Abstract
The prevalence of active drug efflux pump and porin alterations was investigated in Turkish nosocomial strains of Klebsiella pneumoniae exhibiting a multidrug-resistant phenotype. MICs of various antibiotics, including quinolones, chloramphenicol, tetracycline, and β-lactams, for those strains were determined either with or without the efflux pump inhibitor phenylalanine arginine β-naphthylamide (PAβN). Thirty-nine percent of the strains exhibited a PAβN-modulated resistance for quinolones, chloramphenicol, and tetracycline. In these strains, a significant increase of chloramphenicol accumulation was gained in the presence of the efflux pump inhibitor PAβN or with the energy uncoupler carbonyl cyanide m-chlorophenylhydrazone. Moreover, high-level expression of the membrane fusion protein AcrA, which was immunodetected in most of those isolates, suggests that the AcrAB/TolC efflux machinery contributed to their antibiotic resistance. Studies of K. pneumoniae porins indicated that the majority of the strains, including extended-spectrum β-lactamase producers and efflux-positive ones, presented an alteration in their sorbitol-sensitive porin (OmpK35) expression. This is the first report showing the prominent role of active drug efflux in the antibiotic resistance of nosocomial K. pneumoniae strains from Turkey.
Increasing prevalence of multiple-antibiotic resistance among nosocomial strains of gram-negative bacteria is an emerging problem worldwide. The role of target modifications and enzymatic modifications of the drugs in the multiple antibiotic resistance of bacteria has been extensively reported, whereas more rare reports underlined the participation of multidrug efflux pump systems in the multidrug-resistant (MDR) phenotype (1, 5, 12, 17, 20-22, 26). Genes encoding MDR pumps are normal constituents of bacterial chromosomes and thus provide to bacteria the intrinsic potential to develop the MDR phenotype without acquisition of antibiotic resistance genes (5, 6). The activation of multidrug efflux pump genes by mutations or induction caused by stress of exposure to xenobiotics results in overexpression of pumps (1, 5, 6, 22, 24). Thus, bacteria may become resistant to most of the antibiotics that are expelled by these efflux machineries. Among MDR efflux pump mechanisms, AcrAB/TolC in Escherichia coli and several Mex pumps in Pseudomonas aeruginosa have been well studied, such as the resistance-nodulation-division active drug efflux systems (5, 17, 20-22, 24). Similar resistance-nodulation-division pumps are involved in other gram-negative bacterial species, including Klebsiella spp., Enterobacter spp., and Salmonella spp. (5, 16, 18, 26, 27). It is also important to note that in several documented MDR clinical isolates, the efflux mechanism is often associated with a modification of outer membrane permeability via the loss of major porins (5, 21, 22, 25).
The main goal of the present study was to determine the role of the phenotype of drug efflux in several MDR Klebsiella pneumoniae clinical isolates.
MATERIALS AND METHODS
Bacterial strains and media.
Eighteen MDR K. pneumoniae strains were isolated from different patients during 6 months in 2002 at the University Hospital in Istanbul. The origins of the strains are shown in Tables 1 and 2. Identification of strains was performed by using both the VITEK automated system (bioMérieux, Marcy l’Etoile, France) and API 20E (Api-bioMérieux Systems). K. pneumoniae ATCC 11296 was used as the reference strain in determinations of MICs for the various clinical isolates. K. pneumoniae Kp63, a porin-deficient clinical strain (2); K. pneumoniae ATCC 11296; E. coli BW5104 (26), which expresses AcrA at a basal level; and Enterobacter aerogenes ATCC 13048(pJS04) (ompX mutant with its promoter [29]), a strain that overexpresses OmpX, were used as controls for protein analysis. Bacteria were grown either in Mueller-Hinton (MH) broth and agar or in Luria-Bertani (LB) broth or nutrient broth (Difco Laboratories, Detroit, Mich.) at 37°C.
TABLE 1.
Susceptibilities of K. pneumoniae isolates in group A to various antibiotics with and without an efflux pump inhibitor, PAβN
| Strain | Sourced of strain | MIC (μg/ml)a
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OFX | NOR | NAL | CHL | TET | FEP | CAZ | FOX | IPM | MEM | ESBL | ||
| K80 | TAd | 32 | 32 | ≥512 | 128 | ≥128 | 2 | 8 | 16 | 0.125 | ≤0.06 | − |
| K80 + PAβNb | 1 | 8 | 32 | 2 | ≥128 | 1 | ||||||
| K89 | Blood | 32 | 256 | ≥512 | 256 | ≥128 | 2 | 1 | 32 | 0.5 | ≤0.06 | − |
| K89 + PAβN | 0.5 | 4 | 32 | 2 | ≥128 | 0.125 | ||||||
| K2 | Urine | 2 | 2 | 64 | 32 | 8 | 16 | ≥512 | 64 | 1 | 0.25 | + |
| K2 + PAβN | ≤0.06 | 0.25 | 0.25 | 1 | 0.25 | 4 | ||||||
| K32 | Wound | 0.125 | 0.25 | 16 | 16 | 8 | 32 | ≥512 | 64 | 1 | ≤0.06 | + |
| K32 + PAβN | ≤0.06 | 0.125 | 0.25 | 1 | 2 | 8 | ||||||
| K33 | Urine | 1 | 1 | 32 | ≥512 | ≥128 | 2 | 64 | 8 | 0.25 | ≤0.06 | + |
| K33 + PAβN | ≤0.06 | 0.125 | 0.25 | 32 | ≥128 | 1 | ||||||
| K74 | TA | 1 | 1 | 16 | 32 | 8 | 16 | ≥512 | 64 | 1 | ≤0.06 | + |
| K74 + PAβN | ≤0.06 | 0.25 | 0.25 | 0.5 | 4 | 4 | ||||||
| K121 | CSF | 0.125 | 0.5 | 4 | 64 | 4 | 128 | ≥512 | 64 | 1 | 0.125 | + |
| K121 + PAβN | ≤0.06 | 0.125 | 0.25 | 0.5 | 0.125 | NDe | ||||||
| ATCC 11296c | 0.125 | 0.25 | 8 | 8 | 1 | 0.125 | 0.5 | 4 | 0.5 | ND | ND | |
Antibiotics were tested alone or with PAβN. MICs were determined in MH broth according to NCCLS guidelines (19). MICs were obtained from three independent measurements. Abbreviations: OFX, ofloxacin; NOR, norfloxacin; NAL, nalidixic acid; CHL, chloramphenicol; TET, tetracycline; FEP, cefepime; CAZ, ceftazidine; FOX cefoxitin; IPM, imipenem; MEM, meropenem.
PAβN, 50 μM PAβN in each test tube.
K. pneumoniae reference strain.
Abbreviations: TA, tracheal aspiration; CSF, cerebrospinal fluid. ESBL, extended-spectrum β-lactamase.
ND, not determined.
TABLE 2.
Susceptibilities of K. pneumoniae isolates in group B to various antibiotics with and without an efflux pump inhibitor, PAβN
| Strain | Source of strain | MIC (μg/ml)a
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OFX | NOR | NAL | CHL | TET | FEP | CAZ | FOX | IPM | MEM | ESBLe | ||
| K41 | Urine | 128 | ≥512 | ≥512 | ≥512 | 16 | 4 | 128 | 64 | 0.5 | ≤0.06 | + |
| K41 + PAβNb | 16 | 256 | ≥512 | 64 | 4 | 4 | ||||||
| K17 | Urine | 64 | 512 | ≥512 | ≥512 | ≥128 | ≥512 | ≥512 | 64 | 0.25 | ≤0.06 | + |
| K17 + PAβN | 16 | 256 | ≥512 | 128 | ≥128 | ≥512 | ||||||
| K95 | Urine | 32 | 64 | ≥512 | ≥512 | 8 | 256 | ≥512 | 16 | 0.5 | ≤0.06 | + |
| K95 + PAβN | 4 | 64 | ≥512 | 128 | 4 | 256 | ||||||
| K100 | Urine | 32 | 512 | ≥512 | ≥512 | ≥128 | 256 | ≥512 | 32 | 0.5 | 2 | + |
| K100 + PAβN | 8 | 128 | ≥512 | 64 | ≥128 | 256 | ||||||
| K118 | TAc | 32 | 64 | ≥512 | 512 | ≥128 | 64 | 2 | 32 | 0.25 | 0.125 | + |
| K118 + PAβN | 4 | 16 | 256 | 32 | ≥128 | 8 | ||||||
| K116 | Wound | 32 | 64 | ≥512 | ≥512 | ≥128 | 32 | 2 | 32 | 0.5 | ≤0.06 | + |
| K116 + PAβN | 2 | 32 | 256 | 64 | ≥128 | 16 | ||||||
| K216 | Urine | 32 | 128 | ≥512 | 16 | ≥128 | 0.125 | 0.25 | 16 | 0.25 | 0.125 | − |
| K216 + PAβN | 8 | 128 | 512 | 2 | ≥128 | NDd | ||||||
| K192 | Urine | 32 | 256 | ≥512 | 512 | ≥128 | 0.5 | 0.25 | 8 | 0.25 | ≤0.06 | − |
| K192 + PAβN | 4 | 128 | 64 | 64 | ≥128 | ND | ||||||
| K104 | Blood | 4 | 32 | ≥512 | 256 | ≥128 | 0.125 | 0.125 | 4 | 1 | ≤0.06 | − |
| K104 + PAβN | 1 | 16 | 256 | 64 | 32 | ND | ||||||
| K128 | Urine | 4 | 32 | ≥512 | 4 | 4 | ≤0.06 | 0.25 | 4 | 0.5 | ≤0.06 | − |
| K128 + PAβN | 1 | 16 | 256 | 2 | 2 | ND | ||||||
| K132 | Urine | 4 | 16 | ≥512 | ≥512 | ≥128 | ≤0.06 | 0.25 | 16 | 1 | ≤0.06 | − |
| K132 + PAβN | 1 | 16 | 256 | 64 | 16 | ND | ||||||
Antibiotics were tested alone or with PAβN. MICs were determined in MH broth according to NCCLS guidelines (19). MICs were obtained from three independent measurements. Abbreviations: OFX, ofloxacin; NOR, norfloxacin; NAL, nalidixic acid; CHL, chloramphenicol; TET, tetracycline; FEP, cefepime; CAZ, ceftazidine; FOX cefoxitin; IPM, imipenem; MEM, meropenem.
PAβN, 50 μM PAβN in each test tube.
TA, tracheal aspiration.
ND, not determined.
ESBL, extended-spectrum β-lactamase.
Antibiotic susceptibility testing and ESBL detection.
MIC determinations for four structurally unrelated classes of bacterial drugs were carried out using a twofold broth dilution method in MH broth according to the National Committee for Clinical Laboratory Standards (NCCLS) guidelines (19) to evaluate the MDR phenotype. The following antibiotics were purchased from Sigma Chemical Co. (Saint Quentin Fallavier, France): ofloxacin, norfloxacin, nalidixic acid, chloramphenicol, and tetracycline. Cefepime, ceftazidime, cefoxitin, meropenem, and imipenem were provided by Bristol-Myers Squibb (Syracuse, N.Y.), Roche (Neuilly-Sur-Seine, France), Glaxo-Wellcome S.p.A (Verona, Italy), PanPharma S.A. (Fougères, France), Imperial Chemical Industries PLC (London, Great Britain), Merck Sharp and Dohme (Chibert, France), and Sanofi-Synthelabo (Paris, France), respectively. MIC determinations were also carried out with fixed concentrations (50 μM) of the efflux pump inhibitor phenylalanine arginine β-naphthylamide (PAβN) (Sigma Chemical Co.) against quinolones, chloramphenicol, tetracycline, and cefepime (4, 13). The double-disk synergy test was used as a screening test for detecting extended-spectrum β-lactamase (ESBL)-producing strains. Cefotaxime (30-μg), ceftazidime (30-μg), and aztreonam (30-μg) disks were placed on MH agar adjacent to a clavulanate-amoxicillin disk (20 μg of amoxicillin plus 10 μg of clavulanate). Antibiotic-containing disks were from Becton Dickinson Microbiology Systems (Sparks, Md.). The procedures and interpretation of the double-disk synergy test were as described previously (10).
Chloramphenicol accumulation test.
Measurement of [14C]chloramphenicol uptake by intact cells was adapted from methods described in previous studies (2, 4). Exponential-phase bacteria grown in LB broth were pelleted, washed once, and suspended to a density of 1010 CFU/ml in 50 mM sodium phosphate buffer, pH 7, containing 5 mM magnesium chloride. [14C]chloramphenicol (specific radioactivity, 59.46 mCi/mmol) was added to 600 μl of cell suspension at 37°C in a shaking water bath, yielding a final chloramphenicol concentration of 5 μM. At various intervals, 100 μl of the suspension was removed and immediately filtered through GF/C filters (Whatman, Maidstone, Kent, United Kingdom). After three washes with 5 ml of 50 mM sodium phosphate buffer (pH 7) containing 0.1 M lithium chloride, the filters were dried and the radioactivity was measured by using a Packard scintillation counter. Inhibition assays were performed in the presence of an energy uncoupler, carbonyl cyanide m-chlorophenylhydrazone (CCCP), and PAβN at final inhibitor concentrations of 50 and 200 μM, respectively. The MICs of CCCP were above 500 μM for strains ATCC 11296, K80, K89, and K128 (data not shown).
SDS-polyacrylamide gel electrophoresis and immunodetection of AcrA, OmpA, OmpX, and porins.
Cell pellets were prepared from exponential-phase bacteria grown in MH broth and then they were solubilized in loading buffer at 96°C. Samples (0.02 optical density at 600 nm) were loaded on sodium dodecyl sulfate (SDS)-polyacrylamide gels (10% polyacrylamide, 0.1% SDS) and run at 160 V for 1 h (2). Electrotransfer of the resulting bands to nitrocellulose membranes was carried out with 0.05% SDS. After an initial saturating step with Tris-buffered saline (50 mM Tris-HCl, 150 mM NaCl, pH 8) containing 10% skim milk powder at 4°C, nitrocellulose membranes were incubated in Tris-buffered saline containing 10% bovine serum and 0.2% Triton X-100 for 2 h at room temperature in the presence of polyclonal antibodies directed against AcrA or OmpA protein (14, 26). After three washes with the same buffer, detection was performed with alkaline phosphatase-conjugated affinitiPure goat anti-rabbit immunoglobulin G antibodies (Jackson Immuno-Research, West Grove, Pa.). Evaluation of the AcrA and OmpA amounts was made by measuring the intensity of immunoblotting bands exhibited by clinical and control strains. The level of AcrA expression in the control strains (E. coli BW5104 and K. pneumoniae ATCC 11296) was rated as positive (+), and that in the strains overexpressing AcrA protein was recorded as slightly or highly overexpressed (++ and +++, respectively). For OmpX, the same conditions were applied as described for OmpA/AcrA analyses, and protein analysis was performed with a 12% polyacrylamide gel. Polyclonal antipeptide antibodies directed against AcrA, OmpX, and OmpA were used as previously described (4, 14, 26). Evaluation of the OmpX expression levels was made as mentioned for AcrA protein, and E. aerogenes ATCC 13048(pJS04) was used as the positive strain.
Electrophoresis for detection of major K. pneumoniae porins was carried out with an SDS-11% polyacrylamide gel. Cell pellets prepared from both low- and high-osmolality media were used for immunodetection of major porins. The expression of OmpK35 porin has been reported to be downregulated by sorbitol; in contrast, OmpK36 is overexpressed in this high-osmolality medium (9). Our previous studies have shown a strong cross-immunoreactivity between E. coli and K. pneumoniae porins (28). Polyclonal antibodies directed against E. coli OmpC and OmpF porins have been used for the detection of main porins of K. pneumoniae strains (3, 28). The other conditions were the same as those described for OmpA, AcrA, and OmpX immunodetection. Evaluation of the OmpK35 and OmpK36 expressions was made by comparing the immunoblotting signals exhibited by the clinical strains and the control strains (K. pneumoniae Kp63 and K. pneumoniae ATCC 11296).
RESULTS
Antibiotic susceptibility testing.
The 18 K. pneumoniae strains showed a significant degree of multiresistance according to the noticeable resistance level observed with various antibiotics (Tables 1 and 2). For ESBL-producing strains, aztreonam MICs ranged from 256 to ≥512 μg/ml, whereas for non-ESBL producers, the highest aztreonam MIC was 2 μg/ml (data not shown). The isolates could be divided into two major groups according to the results obtained with antibiotic susceptibility tests performed in the presence of the efflux pump inhibitor PAβN (Tables 1 and 2). A noticeable part (39%) of the K. pneumoniae collection exhibited a PAβN-sensitive resistance mechanism (Table 1). In group A isolates (Table 1) PAβN showed its significant effect on MICs of quinolones, chloramphenicol, and/or tetracycline. MICs were reduced by fivefold for at least one of these antibiotic classes tested with this efflux inhibitor (Table 1). Furthermore the strains in group A showed two kinds of susceptibility patterns against different types of quinolones. The MICs of all types of quinolones for K80 and K89 were similarly higher than those for other strains in this group, even when the measure was carried out in the presence of PAβN. For the remaining isolates (K2, K32, K33, K74, and K121) in this group, the MICs of quinolone were lower and the PAβN effect was particularly significant on ofloxacin MICs (Table 1). Concerning group B, the MICs of all antibiotics tested were generally higher for these isolates. In addition, in this group the effect of PAβN on antibiotic susceptibility was strongly reduced compare to group A results (Tables 1 and 2).
Chloramphenicol accumulation test.
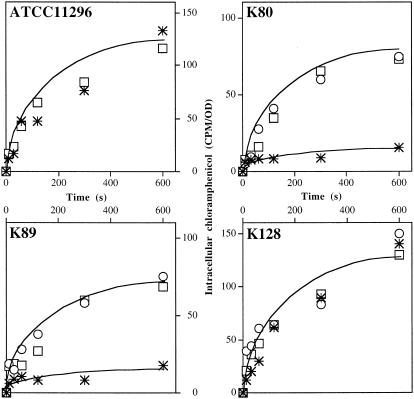
In order to identify the presence of an efflux pump in PAβN-sensitive isolates, measurement of intracellular accumulation of chloramphenicol in the presence of two compounds, the efflux pump inhibitor PAβN and the membrane energy uncoupler CCCP, was performed (13, 20). Several strains belonging to groups A and B were tested for chloramphenicol accumulation capacity with and without PAβN and CCCP. Figure 1 illustrates the results obtained with clinical strains. In group A, K80 and K89 showed a PAβN-sensitive drug phenotype. A low level of intracellular accumulation of chloramphenicol was found in the absence of PAβN or CCCP in these isolates. In contrast, a three- to fourfold increase of intracellular drug concentration was obtained with inhibitors. In the PAβN nonresponding isolate, K128 (group B), the presence of the inhibitors during the incubation time did not lead to any significant change in chloramphenicol accumulation, and similar results were obtained with the ATCC 11296 strain (Fig. 1). These results indicated that an active efflux mechanism contributes to antibiotic resistance in isolates K80 and K89, which belong to group A, by decreasing the intracellular drug concentration. Activity of this efflux mechanism was significantly inhibited by the addition of PAβN or energy uncoupler.
FIG. 1.
Effect of efflux inhibitors on chloramphenicol intracellular accumulation in K. pneumoniae ATCC 11296 and isolates K80, K89, and K128. Exponential-phase bacteria in LB broth were removed, resuspended in sodium phosphate buffer, and incubated with radiolabeled chloramphenicol for various times. The experiments were carried out in the absence (✳) or in the presence (○) of CCCP or in the presence of PAβN (□). Values (expressed as counts per minute/optical density) were obtained from two independent experiments.
Analyses of membrane protein profile.
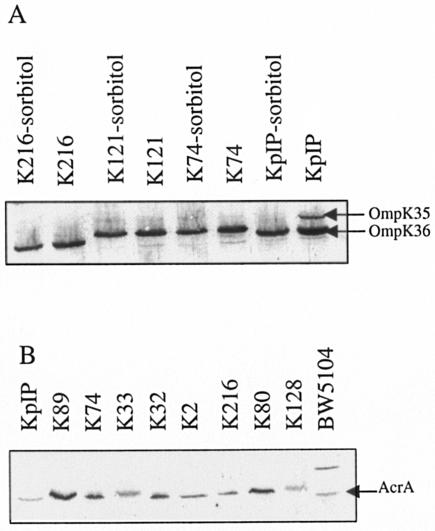
Hernández-Allés et al. have previously shown that sorbitol downregulates the expression of OmpK35 porin in K. pneumoniae (9). Consequently, we investigated the presence of porins in our isolates grown with and without sorbitol. A majority of the strains (12 of 18) did not express OmpK35, even in the low-osmolality medium (Table 3; Fig. 2). In contrast, the expression of OmpK36 porin was elevated in all isolates, and high-osmolality conditions did not modify its biosynthesis level.
TABLE 3.
Expression of various proteins by K. pneumoniae strainsh
| Isolate | Expression of:
|
||||||
|---|---|---|---|---|---|---|---|
| OmpA | AcrA | OmpK35
|
OmpK36
|
OmpX | |||
| S−f | S+g | S− | S+ | ||||
| Group A | |||||||
| K80 | + | +++ | − | − | + | + | + |
| K89 | + | +++ | − | − | + | + | + |
| K2 | + | + | ± | − | + | + | + |
| K32 | + | +++ | ± | − | + | + | + |
| K33 | + | + | + | ± | + | + | + |
| K74 | + | +++ | − | − | + | + | + |
| K121 | + | ++ | − | − | + | + | + |
| Group B | |||||||
| K41 | + | ++ | − | − | + | + | + |
| K17 | + | + | − | − | + | + | + |
| K95 | + | ± | − | − | + | + | + |
| K100 | + | ± | − | − | + | + | + |
| K118 | + | +++ | − | − | + | + | + |
| K116 | + | +++ | − | − | + | + | + |
| K216 | + | + | − | − | + | + | + |
| K192 | + | + | − | − | + | + | + |
| K104 | + | + | + | − | + | + | + |
| K128 | + | ± | + | − | + | + | + |
| K132 | + | + | + | − | + | + | + |
| Control strains | |||||||
| ATCC 11296a | + | + | + | − | + | + | + |
| Kp 63b | + | NDe | − | − | − | − | ND |
| BW5104c | + | + | ND | ND | ND | ND | ND |
| ATCC 13048 (pJS04)d | + | ND | ND | ND | ND | ND | +++ |
K. pneumoniae reference strain.
K. pneumoniae 63 is the control strain used for immunodetection of porins (2).
E. coli BW5104, the control strain used for AcrA immunodetection (26).
E. aerogenes-type ATCC 13048 carrying pJS04 (ompX with his promoter); control strain used for OmpX immunodetection.
ND, not determined.
S−, nutrient broth without sorbitol.
S+, nutrient broth with sorbitol.
Expression of the various proteins was evaluated from immunodetection by Western blotting as previously described (4, 14). Symbols: +++, highly increased signal; ++, increased signal; +, normal signal; ±, weak signal; −, negative signal.
FIG. 2.
Immunodetection of OmpK35, OmpK36, and AcrA in K. pneumoniae strains. Immunoblot assays of whole-cell extracts were carried out with antibodies directed against porins (A) or AcrA (B) as previously described (26, 28). Bacteria, including control strains, were grown with or without sorbitol for the analyses of porin expression. K. pneumoniae ATCC 11296 (KpIP) was used as the control strain in porin immunodetection. E. coli BW5104 (26), a strain expressing AcrA protein at a basal level, was used as a control strain in AcrA immunodetection. Only the relevant part of the immunoblots is shown.
Expression of OmpX has been previously reported to negatively regulate porin synthesis (29). Therefore, we determined its synthesis by immunodetection in our isolates. Interestingly, no significant variation of OmpX expression was detected in any of the strains (Table 3). Immunodetection of OmpA, which plays a key role in the conservation of the membrane architecture, was always positive (Table 3).
Concerning the efflux mechanism, we investigated the synthesis of AcrA, a component of the major pump in Enterobacteriaceae. The overproduction of this efflux protein has been previously reported in E. aerogenes clinical isolates that exhibit an antibiotic efflux mechanism (4, 26). In group A, five strains expressed a high level of AcrA, as illustrated in Fig. 2 and summarized in Table 3. Only two strains (K118 and K116) belonging to group B exhibited an increased AcrA expression level (Table 2; Fig. 2).
DISCUSSION
Turkey is one of the countries with a high prevalence of the MDR phenotype in K. pneumoniae (8, 11, 23). In this study, we have investigated the role of an active drug efflux mechanism in the antibiotic resistance of eighteen MDR K. pneumoniae strains from that country. The activity of the drug efflux pump(s) was identified in 39% of the K. pneumoniae strains. A significant decrease in MICs of quinolones, chloramphenicol, and/or tetracycline was obtained with an efflux pump inhibitor, PAβN, in these strains (Table 1). Concerning group A, we observed two different quinolone susceptibility patterns and also two kinds of response to the PAβN effect. The effect of the efflux inhibitor on high MICs of quinolone for two strains (K80 and K 89) in this group was significant but it was not sufficient to reduce these MICs into the sensitivity ranges. This is the case for norfloxacin and nalidixic acid (Table 1). Mutations in the target proteins, DNA gyrase and topoisomerases IV, are probably involved in the quinolone resistance of those isolates in addition to the expression of efflux pump machinery. PAβN′s effect on the MICs of quinolone for the other strains (K2, K32, K33, K74, and K121) in group A was more significant and sufficient to restore a susceptible profile particularly for nalidixic acid. This result suggested that the major quinolone resistance mechanism found in those strains is a PAβN-sensitive mechanism, namely, a drug efflux mechanism. The results of MICs of drugs plus PAβN were confirmed by chloramphenicol accumulation tests carried out with PAβN or CCCP for certain group A strains (K80 and K89). A large increase in the intracellular concentration of chloramphenicol was generated by the uncoupler CCCP and by the pump inhibitor PAβN in those two strains. In the strain K128 from group B, for which the MICs were nonsensitive to PAβN, no variation in the level of chloramphenicol accumulation was observed after the treatment with these efflux inhibitors. On the basis of these results, we may conclude that the PAβN-sensitive efflux pump system detected in the strains from group A depends on the proton motive force, which is collapsed by the energy uncoupler (20). On the other hand, the high expression level of AcrA protein observed in most of the isolates belonging to group A (five of seven strains) is strong evidence for overexpression of the AcrAB-mediated efflux mechanism in those isolates. This overproduction strongly contributes to their multiple-antibiotic resistance. In E. aerogenes, we have shown previously that the AcrAB/TolC complex participates in chloramphenicol, quinolone, and tetracycline efflux (4, 26) and this pump is inhibited by CCCP and PAβN (4, 14, 15). The results of this study correlated well with recent reports indicating the role of AcrA in ciprofloxacin resistance of K. pneumoniae isolates (18, 27). Taking into account these data and the results presented here, AcrAB/TolC may be the major efflux machinery functioning in MDR K. pneumoniae clinical isolates. It is also important to note that all strains overexpressing the AcrA component also presented alterations in their porin profile and none of them expressed OmpK35 porin in both high- and low-osmolality culture media. On the other hand, OmpK35 deficiency was also detected in most of the other strains, including ESBL producers, in our K. pneumoniae collection. The other major porin, OmpK36, was expressed by all isolates. In all clinical isolates, no variation was detected in OmpX and OmpA synthesis. These observations suggested the absence of a pleitropic alteration impairing the outer membrane protein expression or assembly in these strains (14). Taking into account the simultaneous overproduction of AcrA and alteration of the porin profile, we propose that the complex marA genetic cascade induces the decrease of porin expression via special regulation, such as that of micF, and the activation of efflux pump synthesis in these strains (1, 5, 6, 21, 27).
Finally, the efflux pump mechanism, AcrAB/TolC, significantly contributes to antibiotic resistance in our K. pneumoniae strains. Interestingly, overexpression of efflux pump machinery has been recently shown among nosocomial K. pneumoniae strains during a hospital outbreak (7). Further epidemiologic surveys are necessary to better understand the prevalence of efflux pump activation in the emergence of an MDR phenotype in K. pneumoniae, and the PAβN protocol may be a good indicator in the screening of efflux pump activation.
Acknowledgments
U. Over Hasdemir was supported by a postdoctoral fellowship program from NATO (National Administrator: The Scientific and Technical Research Council of Turkey - TUBITAK). This work was supported by the Université de la Méditerranée and by an AstraZeneca/ESCMID Wall of Resistance Research Grant (J.-M. Pagès and P. Nordmann).
We thank Aventis Hoescht Marion Roussel (Romainville, France) for the gift of radiolabeled chloramphenicol. We thank C. Bollet, A. Davin-Regli, and E. Pradel for fruitful discussions.
REFERENCES
- 1.Alekshun, M. N., and S. B. Levy. 1999. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 7:410-413. [DOI] [PubMed] [Google Scholar]
- 2.Chevalier, J., J.-M. Pagès, A. Eyraud, and M. Malléa. 2000. Membrane permeability modifications are involved in antibiotic resistance in Klebsiella pneumoniae. Biochem. Biophys. Res. Commun. 274:496-499. [DOI] [PubMed] [Google Scholar]
- 3.Dé, E., A. Basle, M. Jaquinod, N. Saint, M. Malléa, G. Molle, and J.-M. Pagès. 2001. A new mechanism of antibiotic resistance in Enterobacteriaceae induced by a structural modification of the major porin. Mol. Microbiol. 41:189-198. [DOI] [PubMed] [Google Scholar]
- 4.Gayet, S., R. Chollet, G. Molle, J.-M. Pagès, and J. Chevalier. 2003. Modification of outer membrane protein profile and evidence suggesting an active drug pump in Enterobacter aerogenes clinical strains. Antimicrob. Agents Chemother. 47:1555-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.George, A. M. 1996. Multidrug resistance in enteric and other Gram-negative bacteria. FEMS Microbiol. Lett. 139:1-10. [DOI] [PubMed] [Google Scholar]
- 6.Grkovic, S., M. H. Brown, and R. A. Skurray. 2001. Transcriptional regulation of multidrug efflux pumps in bacteria. Semin. Cell Dev. Biol. 12:225-237. [DOI] [PubMed] [Google Scholar]
- 7.Gruteke, P., W. Goessens, J. Gils, P. Peerbooms, N. Lemmens-den Toom, M. Santen-Verheuvel, A. Belkum, and H. Verbrugh. 2003. Patterns of resistance associated with integrons, the extended-spectrum β-lactamase SHV-5 gene, and a multidrug efflux pump of Klebsiella pneumoniae causing a nosocomial outbreak. J. Clin. Microbiol. 41:1161-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunseren, F., L. Mamikoglu, and S. Ozturk. 1999. A surveillance study of antimicrobial resistance of gram-negative bacteria isolated from intensive care units in eight hospitals in Turkey. J. Antimicrob. Chemother. 43:373-378. [DOI] [PubMed] [Google Scholar]
- 9.Hernández-Allés, S., S. Albertí, X. D. Álvarez, A. Doménech-Sánchez, L. Martínez-Martínez, J. Gil, J. M. Tomás, and V. J. Benedí. 1999. Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiology 145:673-679. [DOI] [PubMed] [Google Scholar]
- 10.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 11.Koseoglu, O., S. Kocagoz, D. Gur, and M. Akova. 2001. Nosocomial bloodstream infections in a Turkish University hospital: study of gram-negative bacilli and their sensitivity patterns. Int. J. Antimicrob. Agents 17:477-478. [DOI] [PubMed] [Google Scholar]
- 12.Levy, S. B. 2002. Active efflux, a common mechanism for biocide and antibiotic resistance. J. Appl. Microbiol. 92(Suppl.):65S-71S. [PubMed] [Google Scholar]
- 13.Lomovskaya, O., M. S. Warren, A. Lee, J. Galazzo, R. Fronko, M. Lee, J. Blais, D. Cho, S. Chamberland, T. Renau, R. Leger, S. Hecker, W. Watkins, K. Hoshino, H. Ishida, and V. J. Lee. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malléa, M., J. Chevalier, C. Bornet, A. Eyraud, A. Davin-Regli, C. Bollet, and J.-M. Pagès. 1998. Porin alteration and active efflux: two in vivo drug resistance strategies used by Enterobacter aerogenes. Microbiology 144:3003-3009. [DOI] [PubMed] [Google Scholar]
- 15.Malléa, M., J. Chevalier, A. Eyraud, and J.-M. Pagès. 2002. Inhibitors of antibiotic efflux pump in resistant Enterobacter aerogenes strains. Biochem. Biophys. Res. Commun. 293:1370-1373. [DOI] [PubMed] [Google Scholar]
- 16.Martinéz-Martinéz, L., A. Pascual, M. C. Conejo, I. García, P. Joyanes, A. Doménech-Sánchez, and V. J. Benedí. 2002. Energy-dependent accumulation of norfloxacin and porin expression in clinical isolates of Klebsiella pneumoniae and relationship to extended-spectrum β-lactamase production. Antimicrob. Agents Chemother. 46:3926-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazzariol, A., Y. Tokue, T. M. Kanegawa, G. Cornaglia, and H. Nikaido. 2000. High-level fluoroquinolone-resistant clinical isolates of Escherichia coli overproduce multidrug efflux protein AcrA. Antimicrob. Agents Chemother. 44:3441-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazzariol, A., J. Zuliani, G. Cornaglia, G. M. Rossolini, and R. Fontana. 2002. AcrAB efflux systems: expression and contribution to fluoroquinolone resistance in Klebsiella spp. Antimicrob. Agents Chemother. 12:3984-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution susceptibility tests for bacteria grow aerobically, 4th ed. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 20.Nikaido, H. 1998. Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin. Infect. Dis. 27(Suppl. 1):32S-41S. [DOI] [PubMed] [Google Scholar]
- 21.Nikaido, H. 2001. Preventing drug access to targets: cell surface permeability barriers and active efflux in bacteria. Semin. Cell Dev. Biol. 12:215-223. [DOI] [PubMed] [Google Scholar]
- 22.Nikaido, H. 1994. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 264:382-388. [DOI] [PubMed] [Google Scholar]
- 23.Over, U., D. Gur, S. Unal, G. H. Miller, and Aminoglycoside Resistance Study Group. 2001. The changing nature of aminoglycoside resistance mechanisms and prevalence of newly recognized resistance mechanisms in Turkey. Clin. Microbiol. Infect. 7:470-478. [DOI] [PubMed] [Google Scholar]
- 24.Paulsen, I. T., M. H. Brown, and R. A. Skurray. 1996. Proton-dependent multidrug efflux systems. Microbiol. Rev. 60:575-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poole, K. 2003. Overcoming multidrug resistance in gram-negative bacteria. Curr. Opin. Investig. Drugs 4:128-139. [PubMed] [Google Scholar]
- 26.Pradel, E., and J.-M. Pagès. 2002. The AcrAB-TolC efflux pump contributes to multidrug resistance in the nosocomial pathogen Enterobacter aerogenes. Antimicrob. Agents Chemother. 46:2640-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneiders, T., S. G. Amyes, and S. B. Levy. 2003. Role of AcrR and RamA in fluoroquinolone resistance in clinical Klebsiella pneumoniae isolates from Singapore. Antimicrob. Agents Chemother. 47:2831-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simonet, V., M. Malléa, D. Fourel, J. M. Bolla, and J.-M. Pagès. 1996. Crucial domains are conserved in Enterobacteriaceae porins. FEMS Microbiol. Lett. 136:91-97. [DOI] [PubMed] [Google Scholar]
- 29.Stoorvogel, J., M. J. van Bussel, and J. A. van de Klundert. 1991. Biological characterization of an Enterobacter cloacae outer membrane protein (OmpX). J. Bacteriol. 173:161-167. [DOI] [PMC free article] [PubMed] [Google Scholar]