Abstract
Recently, a novel ehrlichial organism was isolated from a raccoon (Procyon lotor) and the isolate (RAC413) was infectious to two naïve raccoons but not laboratory mice, rats or rabbits. In this study, amplification and sequencing of four gene targets (16S rRNA gene, groESL, gltA and rpoB) confirmed that the novel ehrlichial organism was a member of the family Anaplasmataceae and was most closely related to, but distinct from, ‘Candidatus Neoehrlichia mikurensis’ TK4456R and IS58. RAC413 shared the highest sequence similarity with members of the genus Ehrlichia (94.2–95.1, 80.9–83.1, 67.9–71.9 and 39.9–40.7 % similarity for the 16S rRNA gene, groESL, gltA and rpoB, respectively). No sequence variation in three sequences (16S rRNA gene, groESL and gltA) was observed between the RAC413 isolate and five additional sequences amplified from blood of naturally infected raccoons from several geographically isolated populations in the south-eastern USA. Serum samples from four experimentally infected raccoons did not react to Ehrlichia canis, Ehrlichia chaffeensis, Anaplasma marginale or Anaplasma phagocytophilum antigens in an immunofluorescence assay or an Ehrlichia ewingii peptide in an ELISA format. On the basis of the distinctive molecular and serological characteristics and apparent host specificity of this ehrlichial organism, it is proposed that this organism be designated ‘Candidatus Neoehrlichia lotoris’ (reference strain RAC413R).
INTRODUCTION
Members of the family Anaplasmataceae (genera Aegyptianella, Anaplasma, Ehrlichia, Neorickettsia and Wolbachia) are obligately intracellular bacteria that reside within membrane-enclosed vacuoles (Dumler et al., 2001). This family also includes two Candidatus status genera, ‘Candidatus Xenohaliotis’, detected in abalone (Haliotis spp.) (Friedman et al., 2000), and ‘Candidatus Neoehrlichia’, detected in ticks and rodents from Europe and Asia (Schouls et al., 1999; Pan et al., 2003; Brouqui et al., 2003; Sanogo et al., 2003; Kawahara et al., 2004; Beninati et al., 2006; Tabara et al., 2007). ‘Candidatus Neoehrlichia mikurensis’ is distinguished from other genera and species based on sequence analysis of the partial 16S rRNA, citrate synthase (gltA) and groESL genes of several strains (Kawahara et al., 2004; Brouqui et al., 2003). Ultrastructural analysis of a single strain (TK4456R) of this group revealed bacteria in endothelial cells of experimentally infected rats that were consistent with members of the family Anaplasmataceae (Kawahara et al., 2004).
Sequences that were similar to or identical to ‘Candidatus Neoehrlichia mikurensis’ strains TK4456R and IS58 have been detected in Ixodes ricinus ticks in the Netherlands (Ehrlichia-like sp. Shotti variant), I. ricinus in Russia and Italy (called ‘Candidatus Ehrlichia walkerii’) and Ixodes persulcatus and Ixodes ovatus from Japan (Schouls et al., 1999; Alekseev et al., 2001; Brouqui et al., 2003; Sanogo et al., 2003; Pan et al., 2003; Kawahara et al., 2004; Rar et al., 2008). To date, natural infections have been reported from brown or Norway rats (Rattus norvegicus) from China and Japan, small Japanese field mice (Apodemus argenteus), large Japanese field mice (Apodemus speciosus), grey red-backed voles (Myodes rufocanus bedfordiae) and Smith’s red-backed voles (Eothenomys smithii) in Japan, an East European field vole (Microtus rossiaemeridionalis) from Russia and a bank vole (Myodes glareolus) from Italy (Pan et al., 2003; Kawahara et al., 2004; Beninati et al., 2006; Naitou et al., 2006; Tabara et al., 2007; Rar et al., 2008).
In the USA, an ehrlichial organism closely related to ‘Candidatus Neoehrlichia mikurensis’, based on a partial 16S rRNA gene sequence, was previously detected in 53 % of tested raccoons (Procyon lotor) from the Piedmont region of Georgia (Dugan et al., 2005). Recently, this organism was isolated in ISE6 tick cell culture (Munderloh et al., 2007) and the culture was infectious to raccoons but failed to produce a detectable infection in two strains of laboratory mice, laboratory rats or laboratory rabbits (Yabsley et al., 2008a). The organism was commonly detected in tick-infested raccoon populations (prevalence of 46–95 %) but was not found in 137 rodents of 10 species (Yabsley et al., 2008a). Herein, we characterize this organism more fully and propose that it should be designated ‘Candidatus Neoehrlichia lotoris’.
METHODS
Animals and procedures
Six, 10-week-old raccoons (RAC413, RAC-414, RAC974, RAC975 and two negative controls) were acquired from a commercial source (Ruby Fur Farms, New Sharon, IA, USA). Raccoons were infected either by inoculation of infected blood (RAC413 and RAC414) or by inoculation of infected ISE6 tick cells (RAC974 and RAC975) as described previously (Munderloh et al., 2007; Yabsley et al., 2008a). Data on inoculation methods and infection dynamics in these raccoons have been provided previously (Munderloh et al., 2007; Yabsley et al., 2008a).
Serological testing
Plasma samples were tested for antibodies reactive with Ehrlichia chaffeensis, Ehrlichia canis, Anaplasma marginale and Anaplasma phagocytophilum by immunofluorescence assay (IFA) as described previously (Comer et al., 2000; Yabsley et al., 2003; Little et al., 1998) and with Ehrlichia ewingii using an experimental peptide ELISA (IDEXX Laboratories). For IFA, plasma samples were tested at a 1: 64 dilution in 0.01 M PBS. Slides were either purchased commercially from Focus Technologies, (E. chaffeensis and A. phagocytophilum) or made in our laboratory (A. marginale, E. canis and RAC413 organism). To make IFA slides, infected cultures were harvested, washed twice in PBS, applied to slides, allowed to dry and fixed with acetone. Fluorescein isothiocyanate (FITC)-labelled rabbit anti-raccoon immunoglobulin G (Kirkegaard & Perry Laboratories) diluted 1: 50 in PBS was used as a conjugate. During the final wash, slides were counterstained with 1.65 % eriochrome black T. Positive controls for the IFA included serum samples from raccoons experimentally inoculated with either E. chaffeensis (titres of 1: 128 and 1: 1024), E. canis (1: 512) or A. phagocytophilum (1: 256 and 1: 1024) (Yabsley et al., 2008b) and serum from an A. marginale-infected deer (1: 256) (combined with an FITC-labelled anti-deer antibody; Kirkegaard & Perry Laboratories). Serum from a dog experimentally infected with E. ewingii was used as a positive control for the E. ewingii ELISA. Negative control serum samples were collected from young laboratory-raised raccoons, laboratory-raised orphaned fawns and purpose-bred young beagles. Two additional negative serum controls were collected from two raccoons that had been inoculated with 1 × 106 uninfected ISE6 tick cells.
Molecular characterization and phylogenetic analyses
Partial 16S rRNA, groESL, gltA and RNA polymerase β-subunit (rpoB) genes were amplified and sequenced from the culture isolate RAC413 (Inokuma et al., 2001; Taillardat-Bisch et al., 2003; Kawahara et al., 2004). In addition, we investigated potential intraspecific variation by analysis of partial 16S rRNA gene, groESL and gltA sequences from five wild raccoons from geographically diverse locations in Florida (n = 2), Georgia (n = 2) and South Carolina (n = 1). Resulting sequences were aligned with those from related organisms in GenBank using clustal_x. Phylogenetic analyses of the RAC413 sequences and related organisms were conducted using the mega program using the neighbour-joining algorithm with Kimura’s two-parameter model (GenBank accession numbers are provided in Supplementary Table S1, available in IJSEM Online).
RESULTS AND DISCUSSION
All four raccoons, regardless of exposure route or inoculum, became PCR-positive for ‘Candidatus N. lotoris’, and the organism was successfully isolated from each raccoon into ISE6 tick cells (Munderloh et al., 2007; Yabsley et al., 2008a). Plasma samples from each sampling date from the four experimentally infected raccoons did not react with E. chaffeensis, E. canis, E. ewingii, A. marginale or A. phagocytophilum by IFA or ELISA. Beginning on day 16 p.i. (RAC413), day 21 p.i. (RAC414) or day 28 p.i. (RAC974 and RAC975), an antibody reaction to the culture isolate of strain RAC413R was observed at a dilution of 1: 64. Subsequent sampling periods also maintained an antibody response at only a 1: 64 dilution. In positive samples, fluorescence was localized to clusters of intracellular organisms; no fluorescence was observed for samples from raccoons inoculated with uninfected ISE6 tick cells.
The partial 16S rRNA gene sequences (1379 bp) from all six samples of the raccoon ehrlichial agent (isolate RAC413R and samples from five wild raccoons) were identical. The sequences were most similar (98.4–98.8 %) to those of ‘Candidatus N. mikurensis’ strains TK4456R and IS58 (Kawahara et al., 2004) (complete similarity matrix shown in Supplementary Table S2). The level of similarity between the two ‘Candidatus N. mikurensis’ strains was 99.3 %. The highest similarities to members of a currently described genus were with Ehrlichia (94.2–95.1 %), followed by Anaplasma (92.7–93.1 %). The similarities between members of the genera Ehrlichia and Anaplasma ranged from 92.6 to 93 % and the similarity between Aegyptianella pullorum (for the 574 available nucleotides deposited as GenBank accession no. AY125087) and members of the genus Anaplasma ranged from 95.2 to 96.4 % (Rikihisa et al., 2003).
Partial groESL (1226 bp) and gltA (520 bp) sequences from isolate RAC413R and organisms from five wild raccoons were identical. The sequence analysis supported the findings of the 16S rRNA gene sequence analysis; the groESL sequences were most similar (90.3 and 90.6 %, respectively) to ‘Candidatus N. mikurensis’ strains TK4456 and IS58 (see Supplementary Table S3 for complete similarity matrix). The gltA sequence of RAC413R was most similar (84.4 %) to that of Ehrlichia sp. Belluno (‘Candidatus Ehrlichia walkerii’; GenBank accession no. AY098729), which is most closely related to ‘Candidatus N. mikurensis’ by phylogenetic analyses (Yabsley et al., 2008a) (see Supplementary Table S4 for similarity matrix). The genetic similarities between isolate RAC413R and members of the genus Ehrlichia was only slightly higher than the similarities between the genera Ehrlichia and Anaplasma for the groESL (80.9–83.1 and 71.1–76.5 %, respectively) and gltA (67.9–71.9 and 57.1–67.3 %, respectively) genes. The partial rpoB (781 bp) sequence from RAC413R was 39.9–40.7 % identical to those of Ehrlichia species, 36.0–37.7 % identical to those of Anaplasma species and 35.6–36.4 % identical to those of Neorickettsia species; no rpoB sequences were available from ‘Candidatus N. mikurensis’ for comparison.
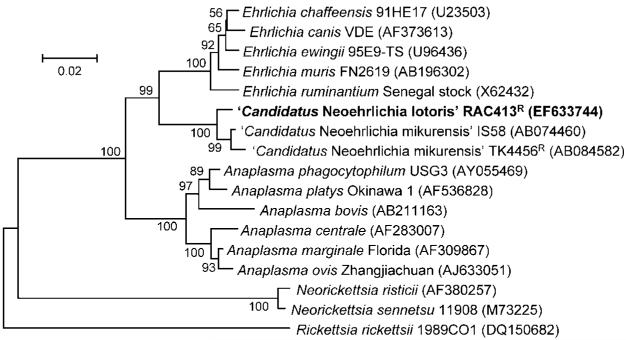
Phylogenetic trees inferred from comparison of the 16S rRNA gene (Fig. 1) and groESL (Fig. 2) sequences provided similar topology and indicated a close relationship of ‘Candidatus N. lotoris’ RAC413R with ‘Candidatus N. mikurensis’. Similar to previous studies (Kawahara et al., 2004; Brouqui et al., 2003), ‘Candidatus N. mikurensis’ was distinct from other genera in the family Anaplasmataceae and formed a well-supported sister clade to the genus Ehrlichia.
Fig. 1.
Phylogenetic tree based on the 16S rRNA gene sequences of ‘Candidatus Neoehrlichia lotoris’ RAC413R and related organisms. Numbers at nodes indicate percentages of bootstrap support based on 1000 replicates. Bar, 0.02 substitutions per nucleotide position.
Fig. 2.
Phylogenetic tree based on the groESL sequences of ‘Candidatus Neoehrlichia lotoris’ RAC413R and related organisms. Numbers at nodes indicate percentages of bootstrap support based on 1000 replicates. Bar, 0.05 substitutions per nucleotide position.
In the current study, we have shown by analysis of several gene targets that an ehrlichial organism recently isolated from a raccoon from the US state of Georgia (Munderloh et al., 2007) is closely related to, but distinct from, ‘Candidatus N. mikurensis’ TK4456R and IS58. 16S rRNA gene and groESL sequences similar to those of ‘Candidatus N. mikurensis’ have been detected from a diverse group of rodents and Ixodes species from Italy, Japan, the Netherlands and Russia (Schouls et al., 1999; Alekseev et al., 2001; Brouqui et al., 2003; Pan et al., 2003; Kawahara et al., 2004; Beninati et al., 2006; Naitou et al., 2006; Rar et al., 2008). All of these current sequences from this group have been lumped together in a single candidate species (‘Candidatus N. mikurensis’). Additional samples from other hosts over a wide geographical range and sequence data from other gene targets are necessary to determine whether ‘Candidatus N. mikurensis’ represents a group of several distinct species or a single, genetically diverse species. In contrast, no differences in sequences of three genes (16S rRNA gene, groESL and gltA) were observed between ‘Candidatus N. lotoris’ RAC413R and five samples from naturally infected raccoons from three US states.
One of the characteristics of ‘Candidatus N. mikurensis’ TK4456R is a lack of serological cross-reactivity with A. phagocytophilum or Ehrlichia muris antigens by IFA (Kawahara et al., 2004). Similarly, no serological cross-reactivity with E. chaffeensis, E. canis, E. ewingii, A. marginale or A. phagocytophilum antigens was noted in four raccoons experimentally infected with ‘Candidatus N. lotoris’ RAC413R. Serological cross-reactions between species of Ehrlichia, Anaplasma and Neorickettsia have been reported (Comer et al., 2000; Rikihisa, 1991), but the level of cross-reactivity is highly variable. The lack of cross-reactivity between ‘Candidatus N. lotoris’ and other ehrlichial organisms may be due to the fact that raccoons develop only a weak immune response to ‘Candidatus N. lotoris’ antigens. Western blot analysis may yield more information about the immunological response of raccoons to ‘Candidatus N. lotoris’.
Members of the family Anaplasmataceae (Aegyptianella, Anaplasma, Ehrlichia, Neorickettsia, Wolbachia, ‘Candidatus Neoehrlichia’ and ‘Candidatus Xenohaliotis’) are obligately intracellular organisms and, although they have similar morphological characters, some genera exhibit unique biological characteristics. Members of the genera Aegyptianella, Anaplasma and Ehrlichia infect a wide range of vertebrate hosts, but all with known life cycles utilize ticks as vectors (Dumler et al., 2001), while members of the genus Neorickettsia infect fish and mammalian hosts and utilize a variety of flukes as vectors (Dumler et al., 2001). In contrast, Wolbachia and ‘Candidatus Xenohaliotis’ do not utilize vectors for transmission; Wolbachia species infect a variety of arthropods and nematodes and are believed to be transmitted primarily transovarially (Dumler et al., 2001), while ‘Candidatus Xenohaliotis californiensis’ is transmitted directly from one host to another (Friedman et al., 2000). Currently, little is known about the natural history of ‘Candidatus N. mikurensis’ and ‘Candidatus N. lotoris’. Many sequences related to, or identical to, ‘Candidatus N. mikurensis’ have been detected in Ixodes ticks, and infections of raccoons with ‘Candidatus N. lotoris’ were associated with tick-infested populations, so these organisms are probably transmitted by ixodid ticks, similar to Ehrlichia and Anaplasma. Future studies should attempt transmission studies and cultivation of ‘Candidatus N. mikurensis’ strains to characterize the proposed genus ‘Candidatus Neoehrlichia’ further.
Description of ‘Candidatus Neoehrlichia lotoris’
‘Candidatus Neoehrlichia lotoris’ [lo.tor’is. L. gen. n. lotoris of a clothes-washer, of Procyon lotor (a raccoon)]. The reference strain is RAC413R. Isolated from a captive raccoon experimentally infected with ‘Candidatus N. lotoris’ by blood inoculation from a naturally infected raccoon from Clarke Co., Georgia, USA (Munderloh et al., 2007). Not infectious to two strains of laboratory mice (Mus musculus, strains BALB/c and C3H), laboratory rats (Rattus norvegicus, Wistar strain) or laboratory rabbits (Oryctolagus cuniculus, New Zealand White) in limited inoculation trials (Yabsley et al., 2008a). Host cells within raccoons are not known. Morphology of organisms in ISE6 tick cells is similar to that of other members of the family Anaplasmataceae. Member of the family Anaplasmataceae based on 16S rRNA gene, groESL and gltA sequences and growth within membrane-bound cytoplasmic vacuoles. Organisms are Gram-negative, highly pleomorphic (compact coccus-like and elongated, wavy forms; 0.5–4 μm), stain light to dark purple with Giemsa and tend to cluster on the side of the endosome nearest the host cell nucleus. Ultrastructurally, the organism has an outer unit membrane separated from the inner cell wall by a thin periplasmic space, characteristic of the family Anaplasmataceae. In some cases, individual bacteria are wrapped in two sets of membranes. Analysis of the 16S rRNA gene sequence revealed a novel 2 bp insertion compared with members of the genus Ehrlichia (AT at base 784 of E. chaffeensis ArkansasT; GenBank accession no. AF416764). Analysis of gltA sequence revealed a novel deletion compared with members of the genera Ehrlichia and Anaplasma (6 bp deletion at base 1011 of E. chaffeensis ArkansasT; GenBank accession no. AF304142) and a 2 bp insertion compared with other members of the family Anaplasmataceae (TTG at base 744 of E. chaffeensis ArkansasT; AF304142). The G+C content of four sequenced genes of ‘Candidatus N. lotoris’ RAC413R is 38.1 mol%. Can be differentiated from ‘Candidatus N. mikurensis’ on the basis of unique 16S rRNA gene, groESL and gltA sequences.
Supplementary Material
ACKNOWLEDGEMENTS
This work was primarily supported by a grant from the Southeast Center for Emerging Biological Threats and the Centers for Disease Control and Prevention to M. J. Y. Additional support was from the University of Georgia Research Foundation. We thank D. Roellig, S. Adams, E. Brown, J. Murdock and M. Savage for animal handling assistance and B. Bonner, N. Seney, T. Cooper and the rest of the Animal Resources staff for excellent animal care. We also thank D. Kavanaugh and J. Smith from APHIS/USDA/WS and K. Pederson of SCWDS for field assistance. We also appreciate the helpful comments of several anonymous reviewers.
Abbreviation
- IFA
immunofluorescence assay.
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA, groESL, gltA and rpoB gene sequences of ‘Candidatus Neoehrlichia lotoris’ RAC413R are EF633744-EF633747, respectively.
Details of reference sequences and similarity matrices for the 16S rRNA gene, groESL, gltA and rpoB sequences are available as supplementary material with the online version of this paper.
REFERENCES
- Alekseev AN, Dubinina HV, Van De Pol I, Schouls LM. Identification of Ehrlichia spp. and Borrelia burgdorferi in Ixodes ticks in the Baltic regions of Russia. J Clin Microbiol. 2001;39:2237–2242. doi: 10.1128/JCM.39.6.2237-2242.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beninati T, Piccolo G, Rizzoli A, Genchi C, Bandi C. Anaplasmataceae in wild rodents and roe deer from Trento Province (northern Italy) Eur J Clin Microbiol Infect Dis. 2006;25:677–678. doi: 10.1007/s10096-006-0196-x. [DOI] [PubMed] [Google Scholar]
- Brouqui P, Sanogo YO, Caruso G, Merola F, Raoult D. Candidatus Ehrlichia walkerii: a new Ehrlichia detected in Ixodesricinus tick collected from asymptomatic humans in Northern Italy. Ann N Y Acad Sci. 2003;990:134–140. doi: 10.1111/j.1749-6632.2003.tb07352.x. [DOI] [PubMed] [Google Scholar]
- Comer JA, Nicholson WL, Paddock CD, Sumner JW, Childs JE. Detection of antibodies reactive with Ehrlichia chaffeensis in the raccoon. J Wildl Dis. 2000;36:705–712. doi: 10.7589/0090-3558-36.4.705. [DOI] [PubMed] [Google Scholar]
- Dugan VG, Gaydos JK, Stallknecht DE, Little SE, Beall AD, Mead DG, Hurd CC, Davidson WR. Detection of Ehrlichia spp. in raccoons (Procyon lotor) from Georgia. Vector Borne Zoonotic Dis. 2005;5:162–171. doi: 10.1089/vbz.2005.5.162. [DOI] [PubMed] [Google Scholar]
- Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, Rikihisa Y, Rurangirwa FR. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol. 2001;51:2145–2165. doi: 10.1099/00207713-51-6-2145. [DOI] [PubMed] [Google Scholar]
- Friedman CS, Andree KB, Beauchamp KA, Moore JD, Robbins TT, Shields JD, Hedrick RP. ‘Candidatus Xenohaliotis californiensis’, a newly described pathogen of abalone, Haliotis spp., along the west coast of North America. Int J Syst Evol Microbiol. 2000;50:847–855. doi: 10.1099/00207713-50-2-847. [DOI] [PubMed] [Google Scholar]
- Inokuma H, Brouqui P, Drancourt M, Raoult D. Citrate synthase gene sequence: a new tool for phylogenetic analysis and identification of Ehrlichia. J Clin Microbiol. 2001;39:3031–3039. doi: 10.1128/JCM.39.9.3031-3039.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara M, Rikihisa Y, Isogai E, Takahashi M, Misumi H, Suto C, Shibata S, Zhang C, Tsuji M. Ultrastructure and phylogenetic analysis of ‘Candidatus Neoehrlichia mikurensis’ in the family Anaplasmataceae, isolated from wild rats and found in Ixodes ovatus ticks. Int J Syst Evol Microbiol. 2004;54:1837–1843. doi: 10.1099/ijs.0.63260-0. [DOI] [PubMed] [Google Scholar]
- Little SE, Stallknecht DE, Lockhart JM, Dawson JE, Davidson WR. Natural coinfection of a white-tailed deer (Odocoileus virginianus) populations with three Ehrlichia spp. J Parasitol. 1998;84:897–901. [PubMed] [Google Scholar]
- Munderloh UG, Yabsley MJ, Murphy SM, Luttrell MP, Howerth EW. Isolation and establishment of the raccoon Ehrlichia-like agent in tick cell culture. Vector Borne Zoonotic Dis. 2007;7:418–425. doi: 10.1089/vbz.2007.0640. [DOI] [PubMed] [Google Scholar]
- Naitou H, Kawaguchi D, Hishimura Y, Inayoshi M, Kawamori F, Masuzawa T, Hiroi M, Kurashige H, Kawabata H. Molecular identification of Ehrlichia species and ‘Candidatus Neoehrlichia mikurensis’ from ticks and wild rodents in Shizuoka and Nagano Prefectures, Japan. Microbiol Immunol. 2006;50:45–51. doi: 10.1111/j.1348-0421.2006.tb03769.x. other authors. [DOI] [PubMed] [Google Scholar]
- Pan H, Liu S, Ma Y, Tong S, Sun Y. Ehrlichia-like organism gene found in small mammals in the suburban district of Guangzhou of China. Ann N Y Acad Sci. 2003;990:107–111. doi: 10.1111/j.1749-6632.2003.tb07346.x. [DOI] [PubMed] [Google Scholar]
- Rar VA, Livanova NN, Panov VV, Kozlova IV, Pukhovskaya NM, Vysochina NP, Tkachev SE, Ivanov LI. Prevalence of Anaplasma and Ehrlichia species in Ixodes persulcatus ticks and small mammals from different regions of the Asian part of Russia. Int J Med Microbiol. 2008;298(Suppl. 1):222–230. [Google Scholar]
- Rikihisa Y. Cross-reacting antigens between Neorickettsia helminthoeca and Ehrlichia species, shown by immunofluorescence and western immunoblotting. J Clin Microbiol. 1991;29:2024–2029. doi: 10.1128/jcm.29.9.2024-2029.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikihisa Y, Zhang C, Christensen BM. Molecular characterization of Aegyptianella pullorum (Rickettsiales, Anaplasmataceae) J Clin Microbiol. 2003;41:5294–5297. doi: 10.1128/JCM.41.11.5294-5297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanogo YO, Parola P, Shpynov S, Camicas JL, Brouqui P, Caruso G, Raoult D. Genetic diversity of bacterial agents detected in ticks removed from asymptomatic patients in north-eastern Italy. Ann N Y Acad Sci. 2003;990:182–190. doi: 10.1111/j.1749-6632.2003.tb07360.x. [DOI] [PubMed] [Google Scholar]
- Schouls LM, Van De Pol I, Rijpkema SG, Schot CS. Detection and identification of Ehrlichia, Borrelia burgdorferi sensu lato, and Bartonella species in Dutch Ixodes ricinus ticks. J Clin Microbiol. 1999;37:2215–2222. doi: 10.1128/jcm.37.7.2215-2222.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara K, Arai S, Kawabuchi T, Itagaki A, Ishihara C, Satoh H, Okabe N, Tsuji M. Molecular survey of Babesia microti, Ehrlichia species and Candidatus Neoehrlichia mikurensis in wild rodents from Shimane Prefecture, Japan. Microbiol Immunol. 2007;51:359–367. doi: 10.1111/j.1348-0421.2007.tb03923.x. [DOI] [PubMed] [Google Scholar]
- Taillardat-Bisch AV, Raoult D, Drancourt M. RNA polymerase β-subunit-based phylogeny of Ehrlichia spp., Anaplasma spp., Neorickettsia spp. and Wolbachia pipientis. Int J Syst Evol Microbiol. 2003;53:455–458. doi: 10.1099/ijs.0.02411-0. [DOI] [PubMed] [Google Scholar]
- Yabsley MJ, Dugan VG, Stallknecht DE, Little SE, Lockhart JM, Dawson JE, Davidson WR. Evaluation of a prototype Ehrlichia chaffeensis surveillance system using white-tailed deer (Odocoileus virginianus) as natural sentinels. Vector Borne Zoonotic Dis. 2003;3:195–207. doi: 10.1089/153036603322662183. [DOI] [PubMed] [Google Scholar]
- Yabsley MJ, Murphy SM, Luttrell MP, Wilcox BR, Ruckdeschel C. Raccoons (Procyon lotor), but not rodents, are natural and experimental hosts for an ehrlichial organism related to “Candidatus Neoehrlichia mikurensis”. Vet Microbiol. 2008a;131:301–308. doi: 10.1016/j.vetmic.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Yabsley MJ, Murphy SM, Luttrell MP, Little SE, Massung RF, Stallknecht DE, Conti LA, Blackmore CGM, Durden LA. Experimental and field studies on the suitability of raccoons (Procyon lotor) as hosts for tick-borne pathogens. Vector Borne Zoonotic Dis. 2008b;8:491–504. doi: 10.1089/vbz.2007.0240. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.