Abstract
Rotavirus G10P[11] strains, which are commonly found in cattle, have frequently been associated with asymptomatic neonatal infections in India. We report the finding of G10P[11] strains associated with severe disease in neonates in Vellore, southern India. Rotavirus strains from 43 fecal samples collected from neonates with or without gastrointestinal symptoms between 1999 and 2000 were genotyped by reverse transcription-PCR. Forty-one neonates (95%) were infected with G10P[11] rotavirus strains, and 63% of the infections were in children who had gastrointestinal symptoms, including acute watery diarrhea. G10P[11] strains were also seen infecting older children with dehydrating gastroenteritis in Vellore. Characterization of the genes encoding VP7, VP4, VP6, and NSP4 of these strains revealed high sequence homology with the corresponding genes of the asymptomatic neonatal strain I321, which in turn is very closely related to bovine G10P[11] strains circulating in India. No significant differences were seen in the sequences obtained from strains infecting symptomatic neonates or children and asymptomatic neonates.
Group A rotaviruses are a major cause of gastroenteritis in children and in the young of most animal species (35). In the developing world, rotavirus diarrhea is a leading cause of mortality in children under the age of 5, and the World Health Organization has recognized the development of rotavirus vaccines as a priority for the reduction of infant mortality in developing countries (43).
Two rotavirus proteins, VP7 and VP4, which constitute the outer layer and the viral spikes, respectively, are known to induce neutralizing antibody responses, and the classification of rotaviruses into G and P types is based on the variability observed in the genes encoding these two proteins (20). At least 14 G types (G1 to G14) and 20 P types (P[1] to P[20]) have been identified to date, of which 10 G types and 5 P types have been found in rotaviruses infecting humans (20, 35). The middle layer of rotaviruses is constituted by a single protein, VP6, arranged in trimers. Among group A rotaviruses, four different subgroups, SGI, SGII, SGI+II, and nonI/nonII, have been distinguished on the basis of VP6 diversity (25), but human rotavirus strains are possibly only from SGI or SGII (29). A nonstructural protein, NSP4, has been identified as a viral enterotoxin capable of inducing age-dependent diarrhea in mice (4). Five different genotypes of NSP4 (A to E) have so far been identified, and all human strains characterized to date are from genotype A or B (7, 30). These four proteins are known to be immunogenic and may be important determinants of pathogenesis. The immunological responses to these proteins may be important for protection from subsequent infection and disease.
Epidemiological studies of rotavirus infections are increasingly revealing a great diversity of rotavirus strains cocirculating in the human population throughout the world (11, 17). Rotaviruses diversify and evolve mainly through two mechanisms: the accumulation of point mutations (genetic drift), which generates genetic lineages and leads to the emergence of antibody escape mutants, and genetic shift through gene reassortment during dual infection of a single cell (28). Zoonotic transmission and gene reassortment between human and animal rotaviruses also contributes to the generation of diversity in rotaviruses infecting humans (28).
Although the correlates of protection are not yet fully understood, it is thought that protection from infection and severe disease may be type specific. Therefore, monitoring the rotavirus types cocirculating at any one time and the emergence of novel rotaviruses is crucial during the implementation of any future vaccination program.
In this study, we describe the detection of pathogenic rotaviruses of G10P[11] genotype in neonates in Vellore, India, associated with disease in almost two-thirds of the infected neonates. Sequence analysis of the genes encoding VP4, VP6, VP7, and NSP4 revealed them to be very closely related to the neonatal asymptomatic strain I321 and to bovine strains commonly circulating in India.
MATERIALS AND METHODS
Clinical specimens.
A total of 43 fecal samples found to be rotavirus positive by enzyme immunoassay and collected between January 1999 and April 2000 from neonates admitted to the neonatal nursery, with or without gastrointestinal symptoms, were characterized for G and P genotypes, subgroup, and NSP4 genotype. Two rotavirus G10P[11] strains isolated from nonneonatal symptomatic children in Vellore in 1998 were also included for comparison.
Rotavirus strain characterization.
G and P genotyping was performed using published methods. Briefly, the rotavirus-positive fecal samples were prepared as 10% suspensions in balanced salt solution. Two hundred microliters of the 10% fecal suspensions was used for nucleic acid extraction with guanidinium isothiocyanate and silica (6), and the extracted nucleic acid was reverse transcribed in the presence of random hexamers (32). The cDNA was then used for G- and P-typing seminested PCRs, using published oligonucleotide primers and methods (23, 24, 34).
First-round amplicons of the genes encoding VP7 of 15 G10 rotavirus strains were obtained using the consensus primers VP7-F (5′ ATGTATGGTATTGAATATACCAC 3′; nt 51 to 71) and VP7-R (5′ AACTTGCCACCATTTTTTCC 3′; nt 914 to 932) and were directly sequenced. First-round amplicons of the genes encoding VP4 were obtained using the oligonucleotide primers Con3 (5′ TGGCTTCGCCATTTTATAGACA 3′; nt 11 to 32) and Con2 (5′ ATTTCGGACCATTTATAACC 3′; nt 868 to 887) (23), which amplify the VP8* region of the VP4-encoding gene. Direct sequencing was performed on 11 rotavirus G10 strains for which P-typing seminested PCR was unsuccessful. In addition, amplicons of the genes encoding the VP6 and NSP4 genes of 12 of these 15 G10 rotavirus strains were sequenced using the oliogonucleotide primers VP6-F (5′ GACGGVGCRACTACATGGT 3′; nt 747 to 766)-VP6-R (5′ GTCCAATTCATNCCGGTGG 3′; nt 1126 to 1106) (29) and NSP4-F (5′ GGCTTTTAAAAGTTCTGTTCCG 3′; nt 1 to 22)-NSP4-R (5′GTCACACTAAGACCATTCC 3′; nt 753 to 732) (7), respectively. Two rotavirus G10P[11] strains isolated from nonneonatal symptomatic children in Vellore in 1998 were also characterized and included in the comparisons.
The amplicons were purified using Qiaquick PCR purification spin columns (Qiagen) prior to sequencing with the same oligonucleotide primers used in the different VP4, VP6, VP7, and NSP4 PCRs and the CEQ2000 Dye Terminator Cycle Sequencing Quick Start kit (Beckman-Coulter). All methods were carried out following the manufacturers' instructions. The sequences were resolved using an automated sequencer (CEQ; Beckman-Coulter). Multiple alignments and phylogenetic analyses were carried out using the Bionumerics (Applied Maths, Kortrijk, Belgium) and BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) software packages. The dendrograms were confirmed by at least two of the following methods; neighbor joining, maximum parsimony, and maximum likelihood. Genotypes and genetic lineages within genotypes were confirmed by bootstrap values of >90% using 1,000 pseudoreplicate runs.
The nucleotide sequences described in this work are freely available upon request.
RESULTS
Of the 43 rotavirus-positive neonates, 27 (63%) had gastrointestinal symptoms: 14 had symptoms of acute gastroenteritis; 7 had necrotizing enterocolitis (NEC); 2 had pneumatosis intestinalis, 1 of them associated with NEC; and 4 had vomiting and/or food intolerance. Sixteen neonates had no gastrointestinal symptoms.
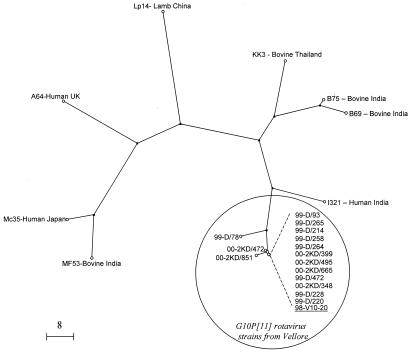
Of the 43 rotavirus-positive samples from which VP7 was characterized, 41 were G10. Sequence alignment and phylogenetic analysis of 15 of these strains showed that the genes encoding VP7 of the G10 strains were very closely related to that of the neonatal asymptomatic strain I321 (>98% homology at the nucleotide level). The sequences of the G10 strains found in this study were also more closely related to the sequences derived from two bovine strains isolated in India (strains B75 and B69) than to other G10 human strains previously found in Japan and the United Kingdom (Fig. 1).
FIG. 1.
Unrooted dendrogram constructed using rotavirus G10 partial VP7 nucleotide sequences (881 bp) and the maximum-parsimony method. VP7 sequences of rotavirus G10 strains available in GenBank/EMBL were used for comparison: Mc35, accession number D14033; A64, accession number X63156; Lp14, accession number L11602; KK3, accession number D01056; B75, accession number AF386919; B69, accession number AF386917; I321, accession number L07658; and MF53, accession number AF386919. Of the strains from Vellore, 99-D/78, 00-2KD/472, 00-2KD/851, 99-D/214, 00-2KD/495, and 99-D/472 were from neonates without gastrointestinal symptoms; strains 99-D265, 99-D/258, 9-D/264, 00-KD/665, 00-KD/399, 00-2KD/348, 99-D/228, and 99-D220 were from neonates with gastrointestinal symptoms; and strain 98-V10-20 was from an infant with severe gastroenteritis. The calibration bar indicates the number of base conversions.
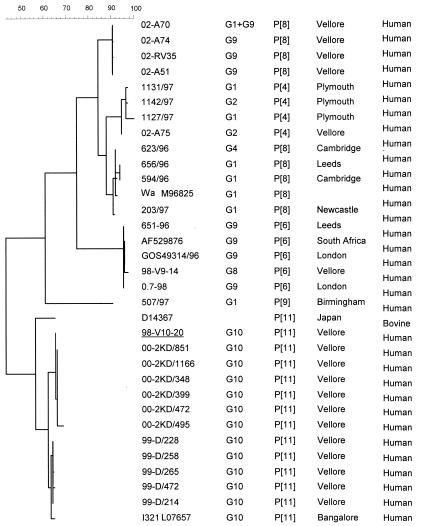
Initially, the P type associated with the majority of the G10 strains could not be determined by reverse transcription (RT)-PCR. Sequencing of the fragment corresponding to the VP8* region of the gene encoding VP4 revealed that these strains were of the P[11] genotype (Fig. 2). The VP8* sequences derived from the strains described in this study were highly conserved and were >90% homologous, at the nucleotide level, to the VP8* sequence of strain I321 and of a bovine rotavirus isolate (D14367) from Japan.
FIG. 2.
Dendrogram constructed using the maximum-parsimony method and rotavirus partial VP4 nucleotide sequences (corresponding to the VP8* region) of different P-type specificities derived from human isolates. The sequence derived from a nonneonatal symptomatic case is underlined. Sequences available in GenBank/EMBL were used for comparison, and the accession numbers are shown in the first column. The calibration bar indicates percent homology.
G10 was found in combination with P[11] in 40 samples and with P[4] in one. A further sample contained multiple rotavirus strains, as indicated by the presence of G9 in combination with P[4] and P[6], and the remaining sample was characterized as G2P[4].
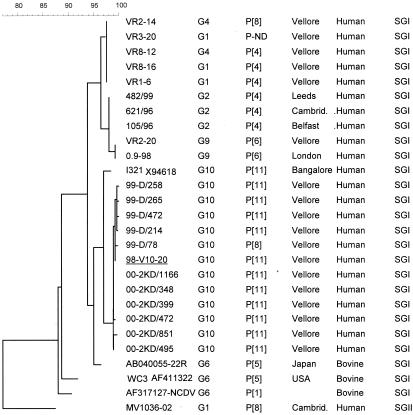
The G10P[11] rotavirus strains were of SGI, according to sequence analysis of a 379-bp fragment of the gene encoding VP6. These VP6 partial sequences clustered in a separate genetic lineage of SGI and were more closely related to the sequences of I321 and bovine strain 22R than to any other VP6 SGI sequences of human rotavirus strains (Fig. 3).
FIG. 3.
Dendrogram constructed using the maximum-parsimony method and rotavirus VP6 partial sequences derived from human strains of SGI associated with different G and P types. Sequences derived from the neonatal asymptomatic strain I321 and of three bovine strains and a human SGII strain were included for comparison. The sequence derived from a nonneonatal symptomatic case is underlined. Sequences available in GenBank/EMBL were used for comparison, and the accession numbers are shown in the first column. The calibration bar indicates percent homology. Cambrid., Cambridge.
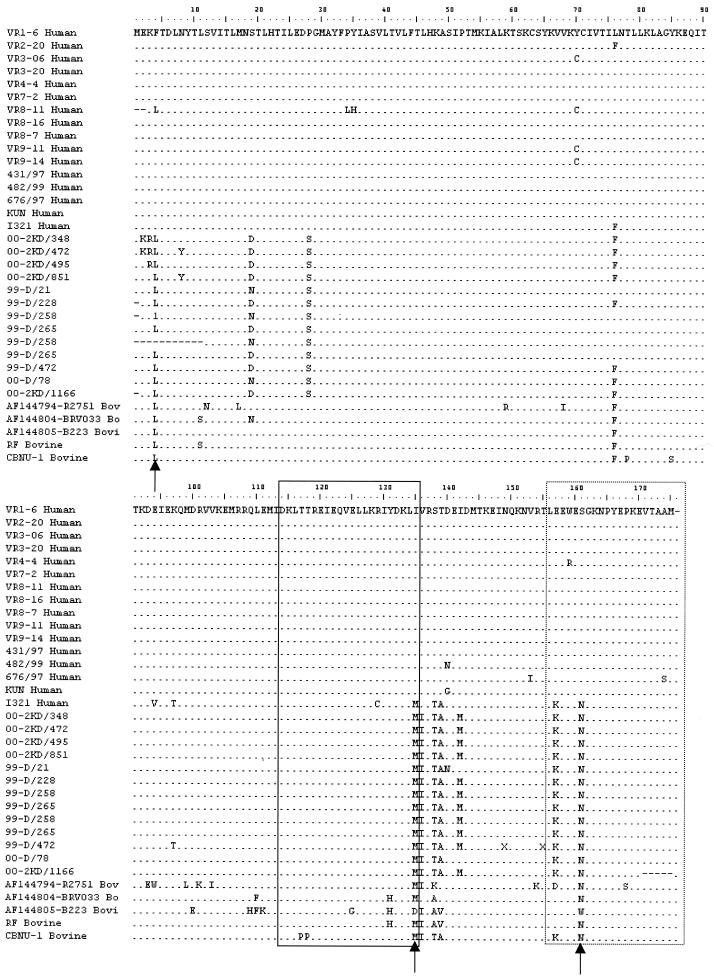
The NSP4 sequences derived from the G10P[11] rotaviruses clustered within NSP4 genotype A but in a separate genetic lineage from other NSP4-A sequences derived from human rotavirus strains (Fig. 4). The NSP4 sequences of the G10P[11] strains were more closely related to the sequences derived from bovine rotavirus strains and were highly homologous to that of the neonatal G10P[11] strain I321 (Fig. 4). Alignment of the deduced amino acid sequences of genotype A NSP4s showed that the G10 strains had amino acid substitutions typical of the NSP4s of bovine strains, amido acid positions 4, 135, 136, 138, and 161 (Fig. 5).
FIG. 4.
Dendrogram constructed using the maximum-parsimony method and rotavirus NSP4 nucleotide sequences derived from human and bovine strains associated with different G and P types. Sequences available in GenBank/EMBL were used for comparison, and the accession numbers are shown in the first column. The calibration bar indicates percent divergence.
FIG. 5.
Alignment of the deduced amino acid sequences of the NSP4 of human and bovine rotavirus strains of NSP4-A genotype specificity. Significant amino acid substitutions between bovine and human NSP4-A lineages are indicated by arrows. The domain between amino acids 114 and 135 associated with the capacity to induce diarrhea in the mouse model and the double-layered particle binding domain (amino acids 156 to 176) are boxed with a continuous and a dotted line, respectively.
No significant differences in the sequences of any of the genes investigated were observed among the strains from neonates with acute gastroenteritis or NEC, older symptomatic children, and those from neonates with no gastrointestinal symptoms.
DISCUSSION
A high incidence of asymptomatic rotavirus infection in neonates has been reported in various studies in India (1, 8, 14, 15, 44). The rotaviruses found associated with neonatal asymptomatic infections exhibit unusual genotypes, such as G9P[11] and G10P[11] (1, 14, 16, 22). It is not yet clear whether the lack of disease is virus or host related and/or dependent on the existence of protecting maternal antibodies. A 7-year study conducted in Bangalore and Mysore, India, showed that I321-like G10P[11] strains were exclusively associated with asymptomatic infections in neonates and estimated the prevalence of asymptomatic infection in this group to be 34% (1).
G10P[11] rotaviruses are common pathogens of cattle in several parts of the world (21, 27, 37, 47), and they are by far the predominant bovine rotavirus throughout India (26, 47). Within this group, there is genetic diversity across the world and within India (26, 47). Zoonotic transmission has been increasingly recognized as an important contributor to the diversity of rotaviruses found in human infections (2, 28, 38, 39). This, coupled with the ability of rotaviruses to reassort during multiple coinfections, results in the exchange of genetic material between human and animal viruses and generates novel viruses (28, 38, 39). Therefore, it is perhaps not surprising that, given the close proximity between cattle and humans in many Indian communities, G10P[11] strains may have been transmitted from cattle to the human population and become an established human pathogen. Where our findings differ from previous reports of infection with G10P[11] strains in India is in the association of these strains with disease in 63% of the neonatal cases analyzed, and also in the identification of G10P[11] rotaviruses causing sporadic infections in older children, associated with disease severe enough to require hospitalization. Rotaviral loads, as determined by quantitative RT-PCR, consistent with symptomatic infection were found in all samples from symptomatic patients, whereas samples from asymptomatic patients had viral loads consistent with low disease severity scores or asymptomatic infection (34). No bacterial infections were detected in any of the cases. These findings indicate that in Vellore, at least, G10P[11] strains are not exclusive to neonatal or asymptomatic infections.
Sequence analysis of the genes encoding VP7, VP6, VP4, and NSP4 has revealed no significant differences between the strains associated with asymptomatic or symptomatic infection in neonates or older children. The sequences of the genes encoding these four proteins were all highly homologous to those of strain I321, including the NSP4 regions between amino acid positions 114 and 135 (Fig. 5), associated with the capacity to induce diarrhea in mice (45). All G10P[11] strains were of SGI and NSP4 genotype A, and both genes were within lineages exclusively comprising bovine rotavirus strains, further strengthening the evidence for zoonotic introduction of these rotaviruses. One of the significant amino acid substitutions observed between the NSP4 genogroup A typical of human strains and those of bovine strains or the human G10P[11] strains from India was located at position 135, but this position was identical in the G10 strains isolated from both asymptomatic and symptomatic cases. A second significant amino acid substitution at position 161 was located within the double-layered particle-binding site. This also provides further evidence of the firm linkage between VP6 and NSP4, as previously observed in common and reassortant human rotaviruses (30).
Complete characterization of the genome of strain I321 through hybridization and/or partial sequence analysis indicated that, with the exception of the genes encoding NSP1 and NSP3, the gene segments were very closely related to those derived from bovine strains (16, 18, 19, 41). Therefore, it was concluded that this strain may have been introduced from cattle and, following reassortment with a human rotavirus strain, became established in the human neonatal population in India (1, 18, 47). NSP1, unlike NSP3, segregated with the species of origin (18, 36, 41), and the finding of an NSP1-encoding gene closely related to those of human rotavirus strains in the context of the otherwise almost entirely bovine I321 strain supported the hypothesis that NSP1 may be an important host restriction factor and hence could be responsible for the spread of this strain in the neonatal population in India (18, 47). Similarly, the NSP1-encoding gene segment of the neonatal strain G9P[11] 199E was shown to be of human origin through total-genome hybridization analysis (12).
Complete characterization and comparison of the genomes of the G10P[11] strains found in symptomatic and asymptomatic infections, including strain I321, and currently circulating bovine G10P[11] strains will show whether there are any significant differences in the remainder of their genetic makeup which may be associated with their ability to induce disease and/or spread from the neonatal population into older children.
The geographical differences observed in the patterns of disease associated with G10P[11] rotaviruses in India may be a consequence of differences in the prevalences of protective maternal antibodies in the different regions. Although currently G10P[11] is the most prevalent rotavirus genotype in cattle throughout India, there are no data preceding 1994 (26, 47). The incidence of infections with G10P[11] rotaviruses in the human population in India may be directly linked to that in cattle, as these strains are probably transmitted from cattle to humans, and symptomatic infection in neonates may reflect the lack of type-specific protective maternal antibodies. In the absence of epidemiological data on the incidences of different viruses in cattle prior to the 1990s, the seroprevalences of G10P[11]-specific antibodies in women of child-bearing age in the different regions of India may identify susceptible populations or groups within populations.
Sporadic symptomatic infections with G10 rotaviruses have been reported in other parts of the world (5, 9, 46). There have been more recent reports from Brazil, where G10 strains have been found in symptomatic infections with incidences varying from 4 to 16% (3, 42). Although the data are scarce, and the characterization of the G10 strains found elsewhere is incomplete, the sequences of the VP7-encoding genes of the strains collected in Vellore are much more closely related to strain I321 and to other bovine strains from India than to the G10 strains found in human infections in Japan and the United Kingdom (5, 15, 46) (Fig. 1). This suggests that the strains found in Japan, the United Kingdom, and India are the result of different zoonotic introductions. However, the lack of diversity within the genes encoding VP7, VP4, VP6, and NSP4 of the Vellore G10P[11] strains and their similarity to I321 suggests that the cases found in India may have arisen from a single transmission event or a cluster of transmission events.
It is important to monitor the spread and evolution of G10 strains in India and in other parts of the world. G9P[11] strains were identified in India around the same period as the G10P[11] strains (14, 15) and were also associated with asymptomatic infections in neonates. Although sporadic cases of G9 strains had also been identified in other parts of the world, the incidence of infections with G9 strains was high only in India. G9P[11] strains were subsequently replaced with G9P[6] (14) strains, which probably originated through reassortment, and these infections were associated with disease. G9P[6] strains spread beyond India in the mid 1990s (10, 13, 40), and a subsequent reassortment event yielded G9P[8], which displaced G9P[6] and successfully spread globally, becoming a common human pathogen on all continents and becoming the most or second most prevalent rotavirus strain in many regions (11, 17, 28, 31). Close monitoring of the evolution and spread of G10 rotaviruses and detailed characterization of representative isolates will be vital for understanding the evolution of rotaviruses and the spread and pathogenesis of novel rotavirus strains and will be of paramount importance for the successful introduction of a vaccination program.
Acknowledgments
Support for this work was obtained through an MRC Research Training Fellowship awarded to Miren Iturriza-Gómara and a Wellcome Trust Trilateral Infectious Disease Initiative grant (no. 063144).
REFERENCES
- 1.Aijaz, S., K. Gowda, H. V. Jagannath, R. R. Reddy, P. P. Maiya, R. L. Ward, H. B. Greenberg, M. Raju, A. Babu, and C. D. Rao. 1996. Epidemiology of symptomatic human rotaviruses in Bangalore and Mysore, India, from 1988 to 1994 as determined by electropherotype, subgroup and serotype analysis. Arch. Virol. 141:715-726. [DOI] [PubMed] [Google Scholar]
- 2.Allen, A., and U. Desselberger. 1985. Reassortment of human rotaviruses carrying rearranged genomes with bovine rotavirus. J. Gen. Virol. 66:2703-2714. [DOI] [PubMed] [Google Scholar]
- 3.Araujo, I. T., A. M. Fialho, R. M. de Assis, M. Rocha, M. Galvao, C. M. Cruz, M. S. Ferreira, and J. P. Leite. 2002. Rotavirus strain diversity in Rio de Janeiro, Brazil: characterization of VP4 and VP7 genotypes in hospitalized children. J. Trop. Pediatr. 48:214-218. [DOI] [PubMed] [Google Scholar]
- 4.Ball, J. M., P. Tian, C. Q. Zeng, A. P. Morris, and M. K. Estes. 1996. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science 272:101-104. [DOI] [PubMed] [Google Scholar]
- 5.Beards, G., L. Xu, A. Ballard, U. Desselberger, and M. A. McCrae. 1992. A serotype 10 human rotavirus. J. Clin. Microbiol. 30:1432-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boom, R., C. J. A. Sol, M. M. M. Salismans, C. L. Jansen, P. M. E. Wertheim-van Dillen, and J. van den Noordaa. 1990. Rapid and simple method for the purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciarlet, M., F. Liprandi, M. E. Conner, and M. K. Estes. 2000. Species specificity and interspecies relatedness of NSP4 genetic groups by comparative NSP4 sequence analyses of animal rotaviruses. Arch. Virol. 145:371-383. [DOI] [PubMed] [Google Scholar]
- 8.Cicirello, H. G., B. K. Das, A. Gupta, M. K. Bhan, J. R. Gentsch, R. Kumar, and R. I. Glass. 1994. High prevalence of rotavirus infection among neonates born at hospitals in Delhi, India: predisposition of newborns for infection with unusual rotavirus. Pediatr. Infect. Dis. J. 13:720-724. [DOI] [PubMed] [Google Scholar]
- 9.Coluchi, N., V. Munford, J. Manzur, C. Vazquez, M. Escobar, E. Weber, P. Marmol, and M. L. Racz. 2002. Detection, subgroup specificity, and genotype diversity of rotavirus strains in children with acute diarrhea in Paraguay. J. Clin. Microbiol. 40:1709-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cubitt, W. D., A. D. Steele, and M. Iturriza. 2000. Characterisation of rotaviruses from children treated at a London hospital during 1996: emergence of strains G9P2A[6] and G3P2A[6]. J. Med. Virol. 61:150-154. [DOI] [PubMed] [Google Scholar]
- 11.Cunliffe, N. A., J. S. Bresee, J. R. Gentsch, R. I. Glass, and C. A. Hart. 2002. The expanding diversity of rotaviruses. Lancet 359:640-642. [DOI] [PubMed] [Google Scholar]
- 12.Cunliffe, N. A., B. K. Das, M. Ramachandran, M. K. Bhan, R. I. Glass, and J. R. Gentsch. 1997. Sequence analysis demonstrates that VP6, NSP1 and NSP4 genes of Indian neonatal rotavirus strain 116E are of human origin. Virus Genes 15:39-44. [DOI] [PubMed] [Google Scholar]
- 13.Cunliffe, N. A., J. S. Gondwe, R. L. Broadhead, M. E. Molyneux, P. A. Woods, J. S. Bresee, R. I. Glass, J. R. Gentsch, and C. A. Hart. 1999. Rotavirus G and P types in children with acute diarrhea in Blantyre, Malawi, from 1997 to 1998: predominance of novel P[6]G8 strains. J. Med. Virol. 57:308-312. [PubMed] [Google Scholar]
- 14.Das, B. K., J. R. Gentsch, H. G. Cicirello, P. A. Woods, A. Gupta, M. Ramachandran, R. Kumar, M. K. Bhan, and R. I. Glass. 1994. Characterization of rotavirus strains from newborns in New Delhi, India. J. Clin. Microbiol. 32:1820-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das, B. K., J. R. Gentsch, Y. Hoshino, S. Ishida, O. Nakagomi, M. K. Bhan, R. Kumar, and R. I. Glass. 1993. Characterization of the G serotype and genogroup of New Delhi newborn rotavirus strain 116E. Virology 197:99-107. [DOI] [PubMed] [Google Scholar]
- 16.Das, M., S. J. Dunn, G. N. Woode, H. B. Greenberg, and C. D. Rao. 1993. Both surface proteins (VP4 and VP7) of an asymptomatic neonatal rotavirus strain (I321) have high levels of sequence identity with the homologous proteins of a serotype 10 bovine rotavirus. Virology 194:374-379. [DOI] [PubMed] [Google Scholar]
- 17.Desselberger, U., M. Iturriza-Gómara, and J. Gray. 2001. Rotavirus epidemiology and surveillance. Novartis Found. Symp. 238:125-147. [DOI] [PubMed] [Google Scholar]
- 18.Dunn, S. J., T. L. Cross, and H. B. Greenberg. 1994. Comparison of the rotavirus nonstructural protein NSP1 (NS53) from different species by sequence analysis and northern blot hybridization. Virology 203:178-183. [DOI] [PubMed] [Google Scholar]
- 19.Dunn, S. J., H. B. Greenberg, R. L. Ward, O. Nakagomi, J. W. Burns, P. T. Vo, K. A. Pax, M. Das, K. Gowda, and C. D. Rao. 1993. Serotypic and genotypic characterization of human serotype 10 rotaviruses from asymptomatic neonates. J. Clin. Microbiol. 31:165-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estes, M. 2001. Rotaviruses and their replication. In D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, 4th ed., p. 1747-1785. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 21.Fukai, K., Y. Maeda, K. Fujimoto, T. Itou, and T. Sakai. 2002. Changes in the prevalence of rotavirus G and P types in diarrheic calves from the Kagoshima Prefecture in Japan. Vet. Microbiol. 86:343-349. [DOI] [PubMed] [Google Scholar]
- 22.Gentsch, J. R., B. K. Das, B. Jiang, M. K. Bhan, and R. I. Glass. 1993. Similarity of the VP4 protein of human rotavirus strain 116E to that of the bovine B223 strain. Virology 194:424-430. [DOI] [PubMed] [Google Scholar]
- 23.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z. Y. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenberg, H. B., V. McAuliffe, J. Valdesuso, R. G. Wyatt, J. Flores, A. R. Kalica, Y. Hoshino, and N. Singh. 1983. Serological analysis of the subgroup protein of rotavirus, using monoclonal antibodies. Infect. Immun. 39:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gulati, B. R., O. Nakagomi, Y. Koshimura, T. Nakagomi, and R. Pandey. 1999. Relative frequencies of G and P types among rotaviruses from Indian diarrheic cow and buffalo calves. J. Clin. Microbiol. 37:2074-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hussein, A. H., E. Cornaglia, M. S. Saber, and Y. el Azhary. 1995. Prevalence of serotypes G6 and G10 group A rotaviruses in dairy calves in Quebec. Can. J. Vet. Res. 59:235-237. [PMC free article] [PubMed] [Google Scholar]
- 28.Iturriza Gómara, M., U. Desselberger, and J. Gray. 2003. Molecular epidemiology of rotaviruses: genetic mechanisms associated with diversity, p. 317-344. In U. Desselberger and J. Gray (ed.), Viral gastroenteritis. Elsevier Science, Amsterdam, The Netherlands.
- 29.Iturriza Gómara, M., C. Wong, S. Blome, U. Desselberger, and J. Gray. 2002. Molecular characterization of VP6 genes of human rotavirus isolates: correlation of genogroups with subgroups and evidence of independent segregation. J. Virol. 76:6596-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iturriza Gómara, M., E. Anderton, G. Kang, C. Gallimore, W. Phillips, U. Desselberger, and J. Gray. 2003. Evidence for genetic linkage between the gene segments encoding NSP4 and VP6 proteins in common and reassortant human rotavirus strains. J. Clin. Microbiol. 41:3566-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iturriza-Gómara, M., B. Isherwood, U. Desselberger, and J. Gray. 2001. Reassortment in vivo: driving force for diversity of human rotavirus strains isolated in the United Kingdom between 1995 and 1999. J. Virol. 75:3696-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iturriza-Gómara, M., J. Green, D. W. Brown, U. Desselberger, and J. J. Gray. 1999. Comparison of specific and random priming in the reverse transcriptase polymerase chain reaction for genotyping group A rotaviruses. J. Virol. Methods 78:93-103. [DOI] [PubMed] [Google Scholar]
- 33.Iturriza-Gómara, M., J. Green, D. W. Brown, U. Desselberger, and J. J. Gray. 2000. Diversity within the VP4 gene of rotavirus P[8] strains: implications for reverse transcription-PCR genotyping. J. Clin. Microbiol. 38:898-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang, G., M. Iturriza-Gomara, J. G. Wheeler, P. Crystal, M. Monica, S. Ramani, B. Primrose, P. D. Moses, C. Gallimore, D. W. Brown, and J. Gray. 2004. Quantitation of Group A rotavirus by real time reverse-transcription polymerase chain reaction: correlation with clinical severity in children in South India. J. Med. Virol. 73:118-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kapikian, A., Y. Hoshino, and R. Chanock. 2001. Rotaviruses, p. 1787-1834. In D. Knipe, P. M. Howley, et al. (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 36.Kojima, K., K. Taniguchi, and N. Kobayashi. 1996. Species-specific and interspecies relatedness of NSP1 sequences in human, porcine, bovine, feline, and equine rotavirus strains. Arch. Virol. 141:1-12. [DOI] [PubMed] [Google Scholar]
- 37.Lucchelli, A., S. Y. Kang, M. K. Jayasekera, A. V. Parwani, D. H. Zeman, and L. J. Saif. 1994. A survey of G6 and G10 serotypes of group A bovine rotaviruses from diarrheic beef and dairy calves using monoclonal antibodies in ELISA. J. Vet. Diagn. Investig. 6:175-181. [DOI] [PubMed] [Google Scholar]
- 38.Nakagomi, O., Y. Isegawa, R. L. Ward, D. R. Knowlton, E. Kaga, T. Nakagomi, and S. Ueda. 1994. Naturally occurring dual infection with human and bovine rotaviruses as suggested by the recovery of G1P8 and G1P5 rotaviruses from a single patient. Arch. Virol. 137:381-388. [DOI] [PubMed] [Google Scholar]
- 39.Nakagomi, O., and T. Nakagomi. 1993. Interspecies transmission of rotaviruses studied from the perspective of genogroup. Microbiol. Immunol. 37:337-348. [DOI] [PubMed] [Google Scholar]
- 40.Ramachandran, M., J. R. Gentsch, U. D. Parashar, S. Jin, P. A. Woods, J. L. Holmes, C. D. Kirkwood, R. F. Bishop, H. B. Greenberg, S. Urasawa, G. Gerna, B. S. Coulson, K. Taniguchi, J. S. Bresee, and R. I. Glass. 1998. Detection and characterization of novel rotavirus strains in the United States. J. Clin. Microbiol. 36:3223-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao, C. D., M. Das, P. Ilango, R. Lalwani, B. S. Rao, and K. Gowda. 1995. Comparative nucleotide and amino acid sequence analysis of the sequence-specific RNA-binding rotavirus nonstructural protein NSP3. Virology 207:327-333. [DOI] [PubMed] [Google Scholar]
- 42.Santos, N., R. C. Lima, C. F. Pereira, and V. Gouvea. 1998. Detection of rotavirus types G8 and G10 among Brazilian children with diarrhea. J. Clin. Microbiol. 36:2727-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh, D. 2003. Initiative will fast-track vaccine for childhood diarrhoea in developing world. Br. Med. J. 326:354.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sukumaran, M., K. Gowda, P. P. Maiya, T. P. Srinivas, M. S. Kumar, S. Aijaz, R. R. Reddy, L. Padilla, H. B. Greenberg, and C. D. Rao. 1992. Exclusive asymptomatic neonatal infections by human rotavirus strains having subgroup I specificity and “long” RNA electropherotype. Arch. Virol. 126:239-251. [DOI] [PubMed] [Google Scholar]
- 45.Tian, P., M. K. Estes, Y. Hu, J. M. Ball, C. Q. Zeng, and W. P. Schilling. 1995. The rotavirus nonstructural glycoprotein NSP4 mobilizes Ca2+ from the endoplasmic reticulum. J. Virol. 69:5763-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urasawa, T., K. Taniguchi, N. Kobayashi, K. Mise, A. Hasegawa, Y. Yamazi, and S. Urasawa. 1993. Nucleotide sequence of VP4 and VP7 genes of a unique human rotavirus strain Mc35 with subgroup I and serotype 10 specificity. Virology 195:766-771. [DOI] [PubMed] [Google Scholar]
- 47.Varshney, B., M. R. Jagannath, R. R. Vethanayagam, S. Kodhandharaman, H. V. Jagannath, K. Gowda, D. K. Singh, and C. D. Rao. 2002. Prevalence of, and antigenic variation in, serotype G10 rotaviruses and detection of serotype G3 strains in diarrheic calves: implications for the origin of G10P11 or P11 type reassortant asymptomatic strains in newborn children in India. Arch. Virol. 147:143-165. [DOI] [PubMed] [Google Scholar]