Abstract
DNA binding by the ETS transcriptional repressor ETV6 (or TEL) is auto-inhibited ~ 50-fold due to an α-helix that sterically blocks its ETS domain binding interface. Using NMR spectroscopy, we demonstrate that this marginally-stable helix is unfolded, and not displaced to a non-inhibitory position, when ETV6 is bound to DNA containing a consensus 5’GGAA3’ recognition site. Although significantly lower in affinity, binding to non-specific DNA is auto-inhibited ~ 5-fold and also accompanied by helix unfolding. Based on NMR chemical shift perturbations, both specific and non-specific DNA are bound via the same canonical ETS domain interface. However, spectral perturbations are smaller for the non-specific complex, suggesting weaker and less well-defined interactions than in the specific complex. In parallel, the crystal structure of ETV6 bound to a specific DNA duplex was determined. The structure of this complex reveals that a non-conserved histidine residue in the ETS domain recognition helix helps establish the specificity of ETV6 for DNA-binding sites containing 5’GGAA3’ versus 5’GGAT3’. These studies provide a unified steric mechanism for attenuating ETV6 binding to both specific and non-specific DNA and expand the repertoire of characterized auto-inhibitory strategies utilized to regulate ETS factors.
Keywords: Inhibitory module, helix unfolding, protein-DNA interface, ETS family, winged helix-turn-helix
Introduction
Auto-inhibition is a powerful ‘on-site’ regulatory mechanism used to modulate a wide variety of biomolecular interactions.1 The ETS family of transcription factors displays several examples of DNA-binding auto-inhibition.2 These proteins are defined by the conserved ETS domain, which recognizes very similar DNA sequences containing a core 5’GGA(A/T)3’ motif.3 One role of controlling DNA binding by auto-inhibition is to provide added specificity for targeting distinct ETS proteins to appropriate transcriptional regulatory sequences. The relief of ETS-1 auto-inhibition though cooperative DNA binding with RUNX1, a frequently occurring partnership in T cell enhancers, exemplifies such added specificity.2, 4
We seek to understand the common and distinct mechanisms of auto-inhibition within the ETS family in order to gain a deeper insight into the evolution of DNA-binding control to achieve biological specificity. In the case of the prototypic member ETS-1, there is an extensive structural and dynamic understanding of its auto-inhibition. A combination of steric and allosteric mechanisms have been uncovered that integrate various signaling events, such as post-translational modifications and partner-protein interactions, resulting in both negative and positive control of ETS-1, respectively.4-6 Briefly, ETS-1 auto-inhibition is mediated by a set of helices that flank its ETS domain. This inhibitory module is distal from the DNA-binding interface, and upon DNA binding, undergoes an allosteric conformational change highlighted by the unfolding of a marginally stable helix.7, 8 Additionally, an adjacent intrinsically disordered serine rich region (SRR) plays a critical role in the Ca+2-dependent phosphorylation-enhanced auto-inhibition of ETS-1 by stabilizing the inhibitory module and by transiently masking its DNA-binding interface.6, 9 A second ETS member, ERG, has also been recently shown to be modestly auto-inhibited by a flexible sequence N-terminal to its ETS domain.10 In contrast, the mechanisms by which most other ETS proteins are auto-inhibited are not well understood.
In this study, we extended our investigation of regulation by auto-inhibition to ETV6 (or TEL, translocation ETS leukemia). Unlike the transcriptional activators ETS-1 and ERG, ETV6 is a repressor.11 Furthermore, it also self-associates due to the presence of a PNT (or SAM) domain, thereby facilitating cooperative binding on tandem ETS DNA-binding sites for repressive activity.12-15 Previously, we mapped an ETV6 inhibitory region C-terminal to its ETS domain.15 This CID (C-terminal inhibitory domain) contains two helices, of which helix H5 sterically blocks the canonical ETS DNA-binding interface and thereby reduces its affinity for specific sequences by ~ 50-fold.15, 16 Similar to ETS-1, the inhibitory helices are marginally stable and their presence dampens dynamics of the ETS domain.16 Preliminary evidence suggesting a conformational change accompanying DNA contact was provided by the relief of ETV6 auto-inhibition due to mutations that potentially disrupt the CID.15 Thus, the central goal of this current study was to determine the conformational changes occurring in the ETV6 ETS domain and the CID upon DNA binding. We also investigated how the CID impacts both specific and non-specific DNA binding, because binding a limited number of specific target sites in the cell occurs against a very high background of non-specific interactions.
We used a set of complementary NMR experiments to demonstrate that residues forming the inhibitory helix H5 are unfolded when ETV6 is bound to DNA. In parallel, with isothermal titration calorimetry (ITC) and NMR spectroscopy, we investigated the impact of CID on the interaction of ETV6 with non-specific DNA lacking the ETS consensus motif. Non-specific binding, which is substantially lower in affinity, is also auto-inhibited and is accompanied by helical unfolding. We further show that ETV6 utilizes a similar binding interface for both specific and non-specific DNA sequences. However, the non-specific complex appears relatively “loose” in comparison to the tight specific complex. To better define this interface, we determined the free and DNA-bound structures of the uninhibited ETV6 ETS domain with NMR spectroscopy and X-ray crystallography, respectively. The DNA-bound structure also helps explain the role of a non-conserved histidine in the preferential binding of ETV6 to the core sequence 5’GGAA3’. Collectively, our studies uncover a unified steric mechanism of auto-inhibition that impacts both non-specific and specific DNA binding by an ETS protein.
Results
Helix H5 in CID unfolds when ETV6 binds to specific DNA
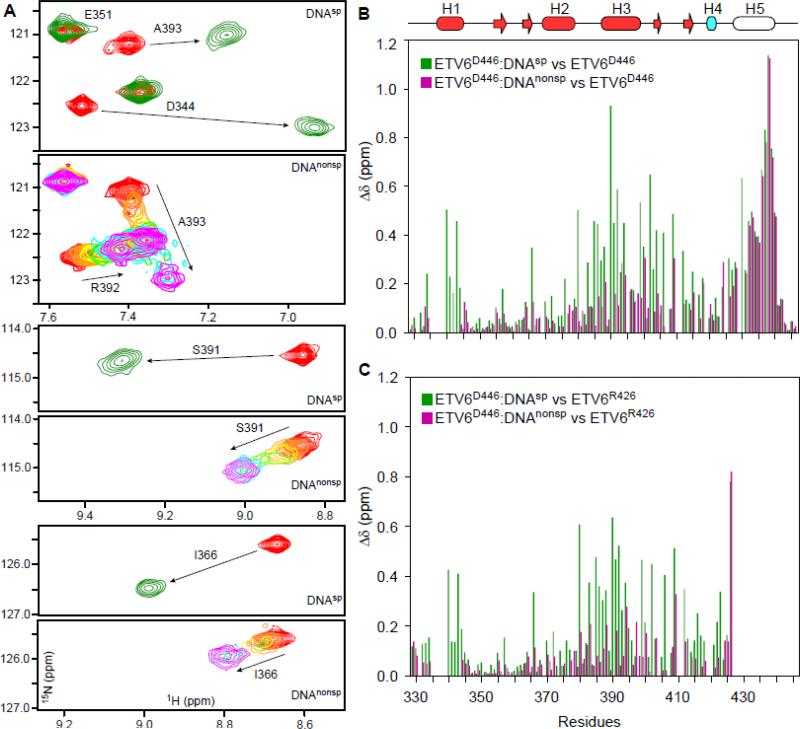
Structures of several ETS proteins show a highly conserved mode of DNA binding whereby the recognition helix H3 inserts into the major groove and makes extensive contacts with the 5’GGA(A/T)3’ motif.2 A similar canonical mode of binding by ETV6 would require that the CID must be either unfolded or otherwise be displaced to remove steric blockage of the ETS domain. We tested this hypothesis by several complementary NMR experiments. Initially, we utilized 15N-HSQC spectroscopy to monitor the interaction of 15N-labeled ETV6D446 with a 15 bp oligonucleotide (DNAsp) containing the ETS consensus motif 5’GGAA3’ (see Table 1 for nomenclature). Upon titration, peaks corresponding to the free protein diminished and a new set of signals corresponding to the bound state emerged (Fig. 1A and Supplemental Fig. S1). Such binding in the slow exchange regime is consistent with the relatively high nanomolar affinity of inhibited ETV6 for the consensus DNA sequence.16 The chemical shifts of the ETV6D446–DNAsp complex were used to predict its backbone structure (Fig. 2A). This indicated that the ETS domain and the CID helix H4 retained the same secondary structure as in the unbound protein.16 In contrast, residues corresponding to helix H5 displayed large 15N-HSQC chemical shift perturbations (CSP's) and resulting chemical shifts diagnostic of a disordered polypeptide chain (Fig. 1B and 2A). In addition, these residues exhibited low order parameters (S2) based on the “random coil index”.17
Table 1.
ETV6 constructs and DNA sequences
| Name | Sequence | Comment |
|---|---|---|
| ETV6R426 | G329-R426 | Uninhibited ETV6a |
| ETV6D446 | G329-D446 | Inhibited ETV6a |
| ETV6D446 | R335-D446 | Inhibited ETV6a |
| ETV6R458 | R335-R458 | Inhibited ETV6a |
| DNAsp | 5′-CAAGCCGGAAGTGAG-3′ | Specific DNAb |
| 3′-GTTCGGCCTTCACTC-5′ | ||
| DNAsp-cryst | 5′-AAAGCCGGAAGTGAG-3′ | Specific DNAb |
| 3′-TTCGGCCTTCACTCT-5′ | ||
| DNAnonsp | 5′-GATGCAGTGTAGTCG-3′ | Non-specific DNA |
| 3′-CTACGTCACATCAGC-5′ | ||
| DNAsp-emsa-1 | 5′-CAAGCCGGAAGTGAG-3′ | Specific DNAb |
| 3′-GTTCGGCCTTCACTC-5′ | ||
| DNAsp-emsa-2 | 5′-CAAGCCGGATGTGAG-3′ | Specific DNAb |
| 3′-GTTCGGCCTACACTC-5′ | ||
The core ETS domain spans Leu337 to Phe415, and Pro419 to Ser424 form helix H4.
The core 5′GGAA3′ or 5′GGAT3′ binding motif is in bold.
Fig. 1. NMR monitored titrations of ETV6 fragments with DNA.
(A) Overlaid regions of the 15N-HSQC spectra of ETV6D446 recorded in the absence (red) or presence of a 1.1 molar ratio of DNAsp (green). High affinity binding occurs the slow exchange regime and only signals from the free or bound protein are detected at intermediate molar ratios (not shown). Also shown are the same spectral regions when ETV6D446 is titrated with DNAnonsp in molar ratios of 0 (red), 0.1, 0.25, 0.5, 0.75, 1, 2, and 3 (magenta). With weaker affinity, binding occurs in the fast exchange regime and signals progressively change from the chemical shift of the free to the DNA-bound state. Some broadening also occurs at intermediate saturation. See Supplemental Figure S1 for the full spectra. (B) Amide CSP's (Δδ = {(ΔδH2 + (0.154ΔδN)2}1/2) for ETV6D446–DNAsp (green) and ETV6D446–DNAnonsp (magenta) with respect to free ETV6D446. The unfolding of helix H5 leads to large CSP's of similar magnitude for residues 430 - 440 in both complexes. In contrast, the CSP's for the ETS domain result from the displacement of helix H5 and the binding of DNA. In (C), the effects of helix H5 unfolding are removed by calculating CSP's relative to unbound ETV6R426, which lacks this helix. Note that these CSP's were calculated using the chemical shifts of ETV6D446 in the presence of a 1.1-fold molar excess of DNAsp and 3-fold molar excess of DNAnonsp, and thus correspond to 99% and 98% saturation, respectively, based on their respective KD values and binding site sizes. The secondary structure (helix, cylinder; strand, arrow) is displayed as cartoon on top with the core ETS domain in red and helix H4 of the CID in cyan. Helix H5, which is unfolded in both complexes, is not colored.
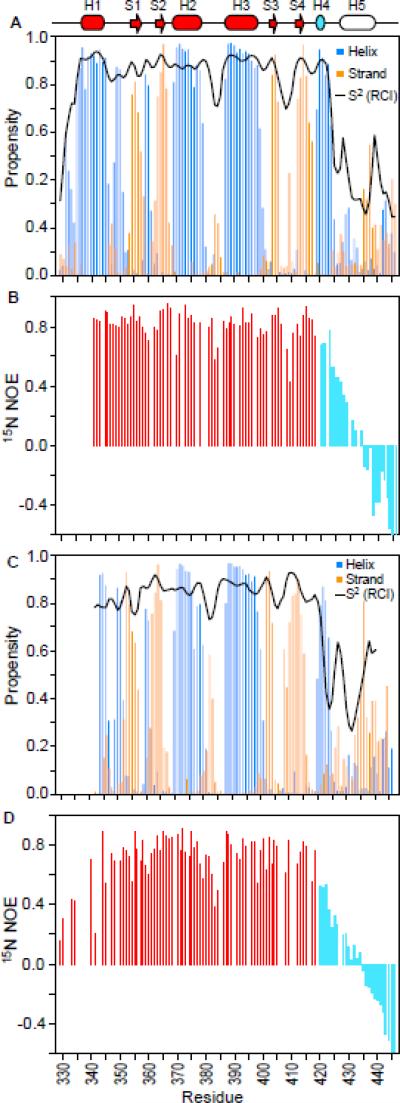
Fig. 2. Helix H5 is unstructured in both specific and non-specific ETV6–DNA complexes.
The normalized secondary structure propensities (helices, blue; strands, orange) and RCI order parameters (S2, black line) for the (A) ETV6D446–DNAsp and (C) ETV6D446–DNAnonsp complexes were calculated from 13Cα, 13Cβ, 15N and 1HN chemical shifts using the program MICS 81. The 15N-NOE data for (B) ETV6D446–DNAsp and (D) ETV6D446–DNAnonsp are shown for the core ETS domain (red) and CID (cyan) residues. Decreasing NOE and S2 values indicate increasing amide mobility on the sub-nsec timescale and thus show that helix H5 is unfolded in both complexes (see also Supplemental Fig. S2). The histogram bars for the two C-terminal residues are truncated, and missing data corresponds to prolines and residues with overlapping or unassigned signals. The top cartoon shows the secondary structural elements (helix, cylinder; strand, arrow) of the free inhibited ETS domain (red) and CID (cyan or not colored).
To more directly characterize the dynamic properties of the ETV6D446–DNAsp complex, we also collected amide 15N T1, T2 and heteronuclear NOE relaxation data (Supplemental Fig. S2). From T1/T2 ratios, the global isotropic tumbling correlation time was determined to be 12.5 ± 0.2 ns. This is consistent with the 23 kDa molecular mass of the ETV6D446–DNAsp complex, 18 and confirms that it is monomeric under the experimental conditions. More importantly, the 15N-NOE values, which are very sensitive to the sub-nsec timescale motions of the amide 15N-1HN bond vector, provide a measure of the fast local backbone dynamics of a protein. These data revealed a well-folded core ETS domain with high 15N-NOE values, yet a highly flexible CID with substantially lower values indicative of pronounced conformational mobility (Fig. 2B). An analysis of the full set of amide 15N relaxation data according to the model-free formalism19, 20 similarly showed that the CID residues are dynamic with substantially lower conventional order parameters than those of the ETS domain (Supplemental Fig. S2).
Finally, we determined the tertiary structure of ETV6D446 bound to DNAsp using NMR spectroscopy. Extensive 1H, 13C and 15N chemical shift assignments, along with backbone dihedral angle and inter-proton NOE distance restraints for the protein component of the ETV6D446–DNAsp complex, were obtained by TROSY-based heteronuclear NMR experiments (Table 2). Although detected, the 1H signals from the unlabeled DNA were not assigned. Using these data, the structural ensemble of the bound protein was calculated (Fig. 3A). Overall, the protein adopts the winged helix-turn-helix (wHTH) fold characteristic of ETS domains, and closely resembles the structure of free auto-inhibited ETV6 ETS domain.16 However, in marked contrast to a well-folded helix H5 blocking the binding interface of the unbound protein, the corresponding CID residues are disordered in the complex with high rmsd values. This is consistent with their dynamic nature and hence lack of any structural restraints. Collectively, chemical shift, 15N relaxation, and structural data clearly demonstrate that helix H5 unfolds upon specific DNA binding, and is not simply displaced as an intact helix to an alternative position that is no longer auto-inhibitory.
Table 2.
NMR refinement statistics for protein structures
| ETV6R426 | ETV6D446 - DNAsp | |
|---|---|---|
| NMR distance and dihedral restraints | ||
| Distance restraints | ||
| Total NOE | 1620 | 1540 |
| Intra-residue | 467 | 464 |
| Inter-residue | 1153 | 1076 |
| Sequential (|i – j| = 1) | 382 | 459 |
| Medium-range (|i – j| <= 4) | 285 | 271 |
| Long-range (|i – j| >= 5) | 486 | 346 |
| Dihedral angle restraints | ||
| ϕ, ψ | 85, 88 | 82, 84 |
| Structure statistics | ||
| Violations (mean ± std. dev,) | ||
| Distance restraints (Å) | 0.06 ± 0.02 | 0.16 ± 0.13 |
| Dihedral angle restraints (°) | 4.72 ± 1.92 | 5.50 ± 2.08 |
| Max. dihedral angle violation (°) | 6.09 | 10.25 |
| Max. distance restraint violation (Å) | 0.42 | 0.32 |
| Deviations from idealized geometry | ||
| Bond lengths (Å) | 0.005 | 0.005 |
| Bond angles (°) | 0.742 | 0.774 |
| Impropers (°) | 2.13 | 2.51 |
| Average pairwise r.m.s. deviationa (Å) | ||
| All heavy atoms | 1.03 ± 0.09 | 1.21 ± 0.12 |
| Backbone only | 0.59 ± 0.08 | 0.74 ± 0.11 |
Pairwise r.m.s. deviation was calculated among 10 refined structures for the residues 337 to 415.
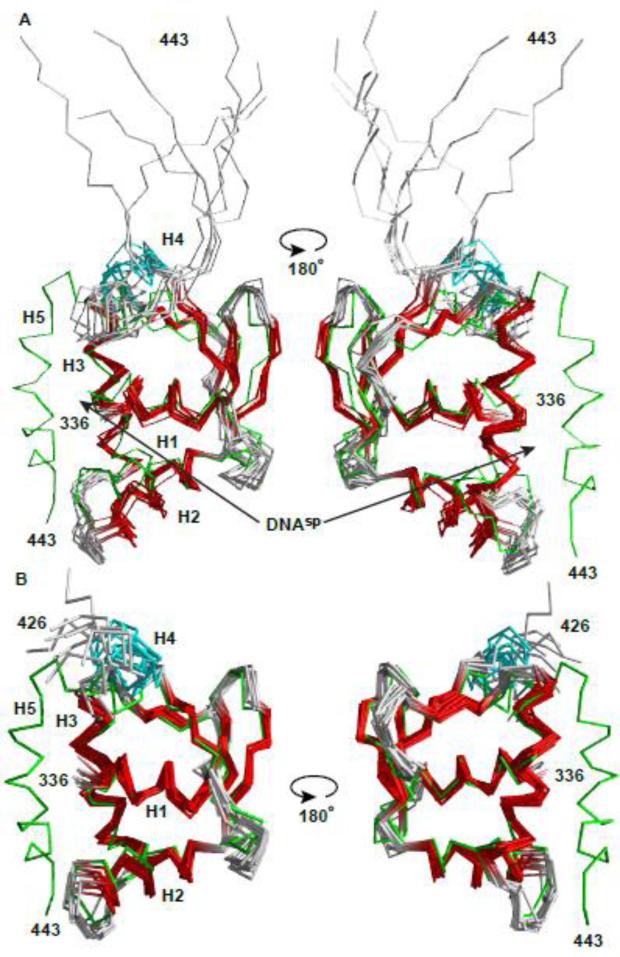
Fig. 3. Structural ensembles of free and bound ETV6 are similar except for the unfolding of helix H5.
The NMR-derived structural ensembles of (A) the ETV6D446–DNAsp complex (ETS domain helices and strands, red; CID helix H4, cyan) and (B) uninhibited ETV6R426 align closely to the lowest energy structure of inhibited ETV6R458 (green).16 CID helix H5, which blocks the DNA-binding interface of ETV6R458, is absent in ETV6R426 and unfolded in the ETV6D446-DNAsp complex. Although present in the latter complex, DNAsp was not included in the structure calculations. The N-terminal Gly-Ser-His-Met and unstructured residues (329-335 and 444-446) are not shown for clarity. Arrows point to the DNA-binding interface along helix H3.
ETV6 binding to non-specific DNA is also auto-inhibited
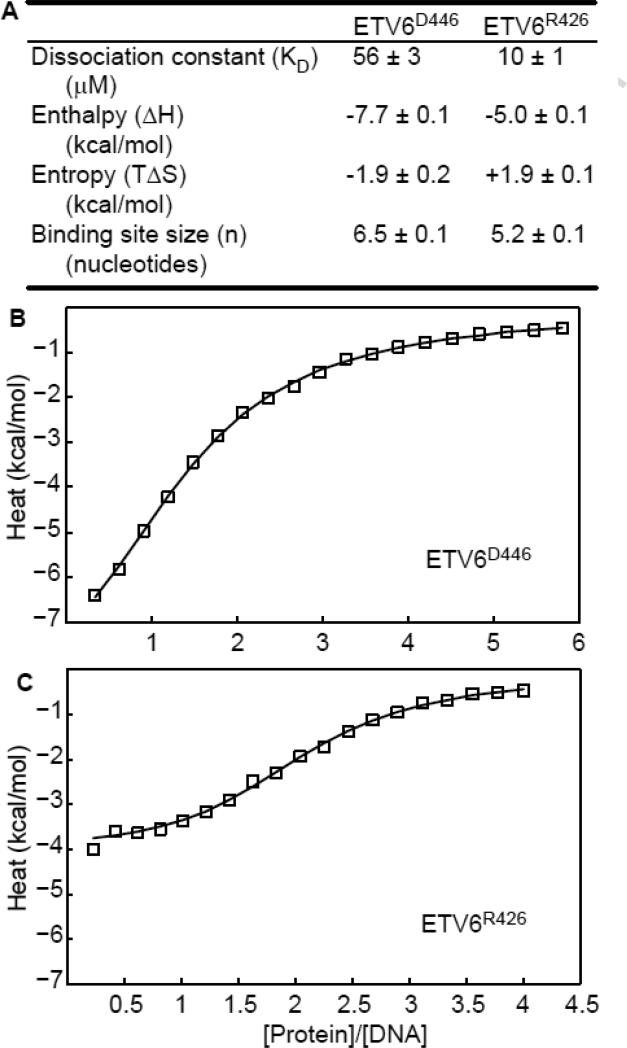
Using isothermal titration calorimetry (ITC) and NMR spectroscopy, we also investigated the interaction of ETV6D446 and ETV6R426 (lacking helix H5) with a 15 bp non-specific DNA oligonucleotide (DNAnonsp, Table 1). In contrast to the case of specific binding, for which there is one single high-affinity site within DNAsp, ETV6 can potentially bind DNAnonsp in numerous positions and in either orientation. Therefore, the measured ITC binding isotherms were fit to a variation of the McGhee-von Hippel model21 developed by Record and co-workers22 for non-specific binding with neighbor exclusion to a finite lattice. As summarized in Figure 4, this yielded the average microscopic dissociation constant (KD), binding enthalpy (ΔH) and the effective binding site size (n). Both ETV6D446 and ETV6R426 bound DNAnonsp with KD values in the micromolar range, which is 103 - 104 fold weaker than their affinities to a specific oligonucleotide.15, 16 Most importantly, the KD value for ETV6D446 is ~ 5-fold higher than that of ETV6R426. Thus, binding to non-specific DNA by the ETV6 ETS domain is also auto-inhibited, albeit to a lesser extent than to specific DNA (~ 50-fold).
Fig. 4. Binding to non-specific DNA is also auto-inhibited.
(A) Summary of the fit ITC data for DNAnonsp, along with the binding isotherms for (B) inhibited ETV6D446 and (C) uninhibited ETV6R426. Raw data are open squares and the best fits to the Record model22 are solid lines.
It is interesting to compare the differences in the binding site size between the specific and non-specific complexes. As described below, the crystal structure of ETV6 bound to specific DNA revealed that the protein contacts ~ 10 nucleotide pairs. In contrast, fitting of the ITC isotherms indicated that ETV6R426 and ETV6D446 effectively bind only 5.2 and 6.5 nucleotide pairs, respectively, in DNAnonsp. These n values reflect the average site size (~ 6 bp) occluded by one ETV6 along the non-specific DNA duplex. Therefore, the first ETV6 molecule could potentially bind in either orientation to the 15 bp oligonucleotide at 10 (= 15-6+1) overlapping sites. However, due to neighbor exclusion, only 2 to 3 protein molecules bind one oligonucleotide under saturating conditions. This could be accommodated by adopting staggered rotated positions along the double helix. Parenthetically, these non-integral values likely arise from the violation of the key premises in the Record model that the DNA lattice consists of identical binding sites and that the protein has only one mode of binding.23 Also, since DNAnonsp contains many potential binding sites, fitting these ITC data to a simple isotherm yielded apparent, overall macroscopic KD values ~ 10-fold lower than the microscopic values presented in Figure 4 (not shown). Importantly, regardless of model, auto-inhibited ETV6D446 binds to DNAnonsp with ~ 5-fold weaker affinity than uninhibited ETV6R426.
Helix H5 also unfolds when ETV6 interacts with non-specific DNA
Complementing ITC, we used NMR spectroscopy to obtain structural details of the ETV6–DNAnonsp interaction. Upon titration with DNAnonsp, numerous amide 15N-1HN signals of ETV6D446 showed progressive chemical shift changes (Fig. 1A and Supplemental Fig. S1). This is diagnostic of fast exchange between the free and bound forms of the protein and is consistent with a micromolar dissociation constant. Furthermore, only one signal per amide was observed in the 15N-HSQC spectrum of the saturated ETV6D446–DNAnonsp complex (Supplemental Fig. S1 and S3). Thus, translocation of the bound protein between the many possible non-specific sites along this oligonucleotide is also fast on the chemical shift timescale.
Insights into the structure of the bound protein could be obtained from its assigned NMR spectra. Based on its mainchain chemical shifts, the ETV6D446 ETS domain retained its wHTH secondary structure when bound to DNAnonsp (Fig. 2C). In contrast, large CSP's occurred for residues corresponding to helix H5, suggesting a significant conformational change (Fig. 1B). Indeed, both chemical shift and 15N relaxation measurements demonstrated that, as with DNAsp, the CID adopted a dynamic random coil conformation in the ETV6–DNAnonsp complex (Fig. 2C,D).
ETV6 binds non-specific DNA via the canonical ETS domain interface
The CSP's also revealed that the ETV6 ETS domain binds non-specific and specific DNA sequences via the same general interface. For this analysis, it is important to recognize that the spectral perturbations reflect both the unfolding of helix H5, which sterically blocks this interface, and interactions with DNA (Fig. 1B and Supplemental Fig. S4). Accordingly, to focus only on the latter, we calculated the CSP's for the ETV6D446–DNAsp and ETV6D446–DNAnonsp complexes relative to free ETV6R426, which lacks the CID helix H5. These CSP's are presented as the magnitude of the combined amide 1HN and 15N changes in Figure 1C, and separately (with upfield or downfield “direction”) for the two nuclei in Supplemental Figure S5. For both complexes, residues in helix H3, the turn between helices H2 and H3, and the wing between S3 and S4 experienced the largest CSP's. When mapped onto the structure of ETV6 (Fig. 5), these residues cluster to the DNA-binding interface that has been well characterized in many ETS domain complexes.24 Amide chemical shifts are exquisitely sensitive to their environment. Thus, CSP's may arise due to proximity to the charged and aromatic moieties in DNA, as well as from local or propagated conformational changes, such as those influencing hydrogen-bonding networks. Regardless of exact cause, the similar patterns of CSP's demonstrate that ETV6D446 uses the same canonical interface to bind both specific and non-specific DNA.
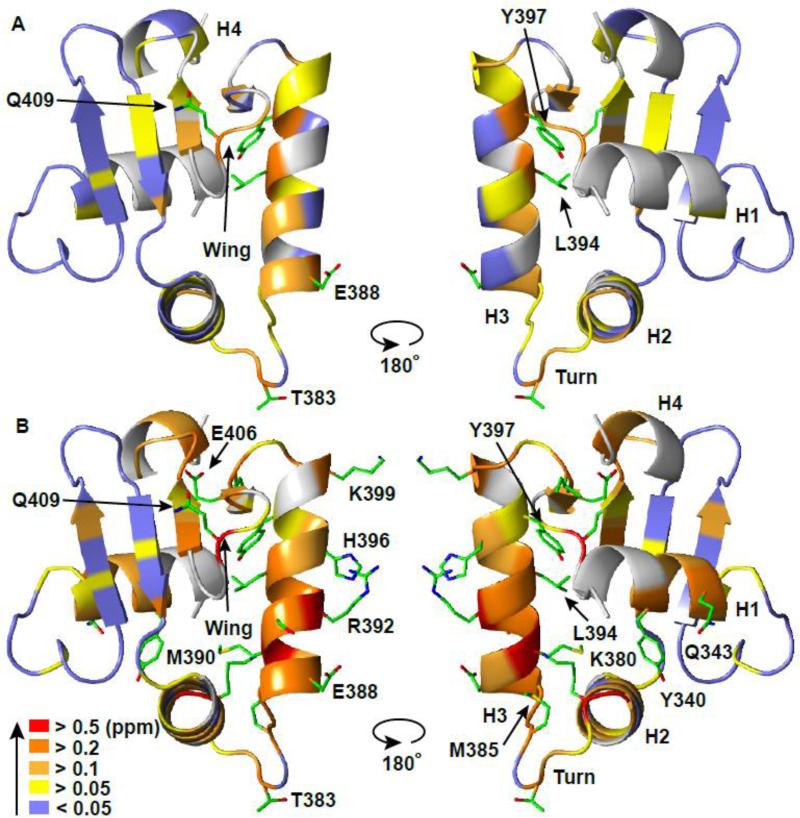
Fig. 5. ETV6 ETS domain binds specific and non-specific DNA via the same canonical interface.
The amide chemical shift perturbations (CSP's, Δδ = {(ΔδH2 + (0.154ΔδN)2}1/2) for (A) ETV6D446–DNAnonsp and (B) ETV6D446–DNAsp with respect to unbound ETV6R426 are mapped onto the crystal structure of ETV6R426-DNAsp-cryst (DNA not shown). Residues (backbone cartoon) are color-coded in the indicated CSP ranges. Prolines and unassigned residues are in grey. Side chains are shown for residues in (A) with CSP > 0.2 ppm and in (B) with CSP > 0.4 ppm. See Figure 1 and Supplemental Figures S1 and S4 for the original data.
Paralleling their relative binding affinities, the CSP's for many residues were in the same approximate direction,25 yet smaller in magnitude, for the DNAnonsp complex than for the DNAsp complex (Figs. 1 and 5, and Supplemental Fig. S5). Comparable patterns of relative NMR spectral changes have been reported for the HMG-box26 and ZNF217 zinc finger27 proteins bound to non-specific versus specific DNA oligonucleotides. This is not a trivial result of incomplete saturation of the ETV6D446–DNAnonsp complex, as the protein was ~ 98 % bound (Supplemental Fig. S3). Rather, this indicates that ETV6D446 forms generally similar, albeit less well-defined, time-averaged interactions with DNAnonsp than with DNAsp. Such interactions likely involve electrostatic contacts between the positively-charged DNA-binding interface of ETV6D446 and the negatively-charged phosphodiester backbone of DNAnonsp, rather than base-specific hydrogen bonds. Also, rapid exchange between binding sites along DNAnonsp should lead to smaller net CSP's due to averaging of potential positive and negative chemical shift changes.
Residue-wise comparison of the CSP's revealed potentially important structural differences between the two complexes (Figs. 1 and 5, and Supplemental Fig. S5). For example, Arg392 and His396, whose side chains interact with DNA bases in the ETV6-DNA crystal structure (see below), experienced substantial CSP's in the specific complex, yet smaller perturbations with DNAnonsp. Five additional lysine and arginine side chains, which interact with the phosphodiester backbone of DNA, also showed large amide CSP's in the specific complex, whereas only two (Lys380 and Arg382) exhibited substantial perturbations in the non-specific complex. In contrast, the 15N chemical shift of Ala393, a residue in helix H3, changed with opposite sign upon binding DNAnonsp relative to DNAsp. We hypothesize that these spectral differences reflect looser, dynamic electrostatic interactions and the lack of direct base contacts in the non-specific DNA complex versus the specific complex. Consistent with this notion, the 15N-NOE values for residues in the turn of the helix(H2)-turn-helix(H3) are slightly lower for ETV6D446 bound to DNAnonsp than to DNAsp, indicating greater fast timescale mobility (Figs. 2C,D).
Insights into the binding interface from structures of free and DNA-bound ETV6R426
To relate the above findings with the atomic details of the ETV6 binding interface, we used X-ray crystallography to determine the structure of its ETS domain bound to DNA. Since disordered regions are not conducive to crystallization and our NMR measurements clearly demonstrated that residues following Arg426 are unstructured in the ETV6–DNAsp complex, we used ETV6R426 for these studies. The crystal structure of this construct bound to a 14bp specific DNAsp-cryst sequence (Table 1) was solved at 2.2 Å resolution (Table 3, Fig. 6A). In parallel, we also used NMR spectroscopy to determine the structural ensemble of free ETV6R426 (Table 2, Fig. 3B). Other than the absence of the folded inhibitory helix H5, the structures of ETV6R426 in its free and DNAsp-cryst-bound states closely resemble that determined previously16 for free inhibited ETV6R458 with an average rmsd of 1.1 Å and 0.9 Å between all corresponding main chain atoms for ordered residues, respectively. Therefore, neither the inhibitory helix H5 nor the DNA measurably alter the average structure of the ETV6 ETS domain. A similar lack of any significant backbone structural changes has been observed for other ETS proteins in their free versus bound states.2
Table 3.
Data collection and refinement statistics for ETV6R426–DNAcryst-sp
| Data collection | |
| Space group | P3121 |
| Cell dimensions | |
| a, b, c (Å) | 57.58, 57.58, 130.48 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) | 50.00-2.20 (2.24-2.20) |
| Rsym or Rmerge | 0.085 (0.711) |
| I / σI | 29.13 (4.34) |
| Completeness (%) | 99.6 (100.0) |
| Redundancy | 7.9 (8.1) |
| Refinement | |
| Resolution (Å) | 26.34-2.20 |
| No. reflections | 13290 |
| Rwork / Rfree | 0.175/0.221 |
| No. atoms | |
| Protein | 813 |
| DNA | 590 |
| Water | 124 |
| B-factors (Å2) | |
| Overall (Wilson) | 47.5 (37.9) |
| Protein | 46.1 |
| DNA | 48.9 |
| Water | 49.6 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.32 |
Fig. 6. Binding interface of ETV6R426–DNAsp-cryst complex.
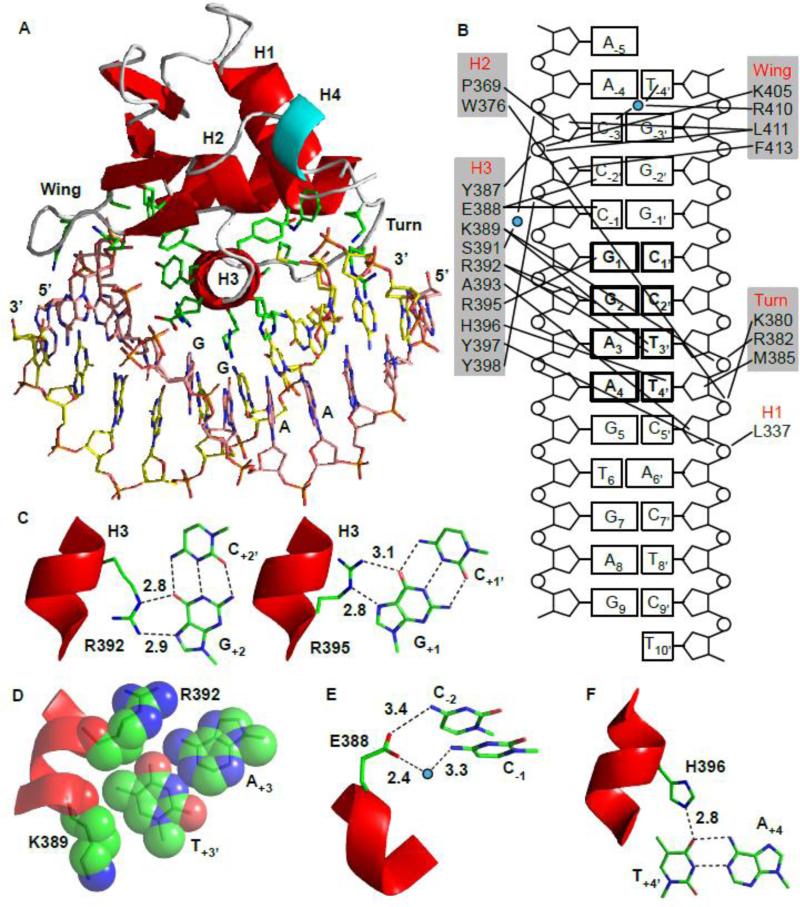
(A) Crystal structure of the ETV6R426–DNAsp-cryst complex. ETS domain residues contacting the DNA are shown in stick representation. Secondary structural elements in the core ETS domain are colored red, loops in grey, and helix H4 in cyan. Corresponding backbone atoms for ordered residues in the crystal ETV6R426–DNAsp-cryst and solution ETV6R426–DNAsp complexes superimposed with an average rmsd of 1.0 Å (not shown). (B) Interfacial interactions with ETV6 residues grouped according to their secondary structure. Water molecules are denoted as blue circles. (C) Direct readout due to the invariant Arg392 and Arg395 interacting with G+2 and G+1, respectively. (D) The methyl group on T+3’ fits a small hydrophobic pocket created by the Arg392 and Lys389 side chains. (E) Glu388 interacts with the C-1 and C-2 bases. (F) Hydrogen bond between His396 and T+4’ provides specificity towards the A+4 of the 5’GGAA3’ motif (Supplemental Fig. S7). (In C-F: carbon, green; oxygen, red; nitrogen, blue.)
The contacts observed in the ETV6R426–DNAsp-cryst interface, as summarized in Figures 6B-F, are highly conserved in the ETS family.24 Direct major groove base readout is mediated by bidentate hydrogen bonding of the invariant Arg392 and Arg395 in the recognition helix H3 to the G+2 and G+1 of the core 5’GGA3’ motif, respectively. In most ETS factors, a tyrosine side chain in helix H3 hydrogen bonds to A+3. However, the corresponding residue in ETV6 is His396, which is unable to provide this interaction. Rather, the specificity towards A+3 is determined via the complementary base T+3’. The methyl group of T+3’ occupies a hydrophobic pocket formed by the side chains of Arg392 and Lys389 (Fig. 6D). This hydrophobic interaction, which is also observed in other ETS domain–DNA structures, allows the Arg392 side chain to adopt the correct rotameric state required to hydrogen bond with the base G+2 and thus influences the overall specificity towards the core 5’GGA3’ motif. The side chain of Glu388 also forms direct and water mediated hydrogen bonds with the amino groups of C-2 and C-1, respectively, thereby establishing the preference of ETV6 for cytosines at these positions.3 Several residues also interact with the phosphodiester backbone of the DNA. The amide NH of Leu337 in helix H1 and Trp376 in helix H2 form highly conserved hydrogen bonds with the phosphate backbone. Residues Lys380 and Arg382 in the turn, Lys389 in H3, and Lys405 and Arg410 in the wing interact electrostatically with phosphate groups. These interactions may provide some specificity in the form of indirect shape readout, 28 or may be relatively non-specific and simply increase the net affinity of ETV6 for DNA.
Histidine 396 determines specificity towards Adenine at the +4 position
Most ETS factors can recognize either adenine or thymine at the +4 position in the core 5’GGA(A/T)3’ sequence.3, 29, 30 In contrast, based on qualitative high-throughput binding assays, ETV6 appears to have a distinct preference for adenine at this position.3 This specificity has been hypothesized to arise from the presence of a non-conserved histidine in helix H3 of ETV6, rather than the more commonly found tyrosine. To confirm these observations, we used an electrophoretic mobility shift assay (EMSA) to measure the KD values of ETV6D446 for DNA binding sites with either a 5’GGAA3’ or 5’GGAT3’ core sequence (Supplemental Fig. S6). Strikingly, ETV6D446 displayed 500-fold higher affinity towards the 5’GGAA3’ –bearing site (KD values of 1.6 ± 0.3 nM versus 800 ± 100 nM). Mutating His396 to a tyrosine reduced this selectivity to only 8-fold (1.9 ± 0.3 nM versus 16 ± 7 nM). Given that both WT- and H396YETV6D446 bound the 5’GGAA3’ oligonucleotide with very similar affinities, the presence of His396 strongly disfavors interactions with a thymine base at the +4 position.
The crystal structure of the ETV6R426–DNAsp-cryst complex provides an explanation for this sequence specificity. As illustrated in Figure 6F, His396 Nε2 is within hydrogen bonding distance (2.8 Å) of the O4 of T4 +4’ on the DNA strand complementary to the 5’GGAA3’. To act as a hydrogen bond donor, the side chain of His396 must be in the positively-charged imidazolium or neutral Nε2H imidazole tautomeric state. When we modeled the A-T nucleotide pair to T-A at position +4, neither base at this position (T+4 or A+4’) can form alternative hydrogen bonds with His396, unless perhaps it adopts the less favored neutral Nδ1H tautomer (Supplemental Fig. S7). In contrast, as shown by crystallographic studies of the ETS protein ELK4, the more common tyrosine can hydrogen bond with an adenine in either the +4 or +4’ position.31 Selectivity for the +4 base has also been observed for the ETS factors PU.1 and PDEF, which have Asn and Gln at the His396 equivalent position, respectively.3 This arises as the Asn and Gln side chains of PU.1 and PDEF form water mediated hydrogen bond with the T+4’ and A+4’ bases, respectively.32-34 In contrast, His396 in ETV6 forms a hydrogen bond directly with the T+4’ base.
Discussion
Steric mechanism of ETV6 auto-inhibition
In this study, we have extended our structural and thermodynamic understanding of the molecular basis of ETV6 auto-inhibition. Initially, we discovered that the CID attenuates specific DNA binding by ~ 50-fold.15 Subsequently, we reported that the CID includes two helices, of which helix H5 packs along the canonical ETS domain DNA-binding interface.16 This immediately predicted that substantial conformational changes must occur in ETV6 to enable its association with DNA. Alternatively, ETV6 might utilize a non-canonical mode of DNA binding, distinct from that of most ETS proteins. Although wHTH proteins typically contact DNA via the recognition helix H3, several different binding mechanisms have been identified.35 Using both NMR spectroscopy and X-ray crystallography, we clearly demonstrated that ETV6 binds specific DNA via the canonical ETS domain interface. Furthermore, multiple lines of experimental evidence, including random coil chemical shifts, low 15N-NOE values and high structural rms deviations, prove that residues forming the inhibitory helix H5 undergo a folded-to-unfolded conformational change in order to form the DNA-bound state. These data also exclude alternative models such as the displacement of the folded helix to a non-inhibitory position. Collectively, this leads to a simple steric mechanism of auto-inhibition in which helix H5 and DNA compete in a mutually exclusive manner for the DNA-binding interface of ETV6. Importantly, amide hydrogen exchange studies revealed that helix H5 is only marginally stable and thus poised to unfold.16 Through thermodynamic linkage, the modest energetic penalty of the requisite unfolding of this helix contributes to the net ~ 50-fold reduction of overall DNA affinity.
In addition to occluding the DNA-binding interface of ETV6, helix H5 also suppresses msec-μsec timescale dynamics in the ETS domain.16 Given that the structures of the ETS domain in ETV6 fragments with or without the inhibitory helix H5 are similar in both the free and DNA-bound states, these motions likely reflect small scale excursions about a common average conformation. This structural plasticity is hypothesized to be a central feature along the protein-DNA recognition pathway, as exemplified by the well-studied lac and CAP repressors.36-38 Although experimentally challenging, delineating the precise roles played by ETV6 backbone and side chain mobility in both DNA binding and its auto-inhibition by the CID is an important future objective.
Non-specific DNA binding and its auto-inhibition
Sequence-specific DNA-binding proteins also have significant affinity for non-specific sequences. In a living cell, such non-specific sites outnumber the specific sites by many orders of magnitude. Although this poses a severe challenge for these proteins to locate their target sites, non-specific binding also greatly helps in this process by both buffering the concentration of free protein and by “facilitated target location”.39, 40 Experimental and theoretical studies indicated that, upon initially encountering a non-specific site, a protein will undergo sliding (one-dimensional diffusion) along the DNA, combined with direct transfer or jumping (three-dimensional diffusion after transient dissociation) between DNA segments in order to search for its specific, high affinity target sites.41-46
A growing number of proteins bound to non-cognate DNA sequences have been characterized by X-ray crystallography (e.g., steroid receptors,47, 48 cro repressor,49 MATα2 homeodomain,50 several restriction enzymes,51-53 and Dam methyltransferase54) and NMR spectroscopy (e.g., lac repressor,36, 37 a HMG-box protein,26 HoxD9 homoedomain,41, 55 the bipartite Oct1,44 papillomavirus E2,56 and the Egr-145 and ZNF21745 zinc fingers). In each of these cases, the same approximate interface is utilized to bind both non-specific and specific DNA. However, at a more detailed level, there are necessarily differences in precise intermolecular orientation, protein-DNA contacts, buried surface area, and so forth, that account for the differences in affinity, and hence specificity, towards various DNA sequences. Broadly speaking, the interaction of sequence-specific DNA-binding proteins with non-specific DNA results primarily from electrostatic interaction between the positively-charged side chains of the protein and the negatively-charged DNA backbone.36, 52 It is also postulated that non-specific complexes are relatively dynamic with a high degree of hydration48, 57 and significant backbone and sidechain motions36, 37, 44, 45, 55 that aid in the efficient scanning for a target sequence. Once the cognate site is located, the protein and DNA undergo conformational changes leading to tight base-specific interactions.58-60
We have characterized the interaction of the ETV6 ETS domain with a non-specific DNA oligonucleotide that lacks the 5’GGAA3’ motif. Consistent with their relative affinities, NMR-monitored titrations show that specific and non-specific interactions are in slow (kex < Δω) and fast (kex > Δω) exchange regimes, respectively, where kex is the exchange rate constant (kex = kon[DNA] + koff) and Δω is the difference in chemical shifts between the free and bound states of ETV6. The faster exchange with non-specific DNA likely reflects a shorter lifetime (1/koff) of the bound state, as would be required for rapid target location. Conversely, the longer lifetime of the specific ETV6-DNA complex at regulatory sites would enable subsequent transcriptional repression. NMR chemical shift measurements confirm that ETV6 binds both specific and non-specific sequences using the same canonical ETS domain interface. However, the non-specific complex appears relatively loose, with weaker, dynamic time-averaged interactions. This conclusion is based on the three to four orders of magnitude higher KD values of various ETV6 constructs for non-specific versus specific DNA's, along with the general pattern of amide 1HN and 15N chemical shifts changing in the same direction, yet with smaller magnitudes, upon binding DNAnonsp versus DNAsp. Since the ETV6 backbone structure is similar in both complexes, switching from a non-specific to specific complex likely involves conformational rearrangements of amino acid side chains in the binding interface without any significant global changes.
We also discovered that the non-specific DNA binding by ETV6 is auto-inhibited and accompanied by the unfolding of helix H5. This is consistent with the need to remove the steric blockage of the ETS domain interface. However, non-specific binding is only attenuated by ~ 5-fold, whereas specific DNA binding is auto-inhibited by ~ 50-fold. This may reflect a contribution of weak non-specific DNA binding via a non-canonical interface not occluded by the CID.50 To the best of our knowledge, this is the first instance where auto-inhibition has been quantified for non-specific DNA binding. However, previous circular dichroism spectroscopic studies had indicated helical unfolding of ETS-1 upon binding to both specific and non-specific DNA.61 Thus, auto-inhibition of non-specific DNA binding is likely a general feature of sequence-specific regulatory proteins such as those of the ETS family.
ETV6 exemplifies further diversity of ETS family auto-inhibitory mechanisms
Although auto-inhibition of DNA binding has been reported for several ETS proteins.2, detailed molecular mechanisms have only been elucidated for ETS-1, ERG and now, ETV6. In the case of ETS-1, unfolding of its helical inhibitory module is linked allosterically to DNA binding. Furthermore, transient “fuzzy” interactions62 by an adjacent disordered SRR increases auto-inhibition in a phosphorylation-dependent manner.6, 9 Auto-inhibition by a flexible sequence in ERG appears akin to that of the SRR in ETS-1.10 In contrast, ETV6 lacks N-terminal inhibitory sequences and contains a helical CID that sterically blocks its ETS domain. Although structurally distinct, ETS-1 and ETV6 auto-inhibition also share several common features, including coupled unfolding of a marginally stable helix with DNA binding and dampening of ETS domain dynamics by the appended inhibitory sequences. The latter dynamic changes were also reported to occur with ERG.10
ETS-1 activity is regulated both positively and negatively via auto-inhibition. In particular, the effect is progressively reinforced due to increasing levels of SRR phosphorylation in response to Ca+2 signaling.6 Conversely, cooperative DNA binding with partner transcription factors, such as RUNX14 and Pax-5,5 relieves auto-inhibition. Although the cellular control of ETV6 is less well characterized, it is reasonable to hypothesize that the CID could also integrate signaling pathways and protein partnerships to modulate DNA binding. Also, the self-association of ETV6 via its PNT domain may compensate for auto-inhibition through cooperative DNA binding to tandem ETS sites.15 The mechanistic insights presented in this manuscript provide a foundation for understanding such potential roles of auto-inhibition in regulating transcriptional repression by ETV6.
Materials and Methods
Protein expression and purification
Three murine ETV6 fragments, with the sole cysteine (Cys334) mutated to serine, were used in this study: ETV6R426 (Gly329-Arg426), ETV6D446 (Gly329-Asp446), and for preliminary experiments, ETV6D446’ (Arg335-Asp446) lacking the first six residues (Table 1). These proteins differ from the previously characterized ETV6R458 (Arg335-Arg458) by the exact N-terminal sequence and by the deletion of unstructured residues C-terminal to helix H5.16 Genes encoding the ETV6 fragments were cloned in pET28b+ vectors and expressed in E. coli BL21 (λDE3) cells. For 15N/13C-labeling, cell cultures were grown in M9 minimal media supplemented with 1 g/L 15NH4Cl and 3 g/L 13C6-glucose (or 13C6/D7-glucose for perdeuterated protein) as the sole nitrogen and carbon source, respectively. For perdeuterated and fractionally deuterated samples, cultures were grown in 99% and 70% D2O M9 media, respectively. After induction for 6 hours at 30 °C, cells were lysed by sonication in the presence of 4 M guanidinium-HCl (H2O). This denaturation step improved net yield and allowed complete back-exchange of the amide protons in the deuterated samples. The His6-tagged proteins were purified by Ni2+ affinity chromatography with on-column refolding before elution. After thrombin cleavage of the His6-tag, the non-native N-terminal residues Gly-Ser-His-Met remained. Using gel filtration chromatography (Superdex 75), the proteins were further purified and exchanged into final sample buffer (20 mM sodium phosphate, 50 mM NaCl, pH 6.5). Protein concentrations were determined by UV absorption using predicted molar absorptivity ε280 values.63
DNA samples for NMR, ITC and crystallographic studies
DNA oligonucleotides were purchased from Integrated DNA Technologies and Sigma-Aldrich (without HPLC purification). Double-stranded DNA duplexes were generated by mixing the relevant single strands at equimolar ratio (determined from predicted molar absorptivity ε260 values), heating to 100°C for 5 mins, and slowly cooling to room temperature. For NMR and ITC titration experiments, gel-filtration chromatography was used to purify and buffer exchange the resulting duplex DNA duplexes. For crystallographic studies, the preformed 1:1 protein-DNA complex was purified by gel-filtration chromatography.
NMR spectroscopy
NMR experiments were performed using TCI-cryoprobe equipped Bruker Avance III 500, 600 and 850 MHz spectrometers at 25 °C. The proteins were 0.3 – 0.6 mM in sample buffer (20 mM phosphate, 50 mM NaCl, pH 6.5) with 6% lock D2O. The collected spectra were processed and analyzed using NMRPipe64 and Sparky,65 respectively. Signals from the 1H, 13C, and 15N nuclei in the backbone of 13C/15N-labeled ETV6D446 and the backbone and side chains of 13C/15N-labeled ETV6R426 were assigned using standard heteronuclear scalar correlation experiments.66 Protein signals from DNAsp complexes of amide-protonated 13C/15N-labeled ETV6D446 with uniformly deuterated, randomly factional (70%) deuterated, or fully protonated side chains were assigned using TROSY-based experiments.67. To ensure saturation, the DNAsp was present in a 1.1 fold molar excess. Protein signals for the ETV6D446-DNAnonsp complex were assigned using 15N-HSQC spectra to monitor the titration of the 15N-labeled ETV6D446 (initial 0.2 mM, final 0.15 mM) with aliquots of a 1.8 mM stock solution of DNAnonsp in sample buffer. The molar ratios of DNAnonsp to ETV6D446 in the titration set were 0, 0.1, 0.25, 0.5, 0.75, 1, 2 and 3, yielding a final protein saturation of ~ 98% based on the data of Figure 4. Binding occurred in the fast exchange limit; thus, amide 1HN and 15N assignments were obtained by tracking shifts relative to the initial free ETV6D446, and 13C assignments were then determined from standard 1H/13C/15N-correlation spectra of the saturated complex.
Amide 15N relaxation
Amide 15N relaxation data (T1, T2, heteronuclear NOE)68 were collected for the specific complex of perdeuterated ETV6D446’ and DNAsp at 25 °C with a 600 MHz NMR spectrometer. Relaxation rate constants were determined with Sparky65 by fitting the peak heights to an exponential decay. Heteronuclear {1H}-15N NOE data were also collected for 15N-labeled ETV6D446 complexed with DNAnonsp. The 15N-NOE value was determined from the ratio of the peaks heights versus a control reference spectrum without 1H saturation. The global tumbling correlation time for the specific complex was calculated using Tensor2.20
NMR structure calculations
NOE-derived distance restraints for 13C/15N-labeled ETV6R426 and ETV6D446–DNAsp were obtained simultaneous 3D 1H-15N/13C-1H NOESY-HSQC (aliphatic/aromatic; τmix = 110 ms), and constant time methyl-methyl and amide-methyl 15N/13C-13C-1H NOESY spectra (τmix = 100 ms).69, 70 The NMR-derived structure ensembles of ETV6R426 and the ETV6D446–DNAsp complex (protein only) were calculated using CYANA 3.071 with chemical shift assignments, dihedral angle restraints from TALOS+,72 NOESY crosspeaks, and manually assigned methyl-methyl distance restraints as input data (Table 2). Structure calculations, combined with automated NOESY spectra assignments, were performed in 7 iterative steps each yielding 100 structures. The 10 lowest energy structures from the final step were further refined with CNS using explicit solvent and molecular dynamics simulations.73 Although present in the ETV6D446–DNAsp complex, the signals from nuclei in DNAsp were not assigned and its co-ordinates were not included in these calculations. Given this lack of chemical shift assignments, any intermolecular NOEs between ETV6D446 and DNAsp in the input NOESY crosspeak list were thus discarded by CYANA. Secondary structure boundaries were determined using DSSP and figures rendered with PyMol.74
Crystallization and structure determination
Low salt buffer containing 20 mM HEPES and 50 mM NaCl at pH 7.5 was used for crystallization. ETV6R426 was mixed with DNAcryst-sp in 1:1.1 molar ratio and the resulting complex was purified by gel filtration chromatography. The purified ETV6R426–DNAcryst-sp was concentrated to ~0.3 mM. Crystallization trials were carried out by the hanging drop vapor diffusion method using 1 ml reservoir solution of 50 mM sodium cacodylate, 100 mM ammonium acetate, 10 mM MgCl2, and 22% PEG 8000 at pH 6.0, and a mixture of 2 μL complex and 2 μL of well solution. Crystals were obtained at room temperature within 5-7 days.
ETV6R426–DNAcryst-sp crystals were soaked stepwise for a few seconds each in mother liquor supplemented with 5% and 10% PEG 8000, and flash frozen using liquid nitrogen. A 2.2 Å resolution dataset was collected at 100 K using the Stanford Synchrotron Radiation Lightsource beam-line 7-1 with 0.9753 Å incident radiation. After data processing with HKL2000,75 MolRep76 was used for initial phase determination using the co-ordinates of the Elk-1 ETS domain (PDB ID: 1DUX) as a starting model. Cycles of structure refinement and building were performed using Phenix,77 Refmac 578 from the CCP4 suite of programs,79 and COOT.80 Water molecules were automatically added using Phenix and manually corrected. The crystal structure of ETV6R426–DNAcryst-sp was determined in space group P3121 with one monomer in the asymmetric unit. Sufficient electron density was observed to build residues 335 to 424 of ETV6R426 and the full duplex oligonucleotide. Ramachandran statistics indicated excellent stereochemistry with 98.8% of residues in the preferred region and no outliers. Data collection and refinement statistics are listed in Table 3.
Isothermal titration calorimetry
ITC measurements were performed at 25 °C with a Microcal ITC200. The protein and DNA samples were buffer-exchanged by gel filtration into 20 mM sodium phosphate and 50 mM NaCl at pH 6.5. Titrations were carried out with 20 μM DNAnonsp in the 200 μL reaction cell and 2 μL injections of ETV6D446 (0.58 mM) or ETV6R426 (0.40 mM) at 3 min intervals for a total of 20 injections. Heat of dilutions, measured by titrating proteins into buffer and buffer into DNA, was subsequently subtracted from the respective titration experiments. The binding of ETV6 fragments to non-specific DNA was analyzed using the Record model22 as described by equations (1-3)
| (1) |
The non-specific binding density νNS = [ETV6]bound/[total DNA bp], [L] is [ETV6]free, KA is the average microscopic association constant to any potential DNA site, n is the number of DNA base-pairs bound by the protein, and N is the total base-pairs in the oligonucleotide. In the ITC experiments, the heat content Qi after each injection ‘i’ is given by:
| (2) |
The reaction cell volume is Vo, binding enthalpy is ΔHo and binding density after injection ‘i’ is νNS,i. The differential heat ΔQi measured by the ITC instrument is given by:
| (3) |
The ITC binding isotherms were fit to these equations with Matlab. Errors in each fitted parameter were estimated from Monte Carlo simulations.
Supplementary Material
Highlights.
An inhibitory helix sterically blocks the ETV6 DNA-binding interface.
The inhibitory helix unfolds when ETV6 binds specific, as well as, non-specific DNA.
ETV6 binds specific and non-specific DNA via the same canonical ETS domain interface.
A non-conserved histidine helps define sequence specificity of ETV6.
A unified mechanism regulates specific and non-specific DNA-binding auto-inhibition.
Acknowledgements
This study was funded by the Canadian Cancer Society Research Institute (CCSRI 2011-700772 to L.P.M.) and the National Institutes of Health (R01GM38663 to B.J.G.). Instrument support was provided by the Canadian Institutes for Health Research (CIHR), the Canada Foundation for Innovation (CFI), the British Columbia Knowledge Development Fund (BCKDF), the UBC Blusson Fund, and the Michael Smith Foundation for Health Research (MSFHR). Funding from the Huntsman Cancer Institute/Huntsman Cancer Foundation is also acknowledged. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393) and the National Center for Research Resources (P41RR001209). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS, NCRR or NIH.
Abbreviations
- ITC
isothermal titration calorimetry
- SRR
serine rich region
- CID
C-terminal inhibitory domain
- NMR
nuclear magnetic resonance
- CSP
chemical shift perturbation
- HSQC
heteronuclear single quantum correlation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
The chemical shift assignments of ETV6R426 have been deposited in the BMRB ID: 19474. The atomic coordinates of ETV6R426 and ETV6R426–DNAcryst-sp have been deposited in the PDB ID: 2MD5 and 4MHG, respectively.
Supplementary Data Supplementary data associated with this article will be included in the online version.
References
- 1.Pufall MA, Graves BJ. Autoinhibitory domains: modular effectors of cellular regulation. Ann. Rev. Cell Dev. Biol. 2002;18:421–462. doi: 10.1146/annurev.cellbio.18.031502.133614. [DOI] [PubMed] [Google Scholar]
- 2.Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Ann. Rev. Biochem. 2011;80:437–471. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, Yan J, Talukder S, Turunen M, Taipale M, Stunnenberg HG, Ukkonen E, Hughes TR, Bulyk ML, Taipale J. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goetz TL, Gu TL, Speck NA, Graves BJ. Auto-inhibition of Ets-1 is counteracted by DNA binding cooperativity with core-binding factor alpha2. Mol. Cell. Biol. 2000;20:81–90. doi: 10.1128/mcb.20.1.81-90.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitzsimmons D, Lukin K, Lutz R, Garvie CW, Wolberger C, Hagman J. Highly cooperative recruitment of Ets-1 and release of autoinhibition by Pax5. J. Mol. Biol. 2009;392:452–464. doi: 10.1016/j.jmb.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pufall MA, Lee GM, Nelson ML, Kang HS, Velyvis A, Kay LE, McIntosh LP, Graves BJ. Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science. 2005;309:142–145. doi: 10.1126/science.1111915. [DOI] [PubMed] [Google Scholar]
- 7.Garvie CW, Pufall MA, Graves BJ, Wolberger C. Structural analysis of the autoinhibition of Ets-1 and its role in protein partnerships. J. Biol. Chem. 2002;277:45529–45536. doi: 10.1074/jbc.M206327200. [DOI] [PubMed] [Google Scholar]
- 8.Lee GM, Donaldson LW, Pufall MA, Kang HS, Pot I, Graves BJ, McIntosh LP. The structural and dynamic basis of Ets-1 DNA binding autoinhibition. J. Biol. Chem. 2005;280:7088–7099. doi: 10.1074/jbc.M410722200. [DOI] [PubMed] [Google Scholar]
- 9.Lee GM, Pufall MA, Meeker CA, Kang HS, Graves BJ, McIntosh LP. The affinity of Ets-1 for DNA is modulated by phosphorylation through transient interactions of an unstructured region. J. Mol. Biol. 2008;382:1014–1030. doi: 10.1016/j.jmb.2008.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Regan MC, Horanyi PS, Pryor EE, Jr., Sarver JL, Cafiso DS, Bushweller JH. Structural and dynamic studies of the transcription factor ERG reveal DNA binding is allosterically autoinhibited. Proc. Natl. Acad. Sci. USA. 2013;110:13374–13379. doi: 10.1073/pnas.1301726110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bohlander SK. ETV6: a versatile player in leukemogenesis. Semin Cancer Biol. 2005;15:162–74. doi: 10.1016/j.semcancer.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Kim CA, Phillips ML, Kim W, Gingery M, Tran HH, Robinson MA, Faham S, Bowie JU. Polymerization of the SAM domain of TEL in leukemogenesis and transcriptional repression. EBOO J. 2001;20:4173–4182. doi: 10.1093/emboj/20.15.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiao F, Song HY, Kim CA, Sawaya MR, Hunter JB, Gingery M, Rebay I, Courey AJ, Bowie JU. Derepression by depolymerization: Structural insights into the regulation of Yan by Mae. Cell. 2004;118:163–173. doi: 10.1016/j.cell.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Graham TG, Vivekanand P, Cote L, Cetera M, Rebay I. Sterile alpha motif domain-mediated self-association plays an essential role in modulating the activity of the Drosophila ETS family transcriptional repressor Yan. Mol. Cell Biol. 2010;30:1158–1170. doi: 10.1128/MCB.01225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green SM, Coyne HJ, 3rd, McIntosh LP, Graves BJ. DNA binding by the ETS protein TEL (ETV6) is regulated by autoinhibition and self-association. J. Biol. Chem. 2010;285:18496–18504. doi: 10.1074/jbc.M109.096958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coyne HJ, 3rd, De S, Okon M, Green SM, Bhachech N, Graves BJ, McIntosh LP. Autoinhibition of ETV6 (TEL) DNA binding: appended helices sterically block the ETS domain. J. Mol. Biol. 2012;421:67–84. doi: 10.1016/j.jmb.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berjanskii MV, Wishart DS. A simple method to predict protein flexibility using secondary chemical shifts. J. Am. Chem. Soc. 2005;127:14970–14971. doi: 10.1021/ja054842f. [DOI] [PubMed] [Google Scholar]
- 18.Daragan VA, Mayo KH. Motional model analyses of protein and peptide dynamics using 13C and 15N NMR relaxation. Prog. Nucl. Mag. Res. Spec. 1997;31:63–105. [Google Scholar]
- 19.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J. Am. Chem. Soc. 1982;104:4559–4570. [Google Scholar]
- 20.Dosset P, Hus JC, Blackledge M, Marion D. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J. Biomol. NMR. 2000;16:23–28. doi: 10.1023/a:1008305808620. [DOI] [PubMed] [Google Scholar]
- 21.McGhee JD, von Hippel PH. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 22.Tsodikov OV, Holbrook JA, Shkel IA, Record MT., Jr. Analytic binding isotherms describing competitive interactions of a protein ligand with specific and nonspecific sites on the same DNA oligomer. Biophys J. 2001;81:1960–1969. doi: 10.1016/S0006-3495(01)75847-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaires JB. Analysis and interpretation of ligand-DNA binding isotherms. Meth. Enzym. 2001;340:3–22. doi: 10.1016/s0076-6879(01)40415-0. [DOI] [PubMed] [Google Scholar]
- 24.Grishin AV, Alexeevsky AV, Spirin SA, Karyagina AS. Conserved structural features of ETS domain-DNA complexes. Mol. Biol. 2009;43:612–619. [PubMed] [Google Scholar]
- 25.Selvaratnam R, Mazhab-Jafari MT, Das R, Melacini G. The auto-inhibitory role of the EPAC hinge gelix as mapped by NMR. Plos One. 2012;7:e48707. doi: 10.1371/journal.pone.0048707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwahara J, Schwieters CD, Clore GM. Characterization of nonspecific protein-DNA interactions by 1H paramagnetic relaxation enhancement. J. Am. Chem. Soc. 2004;126:12800–12808. doi: 10.1021/ja046246b. [DOI] [PubMed] [Google Scholar]
- 27.Vandevenne M, Jacques DA, Artuz C, Nguyen CD, Kwan AHY, Segal DJ, Matthews JM, Crossley M, Guss JM, Mackay JP. New insights into DNA recognition by zinc fingers revealed by structural analysis of the oncoprotein ZNF217. J. Biol. Chem. 2013;288:10616–10627. doi: 10.1074/jbc.M112.441451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of specificity in protein-DNA recognition. Ann. Rev. Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szymczyna BR, Arrowsmith CH. DNA binding specificity studies of four ETS proteins support an indirect read-out mechanism of protein-DNA recognition. J. Biol. Chem. 2000;275:28363–28370. doi: 10.1074/jbc.M004294200. [DOI] [PubMed] [Google Scholar]
- 30.Poon GM, Macgregor RB., Jr. Base coupling in sequence-specific site recognition by the ETS domain of murine PU.1. J. Mol. Biol. 2003;328:805–819. doi: 10.1016/s0022-2836(03)00362-0. [DOI] [PubMed] [Google Scholar]
- 31.Mo Y, Vaessen B, Johnston K, Marmorstein R. Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins. Mol. Cell. 1998;2:201–212. doi: 10.1016/s1097-2765(00)80130-6. [DOI] [PubMed] [Google Scholar]
- 32.Kodandapani R, Pio F, Ni CZ, Piccialli G, Klemsz M, McKercher S, Maki RA, Ely KR. A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex. Nature. 1996;380:456–460. doi: 10.1038/380456a0. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Feng L, Said M, Balderman S, Fayazi Z, Liu Y, Ghosh D, Gulick AM. Analysis of the 2.0 A crystal structure of the protein-DNA complex of the human PDEF ETS domain bound to the prostate specific antigen regulatory site. Biochemistry. 2005;44:7095–7106. doi: 10.1021/bi047352t. [DOI] [PubMed] [Google Scholar]
- 34.Poon GM. Sequence discrimination by DNA-binding domain of ETS family transcription factor PU.1 is linked to specific hydration of protein-DNA interface. J. Biol. Chem. 2012;287:18297–18307. doi: 10.1074/jbc.M112.342345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harami GM, Gyimesi M, Kovacs M. From keys to bulldozers: expanding roles for winged helix domains in nucleic-acid-binding proteins. T.I.B.S. 2013;38:364–371. doi: 10.1016/j.tibs.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 36.Kalodimos CG, Biris N, Bonvin AM, Levandoski MM, Guennuegues M, Boelens R, Kaptein R. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 37.Kalodimos CG, Boelens R, Kaptein R. Toward an integrated model of protein-DNA recognition as inferred from NMR studies on the Lac repressor system. Chem. Rev. 2004;104:3567–3586. doi: 10.1021/cr0304065. [DOI] [PubMed] [Google Scholar]
- 38.Tzeng SR, Kalodimos CG. Allosteric inhibition through suppression of transient conformational states. Nat. Chem. Biol. 2013;9:462–5. doi: 10.1038/nchembio.1250. [DOI] [PubMed] [Google Scholar]
- 39.von Hippel PH, Berg OG. Facilitated target location in biological systems. J. Biol. Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 40.Clore GM. Exploring translocation of proteins on DNA by NMR. J. Biomol. NMR. 2011;51:209–219. doi: 10.1007/s10858-011-9555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iwahara J, Clore GM. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440:1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 42.Givaty O, Levy Y. Protein sliding along DNA: dynamics and structural characterization. J, Mol. Biol. 2009;385:1087–1097. doi: 10.1016/j.jmb.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 43.Blainey PC, Luo GB, Kou SC, Mangel WF, Verdine GL, Bagchi B, Xie XS. Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 2009;16:1224–1234. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takayama Y, Clore GM. Intra- and intermolecular translocation of the bi-domain transcription factor Oct1 characterized by liquid crystal and paramagnetic NMR. Proc. Natl. Acad. Sci. USA. 2011;108:E169–E176. doi: 10.1073/pnas.1100050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zandarashvili L, Vuzman D, Esadze A, Takayama Y, Sahu D, Levy Y, Iwahara J. Asymmetrical roles of zinc fingers in dynamic DNA-scanning process by the inducible transcription factor Egr-1. Proc. Natl. Acad. Sci. USA. 2012;109:E1724–E1732. doi: 10.1073/pnas.1121500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loth K, Gnida M, Romanuka J, Kaptein R, Boelens R. Sliding and target location of DNA-binding proteins: an NMR view of the lac repressor system. J. Biomol NMR. 2013;56:41–49. doi: 10.1007/s10858-013-9723-0. [DOI] [PubMed] [Google Scholar]
- 47.Luisi BF, Xu WX, Otwinowski Z, Freedman LP, Yamamoto KR, Sigler PB. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 48.Gewirth DT, Sigler PB. The basis for half-site specificity explored through a noncognate steroid receptor-DNA complex. Nat. Struct. Biol. 1995;2:386–394. doi: 10.1038/nsb0595-386. [DOI] [PubMed] [Google Scholar]
- 49.Albright RA, Mossing MC, Matthews BW. Crystal structure of an engineered Cro monomer bound nonspecifically to DNA: possible implications for nonspecific binding by the wild-type protein. Protein Sci. 1998;7:1485–1494. doi: 10.1002/pro.5560070701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aishima J, Wolberger C. Insights into nonspecific binding of homeodomains from a structure of MATalpha2 bound to DNA. Proteins. 2003;51:544–551. doi: 10.1002/prot.10375. [DOI] [PubMed] [Google Scholar]
- 51.Winkler FK, Banner DW, Oefner C, Tsernoglou D, Brown RS, Heathman SP, Bryan RK, Martin PD, Petratos K, Wilson KS. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993;12:1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Viadiu H, Aggarwal AK. Structure of BamHI bound to nonspecific DNA: a model for DNA sliding. Mol. Cell. 2000;5:889–895. doi: 10.1016/s1097-2765(00)80329-9. [DOI] [PubMed] [Google Scholar]
- 53.Townson SA, Samuelson JC, Bao Y, Xu SY, Aggarwal AK. BstYI bound to noncognate DNA reveals a “hemispecific” complex: implications for DNA scanning. Structure. 2007;15:449–459. doi: 10.1016/j.str.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 54.Horton JR, Liebert K, Hattman S, Jeltsch A, Cheng X. Transition from nonspecific to specific DNA interactions along the substrate-recognition pathway of dam methyltransferase. Cell. 2005;121:349–361. doi: 10.1016/j.cell.2005.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iwahara J, Zweckstetter M, Clore GM. NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proc. Natl. Acad. Sci. USA. 2006;103:15062–15067. doi: 10.1073/pnas.0605868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown C, Campos-Leon K, Strickland M, Williams C, Fairweather V, Brady RL, Crump MP, Gaston K. Protein flexibility directs DNA recognition by the papillomavirus E2 proteins. Nucl. Acids Res. 2011;39:2969–2980. doi: 10.1093/nar/gkq1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sidorova NY, Rau DC. Differences in water release for the binding of EcoRI to specific and nonspecific DNA sequences. Proc. Natl. Acad. Sci. USA. 1996;93:12272–12277. doi: 10.1073/pnas.93.22.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marcovitz A, Levy Y. Frustration in protein-DNA binding influences conformational switching and target search kinetics. Proc. Natl. Acad. Sci. USA. 2011;108:17957–17962. doi: 10.1073/pnas.1109594108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanchez IE, Ferreiro DU, Dellarole M, de Prat-Gay G. Experimental snapshots of a protein-DNA binding landscape. Proc. Natl. Acad. Sci. USA. 2010;107:7751–7756. doi: 10.1073/pnas.0911734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou HX. Rapid search for specific sites on DNA through conformational switch of nonspecifically bound proteins. Proc. Natl. Acad. Sci. USA. 2011;108:8651–8656. doi: 10.1073/pnas.1101555108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petersen JM, Skalicky JJ, Donaldson LW, McIntosh LP, Alber T, Graves BJ. Modulation of transcription factor Ets-1 DNA binding: DNA-induced unfolding of an alpha helix. Science. 1995;269:1866–1869. doi: 10.1126/science.7569926. [DOI] [PubMed] [Google Scholar]
- 62.Fuxreiter M. Fuzziness: linking regulation to protein dynamics. Mol. Biosyst. 2012;8:168–177. doi: 10.1039/c1mb05234a. [DOI] [PubMed] [Google Scholar]
- 63.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPaSy: The proteomics server for in-depth protein knowledge and analysis. Nucl. Acids Res. 2005;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRpipe - a multidimensional spectral processing system based on UNIX pipes. J. Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 65.Goddard TD, Kneeler DG. Sparky. 3rd ed. USCF; 1999. [Google Scholar]
- 66.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Mag. Res. Spec. 1999;34:93–158. [Google Scholar]
- 67.Yang D, Kay LE. Improved 1HN-detected triple resonance TROSY-based experiments. J. Biomol. NMR. 1999;13:3–10. doi: 10.1023/A:1008329230975. [DOI] [PubMed] [Google Scholar]
- 68.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 69.Pascal SM, Muhandiram DR, Yamazaki T, Formankay JD, Kay LE. Simultaneous acquisition of 15N-edited and 13C-edited NOE spectra of proteins Dissolved in H2O. J. Magn. Reson. Ser. B. 1994;103:197–201. [Google Scholar]
- 70.Zwahlen C, Gardner KH, Sarma SP, Horita DA, Byrd RA, Kay LE. An NMR experiment for measuring methyl-methyl NOEs in 13C-labeled proteins with high resolution. J. Am. Chem. Soc. 1998;120:7617–7625. [Google Scholar]
- 71.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 72.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol NMR. 2009;44:213–23. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Cryst. D Biol Cryst. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 74.DeLano WL. Use of PYMOL as a communications tool for molecular science. Abstracts of Papers of the American Chemical Society. 2004;228:U313–U314. [Google Scholar]
- 75.Otwinowski Z, Minor W. Processing of X-ray diffraction data. Meth. Enzym. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 76.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Applied Cryst. 1997;30:1022–1025. [Google Scholar]
- 77.Adams PD, Afonine PV, Bunkoczi G. b., Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D Biol Cryst. 2004;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. D Biol Cryst. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 79.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A. Overview of the CCP4 suite and current developments. Acta Cryst. D Biol Cryst. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. D Biol Cryst. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 81.Shen Y, Bax A. Identification of helix capping and beta-turn motifs from NMR chemical shifts. J. Biomol. NMR. 2012;52:211–32. doi: 10.1007/s10858-012-9602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.