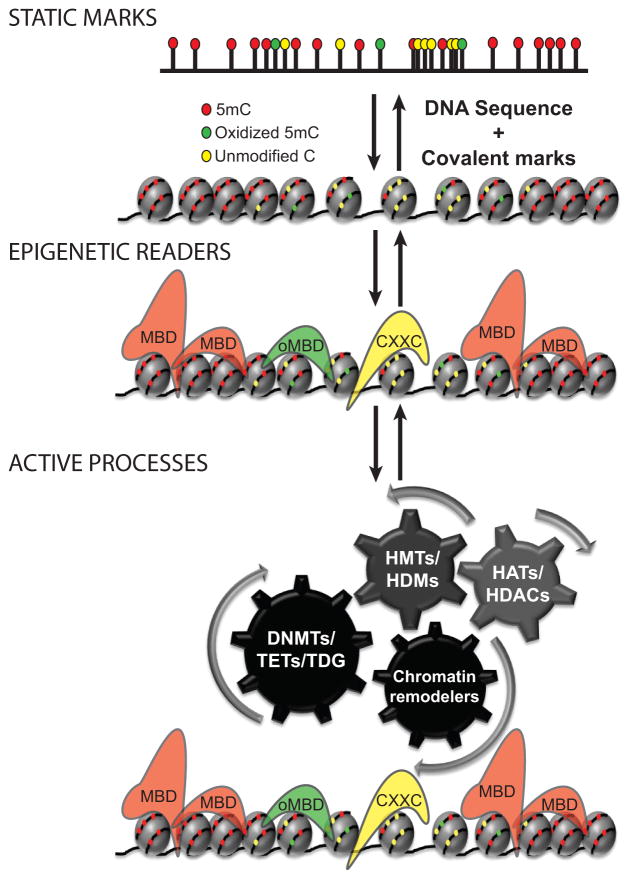
Figure 4. Three components influencing the encoding of epigenetic information linked to DNA methylation.
“Static marks” involve covalent modifications to DNA and histones. CpGs, CpG distributions, and GC skew can influence covalent modifications to DNA. Covalent modifications to chromatin are then read by “epigenetic readers” of those marks, serving adapter-like functions for carrying out regulatory function within the context of chromatin. This includes methyl binding domain (MBD) proteins, proteins with affinity for oxidized 5mC (oxidized methyl-binding domain proteins, oMBD proteins), and methyl-sensitive CXXC domain proteins, Finally, “active processes” regulate the distribution of “static marks” and “epigenetic readers.” This component includes enzymatic modification of both DNA (DNMTs, TETs, and DNA glycosylases such as TDG) and histones (HATs, HDACs, HMTs, HDMs, and others). Also included within this component are ATP-dependent chromatin remodeling complexes.
The “static marks” component of epigenetic codes represents steady-state covalent marks, their distributions genome-wide, and the underlying sequence context. For DNA methylation, sequence contexts include simply CpG, CpG density and distribution, and local GC skew. The static component, however, can be separated according to whether such marks are on histones or DNA.
“Epigenetic readers” are proteins with affinity for or sensitivity to a given covalent modification or epigenetic mark, allowing them to act as adaptors for carrying out various functions associated with each mark or combination of marks. For DNA methylation this would include MBDs, CXXC domains, and other similar types of protein domains that are sensitive to or specific for various DNA methylation states (Fig. 4). Notably, some epigenetic readers are multifunctional proteins that coordinately recognize a DNA methylation state and histone modification. These proteins may be of particular interest in terms of understanding how various epigenetic codes are integrated.
The “active processes” component involves known chromatin-associated enzymes that place, modify, or remove covalent modifications to DNA and histones. Of course, for DNA methylation, DNMTs, TET family proteins, and DNA glycosylases, such as TDG, would be included here. Similarly, histone-modifying enzymes (histone acetyltransferases [HATs], histone deacetylases [HDACs], histone methyltransferases [HMTs], histone demethylases [HDMs], and various other histone modifying enzymes) would represent active processes influencing epigenetic codes. Also included within this component are chromatin-associated proteins that actively maneuver or remove proteins from chromatin, such as ATP-dependent chromatin remodeling complexes.
Are there additional DNA modifications in mammalian genomes?
Given the recent appreciation that there can be as many as five distinct adducts of cytosine in mammalian DNA, a reasonable question is whether all base modifications in mammalian genomes have been identified. Historically, the role of DNA methylation in epigenetic regulation was inferred from bacterial systems, in which more than 100 distinct nucleobase modifications are known. Most of these exist in RNA, with some 15–20 having been described in DNA. Two good examples of these modifications are N-6 methylation of adenine (N-6-meA) and Base J (β-D-glucopyranosyloxymethyluracil). N-6-meA is a common restriction modification, but some DNA adenine methyltransferases (MTases) lack corresponding restriction endonucleases (e.g. Dam and CcrM). Earlier publications have described N-6-meA in a number of organisms, including the eukaryotes protists, fungi, slime mold, algae, and wheat [103]. Another example, Base J, is found in Trypanosoma brucei, all kinetoplastids, and some unicellular flagellates [104]. Its predicted roles include gene silencing and repression of homologous recombination. The presence of 5hmC along with the genes that encode glycosyltransferases in mammalian DNA suggests that Base J could be present. Whether there are additional DNA modifications besides cytosine modifications in mammalian genomes remains to be determined.
Conclusions and future paradigms
Our understanding of epigenetic information encoded by DNA methylation and its dynamic regulation is evolving rapidly with the convergence of data on new base modifications and genome-wide mapping studies. Along with the vast amounts of data being generated for histone modifications, transcription factors, and other chromatin regulators, new paradigms for how various covalent modifications to chromatin encode genomic function are beginning to emerge. With the more recent introduction of concepts related to the dynamic regulation of epigenetic marks, the principles that regulate epigenetic encoding can be separated into three components: static marks, epigenetic readers, and active processes (Fig. 4). Understanding the interplay between these three components and how active processes regulate transitions between static states through epigenetic readers and enzymatic modification of epigenetic marks will be critical to fully grasp the realm of epigenetically encoded information in genomic function.
Acknowledgments
The authors would like to thank Cheryl Strauss for critical reading of the manuscript, and Michael Jin for helping with figures. This work was supported in part by NIH grants NS079625 and HD073162 (P.J.).
Abbreviations
- 5mC
5-methylcytosine
- CGIs
CpG islands
- TSS
Transcription start site
- 5hmC
5-hydroxymethylcytosine
- 5fC
5-formylcytosine
- 5caC
5-carboxycytosine
- WGBS
Whole-genome bisulfite sequencing
- FMR
Fully methylated region
- UMR
Unmethylated region
- LMR
Low-methylated region
- N-6-meA
N-6 methylation of adenine
- Base J
β-D-glucopyranosyloxymethyluracil
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- 1.Riggs ADRV, Martienssen RA. Epigenetic mechanisms of gene regulation. Plainview, NY: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Guo JU, Ma DK, Mo H, Ball MP, Jang MH, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nature neuroscience. 2011;14:1345–51. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–34. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, et al. Global Epigenomic Reconfiguration During Mammalian Brain Development. Science. 2013 doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–7. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szulwach KE, Li X, Li Y, Song CX, Wu H, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nature neuroscience. 2011;14:1607–16. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–32. [PubMed] [Google Scholar]
- 9.Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 10.Wigler M, Levy D, Perucho M. The somatic replication of DNA methylation. Cell. 1981;24:33–40. doi: 10.1016/0092-8674(81)90498-0. [DOI] [PubMed] [Google Scholar]
- 11.Bolden AH, Nalin CM, Ward CA, Poonian MS, Weissbach A. Primary DNA sequence determines sites of maintenance and de novo methylation by mammalian DNA methyltransferases. Mol Cell Biol. 1986;6:1135–40. doi: 10.1128/mcb.6.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruenbaum Y, Cedar H, Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982;295:620–2. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- 13.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, et al. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–4. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 14.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–12. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 15.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–7. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Current opinion in genetics & development. 1993;3:226–31. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 17.Wu H, Coskun V, Tao J, Xie W, Ge W, et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–8. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie M, Hong C, Zhang B, Lowdon RF, Xing X, et al. DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape. Nat Genet. 2013 doi: 10.1038/ng.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. Journal of molecular biology. 1987;196:261–82. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto H, Vertino PM, Cheng X. Molecular coupling of DNA methylation and histone methylation. Epigenomics. 2010;2:657–69. doi: 10.2217/epi.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shirane K, Toh H, Kobayashi H, Miura F, Chiba H, et al. Mouse oocyte methylomes at base resolution reveal genome-wide accumulation of non-CpG methylation and role of DNA methyltransferases. PLoS Genet. 2013;9:e1003439. doi: 10.1371/journal.pgen.1003439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–67. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie W, Barr CL, Kim A, Yue F, Lee AY, et al. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148:816–31. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45:814–25. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome biology. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–80. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–30. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–33. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, et al. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17:637–41. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 34.Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, et al. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23) Cancer Res. 2002;62:4075–80. [PubMed] [Google Scholar]
- 35.He YF, Li BZ, Li Z, Liu P, Wang Y, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–7. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ito S, Shen L, Dai Q, Wu SC, Collins LB, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–3. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. The Journal of biological chemistry. 2011;286:35334–8. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L, Lu X, Lu J, Liang H, Dai Q, et al. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat Chem Biol. 2012 doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, et al. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat Chem Biol. 2012;8:751–8. doi: 10.1038/nchembio.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinney SR, Pradhan S. Ten eleven translocation enzymes and 5-hydroxymethylation in mammalian development and cancer. Adv Exp Med Biol. 2013;754:57–79. doi: 10.1007/978-1-4419-9967-2_3. [DOI] [PubMed] [Google Scholar]
- 42.Koh KP, Rao A. DNA methylation and methylcytosine oxidation in cell fate decisions. Curr Opin Cell Biol. 2013;25:152–61. doi: 10.1016/j.ceb.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341–56. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2011;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 46.Guo JU, Su Y, Zhong C, Ming GL, Song H. Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle. 2011;10:2662–8. doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2011;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–52. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomson JP, Hunter JM, Meehan RR. Deep C diving: mapping the low-abundance modifications of the DNA demethylation pathway. Genome Biol. 2013;14:118. doi: 10.1186/gb-2013-14-5-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–59. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 51.Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–30. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, et al. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40:4841–9. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, et al. Transcriptional and Epigenetic Dynamics during Specification of Human Embryonic Stem Cells. Cell. 2013;153:1149–63. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–5. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 55.Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–48. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2012;44:40–6. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–75. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22:246–58. doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kulis M, Heath S, Bibikova M, Queiros AC, Navarro A, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44:1236–42. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- 60.Li Y, Zhu J, Tian G, Li N, Li Q, et al. The DNA methylome of human peripheral blood mononuclear cells. PLoS biology. 2010;8:e1000533. doi: 10.1371/journal.pbio.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013 doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–12. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic acids research. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 2013;153:678–91. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brinkman AB, Simmer F, Ma K, Kaan A, Zhu J, et al. Whole-genome DNA methylation profiling using MethylCap-seq. Methods. 2010;52:232–6. doi: 10.1016/j.ymeth.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 67.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 68.Lu X, Song CX, Szulwach K, Wang Z, Weidenbacher P, et al. Chemical Modification-Assisted Bisulfite Sequencing (CAB-Seq) for 5-Carboxylcytosine Detection in DNA. Journal of the American Chemical Society. 2013;135:9315–7. doi: 10.1021/ja4044856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–7. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raiber EA, Beraldi D, Ficz G, Burgess HE, Branco MR, et al. Genome-wide distribution of 5-formylcytosine in embryonic stem cells is associated with transcription and depends on thymine DNA glycosylase. Genome biology. 2012;13:R69. doi: 10.1186/gb-2012-13-8-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robertson AB, Dahl JA, Ougland R, Klungland A. Pull-down of 5-hydroxymethylcytosine DNA using JBP1-coated magnetic beads. Nat Protoc. 2012;7:340–50. doi: 10.1038/nprot.2011.443. [DOI] [PubMed] [Google Scholar]
- 72.Robertson AB, Dahl JA, Vagbo CB, Tripathi P, Krokan HE, et al. A novel method for the efficient and selective identification of 5-hydroxymethylcytosine in genomic DNA. Nucleic acids research. 2011;39:e55. doi: 10.1093/nar/gkr051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shen L, Wu H, Diep D, Yamaguchi S, D’Alessio AC, et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–62. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 76.Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–84. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang L, Szulwach KE, Hon GC, Song CX, Park B, et al. Tet-mediated covalent labelling of 5-methylcytosine for its genome-wide detection and sequencing. Nature communications. 2013;4:1517. doi: 10.1038/ncomms2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Serre D, Lee BH, Ting AH. MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic acids research. 2010;38:391–9. doi: 10.1093/nar/gkp992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nature methods. 2010;7:461–5. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Song CX, Clark TA, Lu XY, Kislyuk A, Dai Q, et al. Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine. Nature methods. 2012;9:75–7. doi: 10.1038/nmeth.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–8. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–7. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 83.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–31. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen T. Mechanistic and functional links between histone methylation and DNA methylation. Prog Mol Biol Transl Sci. 2011;101:335–48. doi: 10.1016/B978-0-12-387685-0.00010-X. [DOI] [PubMed] [Google Scholar]
- 85.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 86.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–91. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 87.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–9. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 88.Ayton PM, Chen EH, Cleary ML. Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Molecular and cellular biology. 2004;24:10470–8. doi: 10.1128/MCB.24.23.10470-10478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Birke M, Schreiner S, Garcia-Cuellar MP, Mahr K, Titgemeyer F, et al. The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic acids research. 2002;30:958–65. doi: 10.1093/nar/30.4.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–17. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 91.Terranova R, Agherbi H, Boned A, Meresse S, Djabali M. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc Natl Acad Sci U S A. 2006;103:6629–34. doi: 10.1073/pnas.0507425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, et al. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell. 2010;38:179–90. doi: 10.1016/j.molcel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 94.Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. The Journal of biological chemistry. 2005;280:41725–31. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- 95.Lee JH, Tate CM, You JS, Skalnik DG. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. The Journal of biological chemistry. 2007;282:13419–28. doi: 10.1074/jbc.M609809200. [DOI] [PubMed] [Google Scholar]
- 96.Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–4. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. The EMBO journal. 2013;32:645–55. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vella P, Scelfo A, Jammula S, Chiacchiera F, Williams K, et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol Cell. 2013;49:645–56. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 99.Shi FT, Kim H, Lu W, He Q, Liu D, et al. Ten-eleven translocation 1 (tet1) is regulated by o-linked N-acetylglucosamine transferase (ogt) for target gene repression in mouse embryonic stem cells. The Journal of biological chemistry. 2013;288:20776–84. doi: 10.1074/jbc.M113.460386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gambetta MC, Oktaba K, Muller J. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science. 2009;325:93–6. doi: 10.1126/science.1169727. [DOI] [PubMed] [Google Scholar]
- 101.Kreppel LK, Blomberg MA, Hart GW. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. The Journal of biological chemistry. 1997;272:9308–15. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- 102.Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol. 2012;19:1155–60. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wion D, Casadesus J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nature reviews Microbiology. 2006;4:183–92. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Borst P, Sabatini R. Base J: discovery, biosynthesis, and possible functions. Annual review of microbiology. 2008;62:235–51. doi: 10.1146/annurev.micro.62.081307.162750. [DOI] [PubMed] [Google Scholar]