Abstract
The hemoglobins of the cyanobacteria Synechococcus and Synechocystis (GlbNs) are capable of spontaneous and irreversible attachment of the b heme to the protein matrix. The reaction, which saturates the heme 2-vinyl by addition of a histidine residue, is reproduced in vitro by preparing the recombinant apoprotein, adding ferric heme, and reducing the iron to the ferrous state. Spontaneous covalent attachment of the heme is potentially useful for protein engineering purposes. Thus, to explore whether the histidine–heme linkage can serve in such applications, we attempted to introduce it in a test protein. We selected as our target the heme domain of Chlamydomonas eugametos LI637 (CtrHb), a eukaryotic globin that exhibits less than 50% sequence identity with the cyanobacterial GlbNs. We chose two positions, 75 in the FG corner and 111 in the H helix, to situate a histidine near a vinyl group. We characterized the proteins with gel electrophoresis, absorbance spectroscopy, and NMR analysis. Both T111H and L75H CtrHbs reacted upon reduction of the ferric starting material containing cyanide as the distal ligand to the iron. With L75H CtrHb, nearly complete (> 90%) crosslinking was observed to the 4-vinyl as expected from the X-ray structure of wild-type CtrHb. Reaction of T111H CtrHb also occurred at the 4-vinyl in a 60% yield, indicating a preference for the flipped heme orientation in the starting material. The work suggests that the His–heme modification will be applicable to the design of proteins with a non-dissociable heme group.
Keywords: Truncated hemoglobin, 2/2 hemoglobin, heme post-translational modification, protein engineering, paramagnetic NMR spectroscopy
1. Introduction
Proteins requiring a b heme at their active site hold this essential molecule with multiple interactions. Protein–heme contacts, heme exposure to solvent, coordination of protein residues to the heme iron [1] and apoprotein structure [2] all contribute to the energetics of heme binding. As a consequence, the heme affinity of a given protein depends on solution conditions and properties such as the redox state of the iron [3] and the nature of its exogenous ligands [4, 5]. These last two features can dominate heme affinity. For example, soluble guanylate cyclase, which has an axial histidine ligand, loses heme upon oxidation from the ferrous to the ferric state [6], whereas Dap1p, which has an axial tyrosine ligand, exhibits enhanced heme loss upon reduction from the ferric state to the ferrous state [7]. Various enzymes experience heme loss upon exposure to nitric oxide (NO•) [8].
In part because of the toxicity of free heme, enzymes, gas sensors, and gas or small molecule carriers that use a b heme have evolved to maintain sufficient affinity as they visit the various ligation and redox states required by their physiological role. Uncontrolled heme loss occurs rarely. However, the structural features dictating heme affinity and reactivity are intimately intertwined. Thus, heme retention can be problematic when modifying native heme proteins [9, 10] or designing artificial ones [11]. In those instances, optimization for the desired chemical properties may compromise heme binding and vice versa.
Covalent attachment of the heme to the protein matrix offers a versatile solution to heme loss. Such post-translational modification (PTM) grants several advantages aside from heme retention, notably correct heme positioning and increased thermodynamic stability [12]. In addition, the PTM allows for a departure from the typical composition of a b heme cavity, i.e., the introduction of polar residues and weak axial ligands. The heme group, shown in Fig. 1a, offers several possible points for crosslinking to the protein, the most common of which are the vinyl substituents. In c cytochromes, reductive reaction with cysteines produces thioether bridges [13]. The formation of these linkages in the cell is controlled by dedicated enzymatic machineries that ensure the correct localization of the apoprotein prior to heme binding and modification [14, 15]. This complicated process can be harnessed to prepare artificial cytochromes [16, 17], and thus far the molecular biology procedure to producing the thioether linkages surpasses non-enzymatic methods [18, 19]. For purposes of creating new proteins with non-dissociable heme and studying the effects of heme crosslinking, a chemical approach avoiding the use of cysteines would be advantageous. We have shown previously that the globins from two cyanobacteria (GlbNs from Synechocystis sp. PCC 6803 and Synechococcus sp. PCC 7002) can undergo enzyme-free heme attachment to a histidine, and we now explore the suitability of this reaction for protein engineering applications.
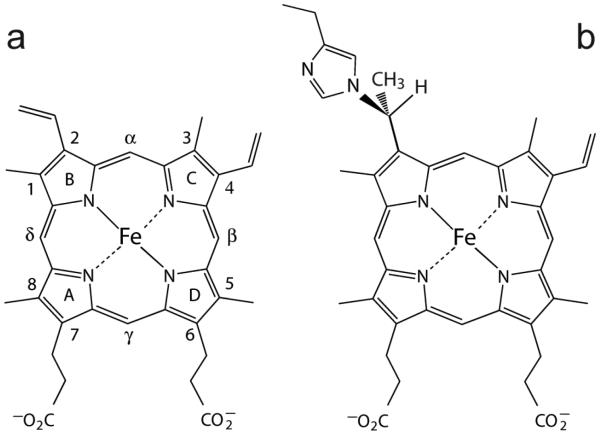
Fig. 1.
(a) The structure and nomenclature of the b heme (iron-protoporphyrin IX). The classic Fischer numbering system is used. Heme orientational isomerism involves a 180° rotation about the α–γ axis. (b) The heme PTM present in GlbN. His117, in the H helix, adds to the 2-vinyl group. Addition can also occur to the 4-vinyl group when the heme is in the flipped orientation [20, 21].
The heme modification in GlbN is the alkylation of histidine Nε by a heme vinyl Cα as shown in Fig. 1b. Several studies have defined the conditions under which the reaction occurs and have amounted to a plausible mechanism [20, 21]. Following reduction to the ferrous state, the key steps are the protonation of the heme vinyl Cβ followed by (or in concert with) a nucleophilic attack by a neutral histidine. In wild-type (WT) ferrous GlbNs, the modification reaches completion in a few seconds at neutral pH and is accelerated by decreasing pH as long as acid denaturation is avoided. We have monitored the reaction in variants of GlbN and proposed that the main determinant of linkage formation is sterics, i.e., whether a productive orientation of the vinyl and histidine ring can be adopted. This is evident in the preparation of a protein containing the WT crosslink (H117 to heme 2-vinyl) and an engineered crosslink (L79H to heme 4-vinyl) [22]. Additionally, we have noted that certain ligands to the ferrous state such as the π-acids CO [21], NO•and O2 (unpublished observations) inhibit the reaction, presumably because of their propensity for a redistribution of electron density of the ferrous heme (e.g., formation of Fe(III)–O2− from Fe(II)–O2, [23]) disfavoring vinyl protonation.
The consequences of the linkage for the reactivity of the heme group have been under investigation as well. A variant of GlbN unable to form the PTM loses the ferrous heme more readily than the ferric heme, which has led to the proposition that the linkage is a physiological necessity [24]. In WT GlbNs, the PTM enhances substantially the thermal stability of the holoprotein [25]. On the other hand, the PTM appears to have only moderate effects on the structure of GlbN and its chemical properties such as electron self-exchange rate, redox potential [26], susceptibility to H2O2-mediated degradation [21], backbone dynamics [25, 27], and response to pressure [28]. In view of these observations, the histidine–heme linkage appears as a mostly benign modification, well suited for protein engineering purposes. However, the general utility of the linkage must be demonstrated by introduction into a protein different from the two original cyanobacterial globins.
The heme domain of Chlamydomonas eugametos LI637, CtrHb, was chosen as a validation protein for several reasons. (1) CtrHb has been the focus of spectroscopic [23, 29, 30] and structural [31] studies. The WT protein is therefore already well characterized. (2) Sequence identity with Synechococcus and Synechocystis GlbNs (Fig. 2) is a modest ~46% with differences distributed throughout the structure, including the heme cavity. (3) In GlbN, the WT reactive histidine is in the H helix (His117, at helical position “H16”) and the engineered reactive histidine is in the FG corner (His79). A superimposition of the three-dimensional structures of cyanomet (ferric state with cyanide bound) CtrHb and post-translationally modified cyanomet Synechocystis GlbN (hereafter GlbN-A) (Fig. 2) reveals that CtrHb has two-residue deletions near the 79 and 117 sites of GlbN. (4) Unlike both GlbNs, CtrHb does not have a histidine that can reversibly coordinate the iron on the distal side. At neutral pH and in the ferrous state, CtrHb is primarily a 5-coordinate species [29]. Thus, CtrHb has distinct heme pocket composition, local backbone geometry, and iron coordination that render it a rigorous test case for the histidine–heme modification of the type depicted in Fig. 1b.
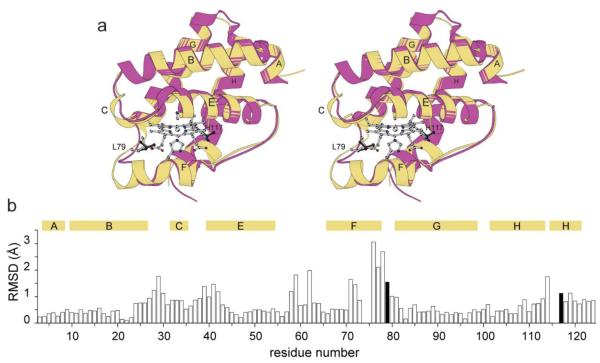
Fig. 2.
Structural comparison of cyanomet C. eugametos L1637 CtrHb (PDB ID: 1DLY) with cyanomet Synechocystis sp. PCC 6803 GlbN-A (PDB ID: 1S69). (a) Stereo view of the superimposition of the two structures. Synechocystis GlbN-A is in yellow with L79 and H117 shown in light gray sticks. Dark gray sticks mark L75 and T111 in CtrHb. (b) Cα rmsd for the superimposition shown in (a). The numbering is that of GlbN. The black vertical bars indicate the position of L79 and H117. The secondary structure of GlbN is indicated with horizontal bars above the rmsd values.
In this report, we inspected the reactivity of CtrHb variants with a histidine facing either the heme 2- or 4-vinyl group on the proximal side of the heme. We demonstrate that the histidine– heme modification can be successfully implanted in CtrHb. Although spontaneous crosslinking does not occur upon reduction to the ferrous state, the addition of cyanide prior to reduction promotes the reaction. The results support that histidine alkylation may be a generalizable reaction for the production of proteins containing a non-dissociable heme.
2. Experimental
2.1 Plasmid construction and protein production
The gene coding for WT CtrHb (UNIPROTKB Q08753, residues 44–164, “H19” in reference [32]) was synthesized with 5’-NdeI and 3’-BamHI restriction sites and inserted in a pJexpress 404 vector by DNA 2.0 (Menlo Park, CA). Because expression levels were low with this plasmid, WT and mutated CtrHb genes were subsequently cloned into a pET3c vector (Novagen, Inc., Madison, WI). Site directed mutagenesis was performed to introduce the T111H or L75H replacement using the QuikChange (Qiagen, Valencia, CA) protocol provided by the manufacturer and primers purchased from Integrated DNA Technologies (Coralville, IA). Restriction enzymes and ligase were from New England Biolabs (Ipswich, MA), and sequencing was performed by GENEWIZ, Inc. (South Plainfield, NJ).
E. coli BL21(DE3) cells (New England Biolabs) were used for overexpression of CtrHb. Cells were grown on M9 medium supplemented with ampicillin (Sigma-Aldrich, St. Louis, MO or Research Products International, Corp., Mount Prospect, IL). Induction was achieved by addition of 1 mM dioxane-free isopropyl β-D-1-thiogalactopyranoside (Santa Cruz Biotechnologies, Santa Cruz, CA). For uniformly labeled 15N protein, 15NH4Cl (Sigma-Aldrich) was used as the sole nitrogen source.
2.2 CtrHb purification
WT and T111H CtrHbs were purified from inclusion bodies as described previously for Synechocystis GlbN [33]. The protocol calls for treatment of the harvested and washed inclusion bodies with 8 M urea, followed with apoprotein refolding by passage over a gel filtration column (Sephadex G-50 Fine, Sigma-Aldrich) equilibrated with 50 mM Tris, 50 μM EDTA pH 8.0 at 4 °C. Further purification was performed by passage through an anion exchange column (DEAE Sephacel, Sigma) either before or after addition of bovine hemin chloride (Fe(III)-protoporphyrin IX, Sigma/Alfa Aesar) to generate the ferric holoprotein. Elution was achieved with a 0–0.4 M NaCl gradient. Although WT and T111H CtrHbs could be lyophilized and resuspended without loss or damage, such was not the case for L75H CtrHb, and solutions of this protein were used immediately upon purification. The yields were ~15 mg/L for L75H CtrHb, ~39 mg/L for T111H CtrHb, and ~50 mg/L for WT CtrHb when the cells were grown in minimal medium.
Apoprotein concentrations were estimated using a calculated extinction coefficient of 10 mM−1 cm−1 at 280 nm. The optical spectra of the WT CtrHb in the ferric, ferrous, cyanomet, and ferrous cyanide states were comparable to those reported in the original work of Guertin and colleagues [29, 32, 34] (Supporting Information Table S1). The T111H and L75H replacements affected the spectral properties to varying degrees, with minimal effect on the cyanomet state (Supporting Information Fig. S1). Ferric holoprotein concentrations were determined on a per-heme basis with an extinction coefficient of 117 mM−1 cm−1 at 410 nm at neutral pH, back-calculated from the published WT cyanomet coefficient [34].
2.3. NMR sample preparation
Lyophilized protein was resuspended in either 100 or 250 mM phosphate buffer in H2O or 100 mM phosphate in D2O. 10% v:v D2O was added to the H2O samples. Protein concentration varied from ~0.6 mM to ~3 mM (final volume of ~300 μL). Sample pH ranged between 7.1 and 7.5. Iron reduction was performed by adding 3- to 13-fold excess sodium dithionite (DT, Sigma-Aldrich). Alternatively, freshly purified protein was concentrated, exchanged in the appropriate buffer, and used without lyophilization. Cyanomet CtrHb was generated by adding a 5-fold excess of potassium cyanide to ferric holoprotein solutions.
2.4 Heme modification
Stocks of ferric WT (as a negative control), T111H, and L75H CtrHbs were prepared either from lyophilized protein, or, in the case of L75H CtrHb, from freshly purified protein. A 5-fold excess of cyanide was added to each sample, followed by 20–30 min incubation at room temperature to ensure saturation of the heme site. An excess of DT (200- to 500-fold for optical samples or 3- to 13-fold for NMR samples) was then added to the solutions. Cyanomet CtrHb samples were incubated for ~3–8 h (WT and T111H) or ~0.5–2 h (L75H) and then oxidized by exposure to air or potassium ferricyanide. Aliquots of the total samples were saved at different stages of the process for optical characterization.
2.5 Hemochromogen and enhanced chemiluminescence assays
Reacted CtrHb samples were analyzed by the hemochromogen assay [35, 36] for rapid assessment of b heme modification. Aliquots were taken from samples used for NMR analysis, diluted to reach an absorbance between 0.3 and 0.6 in the α/β region (500–600 nm), and treated as previously described [20] with basic pyridine followed by DT reduction. Absorbance measurements were performed on an AVIV Spectrophotometer 14DS UV-Vis (AVIV Biomedical Inc., Lakewood, NJ) as described previously [22].
Covalent heme attachment was detected by enhanced chemiluminescence (ECL) [37, 38]. Samples of CtrHb were prepared for DT reduction in pair, one with cyanide present (referred to as “KCN+”) and the other without cyanide (referred to as “KCN−”). CtrHb samples were also prepared with 10-fold excess imidazole (Sigma) or 10 mM azide (Sigma) as alternatives to cyanide as the distal iron ligand. Synechocystis GlbN treated with excess DT to generate GlbN-A [39] was used as a covalently-linked positive control. The ECL assay relies on the peroxidase activity of the heme group. This activity is inhibited by cyanide and high concentrations of either imidazole or azide. Prior to ECL detection, reduced samples containing these ligands were treated by passage over a ~1.3 mL DEAE column. This procedure did not eliminate all exogenous ligand, and the ECL results for these samples are therefore only qualitative.
Electrophoresis was performed with Mini-PROTEAN® 16.5% Tris-Tricine Precast Gel (BIO-RAD, Hercules, CA). The protein samples (5 μM) were denatured by boiling for 10 min with SDS but in the absence of β-mercaptoethanol. After separation, the gels were thoroughly rinsed and exposed to the ECL reagent (Immobilon Western Chemiluminescent HRP (horseradish peroxidase) Substrate, Millipore, Billerica, MA) prepared per manufacturer’s instructions. The ECL reaction was allowed to proceed for 7 min before applying the film (Carestream® Kodak® BioMax® light film, Sigma-Aldrich, St. Louis, MO). Exposure times were 30 s for samples containing no exogenous ligand and 10 s, 30 s, or 7 min for samples partially stripped of exogenous ligand. After the ECL procedure, gels were stained with Coomassie Blue for protein detection.
2.6 Reaction kinetics monitored by absorbance spectroscopy
Samples of WT, T111H, and L75H CtrHb containing ~4 μM protein were prepared in 100 mM phosphate buffer pH 7.0 from concentrated stocks (lyophilized WT and T111H) or freshly prepared protein (L75H). Five-fold excess KCN was added to the samples, and CN− binding was monitored via electronic absorption spectroscopy. Approximately 500-fold excess DT was added to cyanide-saturated samples and reduction was monitored as above. Time course experiments were performed on a Varian Cary 50 Bio UV-Vis spectrophotometer with acquisition parameters similar to those reported previously [22].
2.7 NMR spectroscopy
NMR data were acquired on Bruker AVANCE or AVANCE-II spectrometers operating at a 1H Larmor frequency of 600.13 or 600.53 MHz, respectively, and each equipped with a cryoprobe. Reduction and reaction progress of cyanomet CtrHbs were monitored by 1H 1D NMR spectroscopy with water presaturation. Reactants and products were additionally studied using 1H-1H NOESY (nuclear Overhauser effect spectroscopy), DQF-COSY (double-quantum filtered correlation spectroscopy), TOCSY (total correlation spectroscopy) with TOWNY (total correlation spectroscopy without NOESY) sequence, and natural abundance 1H-13C HMQC (heteronuclear multiple quantum correlation) spectra [40-42]. These data were sufficient to assign most heme resonances and locate many conserved heme pocket residues of each cyanomet CtrHb. Labile protons were identified by acquisition of the same data in 99.9% D2O. 1H-15N HSQC (heteronuclear single quantum correlation) and long-range (lr) histidine selective 1H-15N HMQC spectra [43] were used to confirm the assignments of the engineered and native histidines within WT, T111H and L75H cyanomet CtrHbs [21, 22]. 1H chemical shifts were referenced to DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid) indirectly through the water resonance (4.76 ppm at 298 K); 13C and 15N chemical shifts were referenced as described in [44]. NMR data were processed with TopSpin 2.1 or NMRPipe [45]. Spectral analysis was conducted with Sparky 3 [46].
3. Results
3.1. Choice and preparation of CtrHbs
The selection of sites for histidine introduction was guided by inspection of the CtrHb and GlbN structures (Fig. 2). A significant proportion of CtrHb molecules contain the heme group in a “flipped” orientation that interchanges pyrroles A and B with pyrroles D and C, respectively [31]. These two forms of the protein are referred to as “major” and “minor” heme rotational isomers according to their relative population at equilibrium. The X-ray structure (PDB ID: 1DLY [31]) is that of the major isomer. The two heme conformations effectively double the number of vinyl geometries available for histidine addition. Minimal modeling (amino acid replacement in 1DLY and optimization with UCSF Chimera [47]) suggests that a histidine at position 111, the closest equivalent to H117, can reach the heme 2-vinyl group as positioned in the major heme rotational isomer. The favored rotameric state of H111 points the ring away from the heme toward solvent and generates few clashes. Another potentially reactive site, facing the 4-vinyl group of the major heme rotational isomer, was identified at position 75 as the equivalent of H79 in L79H GlbN. At this position, clashes are predicted regardless of histidine rotameric state. Sites 75 and 111 were chosen despite their suboptimal geometric properties for lack of better candidates.
The apoproteins of WT CtrHb and its L75H and T111H variants aggregated into inclusion bodies during E. coli cell growth. Purification included unfolding in urea, refolding over a size-exclusion column, and reconstitution with ferric heme. For WT and T111H CtrHb, stoichiometric amounts of heme were added prior to a final purification step by anion exchange chromatography. L75H CtrHb had low ferric heme affinity, and passage through this last column resulted in substantial heme loss. To minimize this problem, heme was titrated to a slightly sub-stoichiometric level as the last step of holoprotein preparation, after apoprotein purification by anion exchange chromatography. It is likely that the presence of H75 disrupts the hydrophobic packing near the heme and interferes with heme binding. This was also the proposed explanation for the low heme affinity of the L79H variants of Synechocystis GlbN [22].
3.2. NMR characterization of cyanomet CtrHbs
Ferric CtrHbs to which KCN was added to generate the cyanomet state (S = 1/2) yielded high quality NMR spectra. Fig. 3a, b, and c show the 1H 1D spectrum of cyanomet WT CtrHb and the T111H and L75H variants. In all cases, two sets of resonances are detected per protein. These arise from the heme rotational isomers, which typically exchange slowly on the chemical shift time scale [48, 49]. Assignments of most heme resonances of both isomers in WT CtrHb and the T111H and L75H variants were obtained by established procedures [50]. Portions of spectra with representative through-bond and through-space connectivities are shown in Supporting Information Fig. S2–S11. Heme and protein assignments confirm that in WT, T111H, and L75H cyanomet CtrHbs, the major isomer corresponds to approximately 60–70% of the sample and has the heme positioned as in the crystallographic WT structure (1DLY [31], Supporting Information Fig. S12A–F). NMR data also support that key features of the WT CtrHb heme pocket, such as the presence of Tyr20 (B10) as an H-bond donor to bound cyanide (Supporting Information Fig. S5) and the participation of Gln41 (E7) and Gln45 (E11) in an extended H-bond network (Supporting Information Fig. S6) [31], are largely unaltered by the T111H and L75H replacements (data not shown). Heme 1H chemical shifts of WT and variant cyanomet CtrHbs are listed in Table 1.
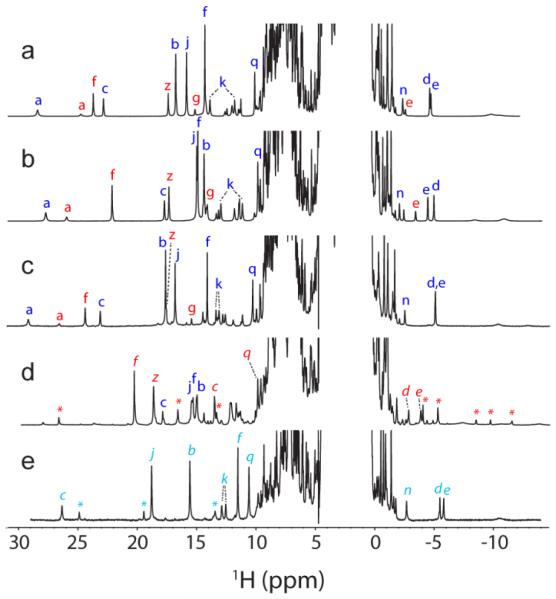
Fig. 3.
Comparison of WT, T111H, and L75H cyanomet CtrHb 1H NMR spectra. (a–c). Spectra collected prior to DT treatment. The major and minor heme orientational isomers are indicated with blue and red letters, respectively. (a) Wild-type cyanomet CtrHb (72% major, 28% minor). (b) T111H cyanomet CtrHb (67% major, 33% minor). (c) L75H cyanomet CtrHb (62% major, 38% minor). (d–e). Spectra collected after DT treatment and reoxidation. (d) T111H cyanomet CtrHb-A4 product mixture. The predominant form (~60%, H111 reaction with the 4-vinyl group) is marked with red italics. The spectrum also contains 25% of the unreacted major isomer (starting material, b, blue letters), and a small amount of a third species (marked with red *). (e) L75H cyanomet CtrHb-B product (~90%, H75 reaction with the 4-vinyl group). A second product species (10%, likely H75 reaction with the 2-vinyl group) is marked with cyan *. Peak labeling: a, Tyr20 OηH; b, heme 1-CH3; c, heme 2-vinyl Hα; d, e, heme 2-vinyl Hβcis, Hβtrans; f, heme 3-CH3; g, heme 4-vinyl Hα; j, heme 5-CH3; k, heme 6-propionate Hα, Hα’; n, heme 7-propionate Hβ; q,Tyr20 CεHs; z, heme 8-CH3.
Table 1.
Heme b and engineered histidine chemical shifts of WT, T111H, and L75H cyanomet CtrHbsa.
| WT | WT* | T111H | T111H*b | L75H | L75H* | |
|---|---|---|---|---|---|---|
| 1-CH3 | 16.77 | 3.2 | 14.38 | 3.13? | 17.64 | |
| 2-v α | 22.87 | 12.46 | 17.75 | 13.20 | 23.14 | 12.85 |
| 2-v βc, βt | −4.55, −4.65 | −1.69, −2.49 | −4.84, −4.34 | −2.34, −3.32 | −5.10, −5.11 | −1.11, −1.42 |
| 3-CH3 | 14.35 | 23.71 | 14.97 | 22.08 | 14.17 | 24.42 |
| 4-v α | 6.68 | 15.17 | 7.04 | 14.23 | 6.59 | 15.46 |
| 4-v βc, βt | 0.29, 0.58 | −1.53, −0.12 | 0.38, 0.72 | −1.13, −0.05 | 0.33, 1.12 | −1.81, −0.47 |
| 5-CH3 | 15.87 | 5.65 | 15.02 | 6.02? | 16.83 | 6.17? |
| 6-p α, α’ | 13.88, 11.76 | 9.35, 2.75 | 12.96, 11.16 | 9.28, 2.77 | 13.35, 13.08 | 8.69, 3.60 |
| 6-p β, β’ | 0.32, −0.98 | −0.84, −1.53 | 0.30, −0.99 | −1.63, −0.89 | 0.09, −1.06 | −1.27, −2.08 |
| 7-p α, α’ | 7.84, 3.68 | 14.21, 11.38 | 7.97, 4.57 | 14.46, 11.44 | 7.06, 3.66 | 12.79, 11.90 |
| 7-p β, β’ | −1.34, −2.31 | 1.00, −0.22 | −1.99, −1.08 | 0.59, −0.68 | −1.56, −2.53 | 0.62, −0.42 |
| 8-CH3 | 6.17 | 17.42 | 7.49 | 17.36 | 5.91 | 17.64 |
| H111 Hε1, Hδ2 | n/a | n/a | 7.86, 7.28 | 7.91, 7.32 | n/a | n/a |
| H75 Hε1, Hδ2 | n/a | n/a | n/a | n/a | 9.43, 6.29 | |
| H75 Nδ1, Nε2 | n/a | n/a | n/a | n/a | 252.1, 163.0 |
pH 7.2, 298 K.
A
refers to the minor heme orientational isomer.
A ? indicates a tentative assignment.
Inspection of Fig. 3 shows that at neutral pH the histidine replacement at position 111 perturbs the spectrum more profoundly than at position 75. In the major heme isomer of T111H CtrHb, the substituents of the B pyrrole undergo a relatively large chemical shift perturbation, apparently related to the reorientation of the 2-vinyl group from a “cis”-like conformation in WT CtrHb (i.e., with Cβ out of the heme plane and pointing toward the 3-CH3, Supporting Information Fig. S2) to a “trans”-like conformation (i.e., Cβ pointing toward the 1-CH3, Supporting Information Fig. S7). NOEs are detected between H111 Hδ2 and the heme 1-CH3, whereas H111 Hε1 makes dipolar contact with F48 Hζ. The geometry is consistent with an outward position of the histidine Nε (g− rotameric state) and forecasts a low probability for reaction with the 2-vinyl Cα. In the minor isomer, H111 Hδ2 contacts the 4-vinyl Hβcis and exhibits the same NOE to F48, in support of a similar conformation for H111 in both major and minor isomers. 1H-15N lr-HMQC data collected at neutral pH do not reveal the H111 spin system (Supporting Information Fig. S13), presumably because of broadening due to exchange between neutral and protonated states.
The heme chemical shifts of cyanomet L75H CtrHb form a pattern similar to that of WT CtrHb. The side chain of H75 in the major isomer was identified by comparison with WT spectra; the unusual Hε1 and Hδ2 chemical shifts (9.30 ppm and 6.15 ppm, respectively) reflect paramagnetic and heme ring current contributions. NOEs to the heme and proximal histidine further define the position of the imidazole ring (not shown). Observed contacts are between H75 Hε1 and H68 (proximal histidine) Hβs, and between H75 Hδ2 and the heme 3-CH3 and 4-vinyl. The 4-vinyl has a trans-like orientation (i.e., Cβ pointing toward the 3-CH3, Supporting Information Fig. S10, and analogous to the WT and T111H variant, Supporting Information Fig. S2 and S7, respectively). The data are consistent with a g− rotameric state, which orients the imidazole ring toward the heme and may favor reaction. Comparison of WT and L75H 1H-15N lr-HMQC spectra confirms the histidine assignments and further indicates that, at neutral pH, the engineered H75 side chain is in the Nε2H tautomeric state (Supporting Information Fig. S13).
3.3. Heme modification in T111H and L75H CtrHb
The first attempt at producing CtrHbs with covalently attached heme was based on results obtained with Synechococcus and Synechocystis GlbNs. For these proteins to undergo the reaction, it suffices to reduce the heme iron [39]. Ferric T111H and L75H CtrHb were incubated with excess DT for variable periods of time, up to ~2 h, oxidized with potassium ferricyanide, and then exposed to an excess of KCN for NMR comparison with their respective untreated form. Absence of reaction is hinted at by unchanged optical spectra and made apparent by ECL assay of denaturing gels, which detects heme only in the dye front of DT-treated CtrHbs (Fig. 4a, lanes 1–3). In this assay Synechocystis GlbN-A shows a strong signal at the protein’s molecular weight (Fig. 4a, lane 4), as expected. The NMR spectra of the treated CtrHbs are not presented as they are identical to those of the non-reduced starting material.
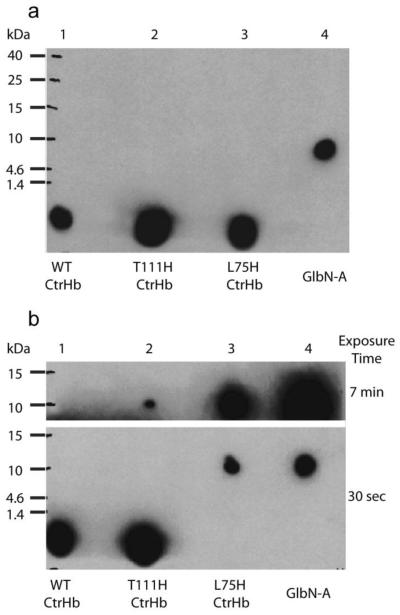
Fig. 4.
ECL SDS-PAGE detection of covalently bound heme in CtrHbs. (a) Ferric samples were reduced with 2 mM DT for ~30 min then re-oxidized with air before being subjected to electrophoresis. The image was obtained with a 30-s exposure. The heme of WT CtrHb and the T111H and L75H variants (lane 1-3) migrates at the dye front. WT GlbN-A (lane 4) is included as a positive control for covalent linkage. (b) Ferric WT and variant CtrHbs were first saturated with cyanide and then reduced for ~30 min. The figure shows images of film that was exposed for either 30 s or 7 min. The heme from WT CtrHb (lane 1) migrates with the dye front. The T111H CtrHb lane (lane 2) shows incomplete heme attachment. The longer exposure time was necessary to observe the faint crosslinked T111H CtrHb band. The L75H CtrHb lane (lane 3) shows complete crosslinking as in the GlbN-A control (lane 4).
Failure to react can be due to various factors, including heme dissociation, incorrect histidine–vinyl geometry or unfavorable electron distribution of the porphyrin ring [22]. Binding of a suitable (permissive) ligand on the distal side may alleviate some of these problems. We therefore chose to initiate reduction from the stable and well-folded cyanomet state. The resulting ferrous–cyanide complex has a distinctive optical spectrum [34] (Supporting Information Fig. S1A-D), and although it is not expected to be as stable as its ferric counterpart [51], it may persist long enough to assist in the reaction.
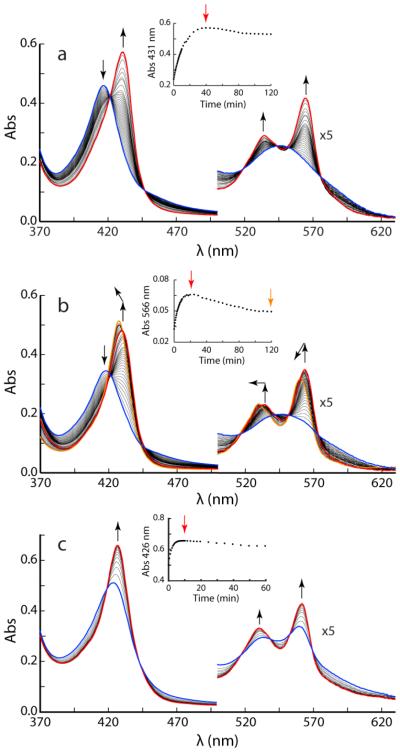
Reduction of cyanomet WT CtrHb with 500-fold excess DT at pH 7.0 was monitored optically (Fig. 5a). Over a period of 40 min, the Soret maximum shifts from 416 nm to 431 nm and sharpens considerably. The end spectrum displays resolved α and β bands and corresponds to that of the cyanide adduct of ferrous CtrHb reported by Milani and coworkers [34] (Supporting Information Table S1). The transition displays isosbestic points, which is an indication of an apparent two-state process. At longer incubation times, a slow but nearly uniform decay in absorbance is observed and attributed to DT-mediated heme bleaching. Reduction of the WT protein without exogenous ligand but under otherwise identical conditions is considerably faster (complete within 2 min) and leads to a broad Soret band at neutral pH (Supporting Information Fig. S1B). The WT CtrHb behavior was used to gauge the reaction of the variants.
Fig. 5.
DT-mediated reduction of cyanomet CtrHbs monitored by UV-vis spectrophotometry. In each reaction, spectra were taken every 30 s for the first 10 min, every 2 min for the following 20 min, and every 5 min for the remaining 1.5 h. (a) Reduction of cyanomet WT CtrHb. The reduction progresses over 40 min from the blue spectrum to the red spectrum. Inset: kinetic trace at 431 nm; the decrease in signal after 40 min (time marked by red vertical arrow) is attributed to damage by DT. (b) Reduction of cyanomet T111H CtrHb. The initial response is similar to WT (blue to red spectrum) but occurs within 25 min. An additional blue shift of the spectrum is distinguished after this first phase (red to orange spectrum). Inset: kinetic trace at 566 nm to show the biphasic nature of the spectral evolution. The red and orange arrows mark the time of the red and orange spectra. (c) Reduction of cyanomet L75H CtrHb. The Soret band shifts to ~426 nm within the ~10-s manual mixing dead time. The reduction progresses over 10 min from the blue spectrum to the red spectrum, which is distinct from ferrous L75H CtrHb and corresponds to crosslinked L75H. Inset: kinetic trace at 426 nm, the decrease in signal after 10 min (time marked by red vertical arrow) is attributed to damage by DT. Note that only the first 60 min are shown.
Cyanomet T111H CtrHb shows a response similar to that of cyanomet WT CtrHb, albeit on a slightly faster time scale for reduction (complete within 25 min under the same conditions as WT CtrHb). In addition, after 2 h of incubation with excess DT, the Soret maximum has moved from 430 nm to 428 nm (Fig. 5b, red to orange traces). Similar changes are observed for the α and β bands. These blue shifts suggest a decreased degree of conjugation of the macrocycle as caused by crosslink formation.
Reduction of cyanomet L75H CtrHb gives distinct results (Fig. 5c). Addition of DT leads to a rapid (< 2 min) sharpening of the Soret maximum with a shift to 426 nm. The intensity is at first lower than expected based on the WT extinction coefficient and increases over time while remaining at 426 nm. The final spectrum (Supporting Information Fig. S1D) differs from that of the ferrous protein (Supporting Information Fig. S1B) and is inconsistent with a simple loss of cyanide from the reduced state.
The hemochromogen assay was performed to judge heme integrity. Both T111H and L75H CtrHbs treated with DT in the cyanomet state exhibit a blue shift of the hemochrome α and β maxima compared to WT CtrHb (data not shown) in support of a modified heme. Furthermore, Fig. 4b illustrates that ECL staining of treated cyanomet L75H CtrHb detects heme only where the protein has migrated, similar to WT GlbN-A and in support of covalent attachment. ECL staining of treated cyanomet T111H CtrHb showed the presence of heme associated with the protein as well as free heme migrating with the dye front.
The changes observed in the optical spectra upon reduction of cyanomet T111H and L75H CtrHbs, along with the ECL results, indicated the presence of a covalent bond between the heme and protein in treated samples, but did not reveal the nature of the bond. Thus, these data warranted a detailed examination of the products by NMR spectroscopy. For both variants, the ferric cyanomet form was first generated by incubation with excess potassium cyanide and upon saturation, reacted with excess DT and monitored kinetically by 1H NMR spectroscopy for at least 2 h (Supporting Information Fig. S14–16). At the end of this period, the samples were exposed to potassium ferricyanide or air and completely reoxidized to generate the S = 1/2 cyanomet state. WT cyanomet CtrHb, when subjected to this treatment shows signs of modification. Reaction extent is small but reproducible, clearly detectable by NMR spectroscopy (Supporting Information Fig. S14C), but does not yield a covalently attached heme (Fig. 4b, lane 1). This illustrates the potential for protein damage, likely due to the production of O2− or H2O2, when using DT to generate the crosslink. Comparison of the cyanomet 1H-1D NMR spectra of T111H and L75H CtrHbs before and after treatment shows significant differences (Fig. 3b–e).
3.4. NMR Identification of the heme modification in T111H CtrHb
The T111H CtrHb spectrum resulting from the treatment (Fig. 3d) contains a multitude of hyperfine shifted lines with intensities suggesting a mixture of at least three species. The new predominant set of resonances corresponds to a major product accounting for ~60% of the sample. The inability for DT to reduce cyanomet T111H CtrHb completely under these NMR conditions can be seen from Supporting Information Fig. S15 and explains the second set of peaks, which corresponds to the cyanomet major heme orientational isomer of the starting material (~25%). Finally, a third set corresponds to a minor product that may be due to a different modification of the heme or DT-damaged protein. Notably, after treatment, peaks corresponding to the minor cyanomet isomer of the T111H starting material have vanished. We therefore hypothesize that the primary product contains a covalent linkage between H111 and the heme 4-substituent in an orientation analogous to the minor isomer starting material. We refer to this product as T111H CtrHb-A4 (Supporting Information Fig. S20A).
Signals corresponding to heme methyl groups of T111H CtrHb-A4 are assigned on the basis of their high intensity and characteristic paramagnetic downfield 1H and upfield 13C shifts. DQF-COSY spectra reveal one vinyl and both propionate spin systems. Intra-heme NOEs between the 1-CH3 and 8-CH3, along with contacts between the heme 1- and 2-substituents indicate that the heme 2-vinyl group has remained intact (Supporting Information Fig. S17). Cyanomet T111H CtrHb 1H-15N lr-HMQC data collected before and after DT treatment and compared with WT data indicate the presence of a peculiar imidazole system [52] in the T111H CtrHb-A4 product, exhibiting two downfield shifted 15N signals J-coupled to a proton at 8.36 ppm, itself coupled to a proton at 6.48 ppm. These resonances are attributed to H111 (Table 2). Restraints defining the position of the side chain include: H111 Hε1 to heme 3-CH3 and F48 Hζ and Hε protons, and H111 Hδ2 to the heme 3-CH3 and V115 side chain methyls. Both H111 Hε1 and Hδ2 protons have shared NOEs (strong and weak, respectively) to a methyl group at 3.39 ppm, which is assigned to the modified heme 4-CβH3 group. Accordingly, a scalar connectivity is observed between the 4-CβH3 and a single proton at 6.15 ppm (Supporting Information Fig. S17) corresponding to the modified heme 4-CαH. This proton, like the 4-CβH3, is in dipolar contact with H111 Hε1. Although JHN-coupling between H111 15Nε2 and the newly produced 4-CβH3 or 4-CαH was not detected in the long-range data, the observed NOEs, heme methyl 1H chemical shift dispersion pattern [53], propionate shifts, and unusual H111 15N shifts are all consistent with modification occurring at H111 and the heme 4-substituent (Supporting Information Fig. S20A).
Table 2.
Reacted heme and alkylated histidine chemical shifts of T111H-A4 and L75H-B cyanomet CtrHbsa
| T111H-A4 | L75H-B | |
|---|---|---|
| 1-CH3 | 2.02 | 15.57 (−26.3) |
| 2-v α | 13.47 | 26.44 |
| 2-v βc, βt | −2.83, −3.84 | −5.46, −5.78 |
| 3-CH3 | 20.26 | 11.55 (−31.1) |
| 4-α | 6.13 | 0.54 |
| 4-β-CH3 | 3.45 | −0.49 (25.2) |
| 5-CH3 | 5.88 | 18.81 (−34.8) |
| 6-p α, α’ | 8.50, 1.87 | 12.91, 12.56 |
| 6-p β, β’ | −1.03, −1.84 | −0.27, −1.57 |
| 7-p α, α’ | 15.46, 12.19 | 7.74, 3.89 |
| 7-p β, β’ | 0.53, −0.81 | −1.75, −2.66 |
| 8-CH3 | 18.65 | 5.66 (−12.7) |
| H111 Hε1, Hδ2 | 8.36, 6.48 | n/a |
| H111 Nδ1, Nε2 | 250.6, 220.7 | n/a |
| H75 Hε1, Hδ2 | n/a | 7.12, 5.42 |
| H75 Nδ1, Nε2 | n/a | 254.6, 198.6 |
pH 7.2, 298 K.
Values in parentheses are 13C shifts.
3.5. NMR Identification of the heme modification in L75H CtrHb
Unlike WT and T111H cyanomet CtrHbs, which are reduced slowly and with different efficiencies between the major and minor forms (Supporting Information Fig. S14–15), DT reduction of L75H cyanomet CtrHb leads to complete and rapid disappearance of all cyanomet resonances (Supporting Information Fig. S16). Reoxidation results in a spectrum (Fig. 3e) distinct from the starting material (Fig. 3c) and consistent with high yield of one major product (> 90 %, henceforth L75H CtrHb-B). Only one heme vinyl group can be identified in DQF-COSY data of the product (Supporting Information Fig. S18). The intact vinyl Hα exhibits an NOE to a heme methyl, itself having a weak intraheme connectivity to a second heme methyl. The vinyl and methyl chemical shifts (Table 2) and NOE pattern are analogous to the WT 2-vinyl ↔ 1-CH3 ↔ 8-CH3 sequence and suggest that the reaction occurs on pyrrole C. Also similar to the WT, characteristic 1-CH3 ↔ F48 and 2-vinyl Hα ↔ F80 contacts are observed. Of the 2-vinyl βcis and βtrans protons, the latter is oriented toward a third heme methyl, assigned as the heme 3-CH3 (Supporting Information Fig. S18C). This methyl shows distinctive dipolar contacts, including strong NOEs to the side chain methyls of V83 and the ring of F80. In addition, the heme 3-CH3 displays an NOE to a methyl group, the latter (−0.49 ppm, Supporting Information Fig. S18D) J-coupled to a proton resonating at 0.54 ppm. The relative intensity of the 0.54 ppm and −0.49 resonances is consistent with a 4-CαH-CβH3 heme moiety. The 3:1:1:3 splitting pattern observed in the 13C dimension of the 1H-coupled natural abundance 13C spectrum confirms that the signal at −0.49 ppm corresponds to a methyl group (Supporting Information Fig. S19). The 4-CαH is oriented towards the heme 5-CH3 and the 4-CβH3 is proximal to the heme 3-CH3, as in the starting material L75H major isomer.
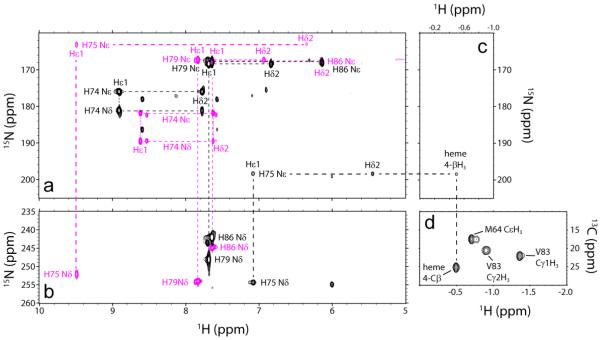
Because WT CtrHb does not form a crosslink, H75 is the logical candidate for modification. 1H-15N lr-HMQC spectra collected on cyanomet WT and L75H CtrHbs before and after treatment demonstrate that, in L75H CtrHb-B, signals corresponding to intact H75 are missing and a new set of shifted cross peaks are detected. Both H75 imidazole nitrogens exhibit J-coupling to a proton at 7.12 ppm (Hε1). The ring 15Nε2 is also J-coupled to a proton at 5.42 ppm (Hδ2) and to the heme 4-CβH3 (−0.49 ppm, Fig. 6). NOE connectivities define the position of the modified histidine: H75 Hδ2 has strong dipolar interactions with both the heme 3- and 4-CβH3 groups, whereas H75 Hε1 is oriented towards the 5-CH3 group (data not shown). Together, these data support that the H75 ring undergoes a rotation from its original position and forms the anticipated crosslink, with R stereochemistry at the heme 4-Cα, as depicted in Supporting Information Fig. S20B. Interestingly, for both T111H and L75H CtrHbs, the engineered histidine reorients in order to react with the heme.
Fig. 6.
Histidine assignment in cyanomet L75H CtrHb and product CtrHb-B by NMR spectroscopy (pH 7.1–7.2, 298 K). (a) Overlay of 1H-15N lr-HMQC data (15N upfield region). Signals from all histidine residues (except proximal H68) were detected. Reactant (magenta) and product (black) L75H histidine spin systems are connected with dashed lines. (b) Overlay of 1H-15N lr-HMQC data (15N downfield region). Colors are as in (a). Note that the similarity of the H86, H74, and H79 imidazole signals (a, b) in CtrHb and CtrHb-B. Conversely, H75 Nε2, along with Hε1 and Hδ2, undergoes significant changes. (c) 1H-15N lr-HMQC data (1H far-upfield region). In L75H cyanomet CtrHb-B, H75 Nε2 has a cross peak at −0.49 ppm, indicative of histidine N-alkylation. (d) Portion of a natural abundance 1H-13C HMQC spectrum collected on L75H cyanomet CtrHb-B. The coupled 1H-13C HSQC spectrum (Supporting Information Fig. S19) confirms that the resolved peak at a proton shift of −0.49 ppm is a methyl group.
3.6 Mechanistic considerations
The data accumulated with GlbN [20-22, 26] has provided a set of criteria for the addition of histidine to the ferrous heme. The first is proximity. The histidine must be within reach of a heme vinyl, and sp3 geometry at the vinylic Cα atom must be sterically possible; however, the histidine need not be optimally oriented in the starting material as long as the barrier to adopt the productive state can be overcome thermally. The second is the ionization state of the histidine. In GlbN, data are consistent with the histidine providing the proton to the vinyl Cβ. Protonation is the rate determining step and the reaction should be carried out at a sufficiently low pH. A third criterion is the nature of the iron distal ligand. These are either permissive (histidine, cyanide, water/5-coordinate) [20] or inhibiting (O2 and NO, data not shown; CO) [21, 26]. The inhibiting ligands have a slow dissociation rate constant, and their electron-withdrawing property is presumably responsible for the inhibition. In GlbN, the permissive ligands are not tightly bound and may or may not be released prior to protonation and nucleophilic attack.
The behavior of the CtrHb variants brings new insights into the mechanism and limitations of the histidine addition. Ferrous WT CtrHb at neutral pH is reported to be a mixture of 4-, 5-, and 6-coordinate species, with equilibrium favoring the 5-coordinate high-spin complex [29]. In preliminary experiments, this protein was found to lose heme rapidly to apomyoglobin (half-life of ~8 min at pH 7.7, data not shown). Optical spectra suggest the same mixture of species in reduced T111H CtrHb and a slight increase in the population of a 6-coordinate species in L75H CtrHb (Supporting Information Fig. S1B). The failure of ferrous T111H and L75H CtrHbs to react therefore appears related to absent or fleeting heme–protein contacts.
The modification occurs when reducing the well-folded cyanomet state. A priori, steric (i.e., correct seating of the heme), electrostatic, or heme electronic factors can all contribute to the reactivity. Attempts to crosslink in the presence of azide as an alternative anionic ligand were not successful (Supporting Information Fig. S21) and imidazole coordination led only to partial crosslinking (Supporting Information Fig. S22). The possibility that cyanide promotes the covalent modification by stabilizing the protonated vinyl seems unlikely given that binding of this anionic ligand is not expected to perturb significantly the electron distribution of the ferrous heme [54]. Thus, the data point to steric effects as the major determinants of reactivity in the absence of inhibiting ligands.
The protein conformation adopted when cyanide is bound may influence the reaction via subtle effects on the vinyl groups. We showed above that in the cyanomet complex, the major heme orientational isomer of WT CtrHb has the 2-vinyl group out of the heme plane and in a cis-like orientation, whereas T111H CtrHb undergoes a rearrangement to a trans-like orientation (Supporting Information Fig. S11). The stereochemistry of the products indicates that the reactive vinyl is in a trans-like conformation. This conformation has a higher degree of conjugation with the porphyrin ring than cis-like conformations [55] and may undergo protonation more readily. Whether a trans vinyl configuration is in general a necessary condition for reaction is not clear. Regardless, the lack of reaction between H111 and the 2-vinyl (major isomer) in T111H CtrHb shows that a trans vinyl near a histidine is an insufficient feature for reaction. Facile heme reorientation associated with low heme and exogenous ligand affinity in the ferrous state presents H111 with the 4-vinyl group as a competitive alternative to the 2-vinyl group. Thus, the yield of T111H CtrHb-A4 exceeds the minor isomer content in the cyanomet starting material. Likewise, the yield of L75H CtrHb-B exceeds the initial major isomer content. As anticipated, heme reorientation on a relatively rapid time scale emerges as a significant advantage for reaction.
In natural heme proteins, the vinyl groups are often surrounded by hydrophobic side chains. In prior work, we surveyed over 300 structures for the presence of leucines with Cα atom within 7 Å of a vinyl Cα atom and Cβ atom within 6 Å of the same vinyl Cα atom [15]. These criteria identified L79 in GlbN and L75 in CtrHb as candidates for histidine replacement and heme modification. Many other leucines were found. Examples include L106 in nitrophorin 2 (1PEE), L211 in chlorite dismutase (2VXH), L37 in ascorbate peroxidase (1APX), and L71 in cytochrome b5 (3NER), chosen to highlight proteins that have architectures different from that of globins. Many isoleucines, filtered by the same distance criteria, also occupy positions where a histidine may react. For in vitro applications, screening of pH conditions and permissive exogenous ligands that seat the heme properly can be systematically explored. Thus, it is reasonable to expect that many natural heme proteins can be modified to undergo the reaction, or artificial proteins can be designed to present a histidine with appropriate geometry to the heme.
4. Conclusion
The present work has several implications for the chemistry of heme proteins. (1) The existence of the PTM in GlbN [40] and its engineered presence in GlbN [22] and CtrHb cautions that the PTM may occur in additional heme proteins, not only wild-type but also unwittingly in His-tagged versions, (histidine) variants, or designed proteins. (2) In preparing the histidine Nε2–heme vinyl Cα crosslink, care must be exercised in the choice of a reducing agent to avoid heme and protein damage, as occasionally caused by the oxidative by-products of aerobic DT reduction. Anaerobic DT treatment or reduction with alternative agents such as a ferredoxin system [56] if sufficiently powerful may be preferable. (3) It is possible to engineer the crosslink in a b heme protein using relatively mild reaction conditions. Although detailed kinetic analysis was not performed on CtrHb variants, the products are consistent with the mechanism described previously [21], in which case the pKa of the reactive histidine, in addition to its ability to adopt the geometry of the product, is an important factor. (4) The necessity of using the cyanomet adduct as the species to be reduced rather than the ligand free protein (as in GlbNs) is likely related to the conformation of the starting material. In view of the CtrHb results, we expect that successful application to other proteins will depend principally on the position of the engineered histidine, the local flexibility of the supporting structure, the absence of inhibiting distal ligands, and possibly the orientation of the heme vinyl group.
The ability to prepare non-natively crosslinked versions of heme proteins extends the range of heme chemistry questions that can be addressed by biophysical methods. For example, to what extent does protonation of the alkylated histidine influence the heme reduction potential? How do the histidine-heme linkages differ from the cysteine–heme thioether linkages of c-type cytochromes? What is the electronic influence of the distal ligand? Practical usage of the crosslinking reaction includes the formulation of artificial enzymes or oxygen transporters with non-dissociable heme; such systems are expected to have an elongated lifetime, have increased resistance to proteolytic cleavage, and may be capable of functioning over a wide range of conditions (e.g. high temperature, low pH) for industrial or biomedical applications.
Supplementary Material
Highlights.
Control of both heme reactivity and affinity in designed proteins is a challenge.
Addition of a histidine to a heme vinyl is proposed as a tool to prevent heme loss.
Proof of concept is offered with two histidine variants of a Chlamydomonas Hb.
NMR spectroscopy is essential to the characterization of the products.
Acknowledgement
This study was supported by National Science Foundation grant MCB-1330488. Data were collected at the Johns Hopkins Biomolecular NMR Center with assistance from Dr. Ananya Majumdar.
Abbreviations
- 1D
one-dimensional
- CtrHb
heme domain of Chlamydomonas eugametos hemoglobin LI637
- DQF-COSY
double-quantum filtered correlation spectroscopy
- DSS
4,4-dimethyl-4-silapentane-1-sulfonic acid
- DT
sodium dithionite
- ECL
enhanced chemiluminescence
- HMQC
heteronuclear multiple quantum correlation
- HSQC
heteronuclear single quantum correlation
- lr
long-range
- NOESY
nuclear Overhauser effect spectroscopy
- PAGE
polyacrylamide gel electrophoresis
- PTM
post-translational modification
- TOWNY
total correlation spectroscopy without NOESY
- TOCSY
total correlation spectroscopy
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This paper is dedicated to the memory of James D. Satterlee, friend and mentor.
References
- [1].Hargrove MS, Barrick D, Olson JS. Biochemistry. 1996;35:11293–11299. doi: 10.1021/bi960371l. [DOI] [PubMed] [Google Scholar]
- [2].Landfried DA, Vuletich DA, Pond MP, Lecomte JTJ. Gene. 2007;398:12–28. doi: 10.1016/j.gene.2007.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cowley AB, Kennedy ML, Silchenko S, Lukat-Rodgers GS, Rodgers KR, Benson DR. Inorg. Chem. 2006;45:9985–10001. doi: 10.1021/ic052205k. [DOI] [PubMed] [Google Scholar]
- [4].Traylor TG, Sharma VS. Biochemistry. 1992;31:2847–2849. doi: 10.1021/bi00126a001. [DOI] [PubMed] [Google Scholar]
- [5].Gullotti M, Santagostini L, Monzani E, Casella L. Inorg. Chem. 2007;46:8971–8975. doi: 10.1021/ic700852c. [DOI] [PubMed] [Google Scholar]
- [6].Fritz BG, Hu X, Brailey JL, Berry RE, Walker FA, Montfort WR. Biochemistry. 2011;50:5813–5815. doi: 10.1021/bi200794c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Thompson AM, Reddi AR, Shi X, Goldbeck RA, Moënne-Loccoz P, Gibney BR, Holman TR. Biochemistry. 2007;46:14629–14637. doi: 10.1021/bi7013739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kim YM, Bergonia HA, Muller C, Pitt BR, Watkins WD, Lancaster JR., Jr. J. Biol. Chem. 1995;270:5710–5713. doi: 10.1074/jbc.270.11.5710. [DOI] [PubMed] [Google Scholar]
- [9].Kundu S, Hargrove MS. Proteins. 2003;50:239–248. doi: 10.1002/prot.10277. [DOI] [PubMed] [Google Scholar]
- [10].Alayash AI. Trends Biotechnol. 2014;32:177–185. doi: 10.1016/j.tibtech.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gibney BR, Rabanal F, Reddy KS, Dutton PL. Biochemistry. 1998;37:4635–4643. doi: 10.1021/bi971856s. [DOI] [PubMed] [Google Scholar]
- [12].Liu J, Chakraborty S, Hosseinzadeh P, Yu Y, Tian S, Petrik I, Bhagi A, Lu Y. Chem. Rev. 2014;114:4366–4469. doi: 10.1021/cr400479b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kranz RG, Richard-Fogal C, Taylor JS, Frawley ER. Microbiol. Mol. Biol. Rev. 2009;73:510–528. doi: 10.1128/MMBR.00001-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bowman SE, Bren KL. Nat. Prod. Rep. 2008;25:1118–1130. doi: 10.1039/b717196j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mavridou DA, Ferguson SJ, Stevens JM. IUBMB Life. 2013;65:209–216. doi: 10.1002/iub.1123. [DOI] [PubMed] [Google Scholar]
- [16].Braun M, Rubio IG, Thöny-Meyer L. Appl. Microbiol. Biotechnol. 2005;67:234–239. doi: 10.1007/s00253-004-1804-2. [DOI] [PubMed] [Google Scholar]
- [17].Braun M, Thöny-Meyer L. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12830–12835. doi: 10.1073/pnas.0402435101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Barker PD, Ferrer JC, Mylrajan M, Loehr TM, Feng R, Konishi Y, Funk WD, MacGillivray RT, Mauk AG. Proc. Natl. Acad. Sci. U. S. A. 1993;90:6542–6546. doi: 10.1073/pnas.90.14.6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Daltrop O, Allen JW, Willis AC, Ferguson SJ. Proc. Natl. Acad. Sci. U. S. A. 2002;99:7872–7876. doi: 10.1073/pnas.132259099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nothnagel HJ, Love N, Lecomte JTJ. J. Inorg. Biochem. 2009;103:107–116. doi: 10.1016/j.jinorgbio.2008.09.009. [DOI] [PubMed] [Google Scholar]
- [21].Nothnagel HJ, Preimesberger MR, Pond MP, Winer BY, Adney EM, Lecomte JTJ. J. Biol. Inorg. Chem. 2011;16:539–552. doi: 10.1007/s00775-011-0754-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Preimesberger MR, Wenke BB, Gilevicius L, Pond MP, Lecomte JTJ. Biochemistry. 2013;52:3478–3488. doi: 10.1021/bi400289e. [DOI] [PubMed] [Google Scholar]
- [23].Das TK, Couture M, Ouellet Y, Guertin M, Rousseau DL. Proc. Natl. Acad. Sci. U. S. A. 2001;98:479–484. doi: 10.1073/pnas.98.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hoy JA, Smagghe BJ, Halder P, Hargrove MS. Protein Sci. 2007;16:250–260. doi: 10.1110/ps.062572607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Vuletich DA, Falzone CJ, Lecomte JTJ. Biochemistry. 2006;45:14075–14084. doi: 10.1021/bi061532g. [DOI] [PubMed] [Google Scholar]
- [26].Preimesberger MR, Pond MP, Majumdar A, Lecomte JTJ. J. Biol. Inorg. Chem. 2012;17:599–609. doi: 10.1007/s00775-012-0880-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pond MP, Majumdar A, Lecomte JTJ. Biochemistry. 2012;51:5733–5747. doi: 10.1021/bi300624a. [DOI] [PubMed] [Google Scholar]
- [28].Dellarole M, Roumestand C, Royer C, Lecomte JTJ. Biochim. Biophys. Acta. 2013;1834:1910–1922. doi: 10.1016/j.bbapap.2013.04.016. [DOI] [PubMed] [Google Scholar]
- [29].Couture M, Das TK, Lee HC, Peisach J, Rousseau DL, Wittenberg BA, Wittenberg JB, Guertin M. J. Biol. Chem. 1999;274:6898–6910. doi: 10.1074/jbc.274.11.6898. [DOI] [PubMed] [Google Scholar]
- [30].Das TK, Couture M, Lee HC, Peisach J, Rousseau DL, Wittenberg BA, Wittenberg JB, Guertin M. Biochemistry. 1999;38:15360–15368. doi: 10.1021/bi991237e. [DOI] [PubMed] [Google Scholar]
- [31].Pesce A, Couture M, Dewilde S, Guertin M, Yamauchi K, Ascenzi P, Moens L, Bolognesi M. EMBO J. 2000;19:2424–2434. doi: 10.1093/emboj/19.11.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Couture M, Guertin M. Eur. J. Biochem. 1996;242:779–787. doi: 10.1111/j.1432-1033.1996.0779r.x. [DOI] [PubMed] [Google Scholar]
- [33].Lecomte JTJ, Scott NL, Vu BC, Falzone CJ. Biochemistry. 2001;40:6541–6552. doi: 10.1021/bi010226u. [DOI] [PubMed] [Google Scholar]
- [34].Milani M, Ouellet Y, Ouellet H, Guertin M, Boffi A, Antonini G, Bocedi A, Mattu M, Bolognesi M, Ascenzi P. Biochemistry. 2004;43:5213–5221. doi: 10.1021/bi049870+. [DOI] [PubMed] [Google Scholar]
- [35].de Duve C. Acta Chem. Scan. 1948;2:264–289. doi: 10.3891/acta.chem.scand.02-0264. [DOI] [PubMed] [Google Scholar]
- [36].Berry EA, Trumpower BL. Anal. Biochem. 1987;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- [37].Dorward DW. Anal. Biochem. 1993;209:219–223. doi: 10.1006/abio.1993.1110. [DOI] [PubMed] [Google Scholar]
- [38].Vargas C, McEwan AG, Downie JA. Anal. Biochem. 1993;209:323–326. doi: 10.1006/abio.1993.1127. [DOI] [PubMed] [Google Scholar]
- [39].Vu BC, Vuletich DA, Kuriakose SA, Falzone CJ, Lecomte JTJ. J. Biol. Inorg. Chem. 2004;9:183–194. doi: 10.1007/s00775-003-0512-1. [DOI] [PubMed] [Google Scholar]
- [40].Vu BC, Jones AD, Lecomte JTJ. J. Am. Chem. Soc. 2002;124:8544–8545. doi: 10.1021/ja026569c. [DOI] [PubMed] [Google Scholar]
- [41].Falzone CJ, Lecomte JTJ. J. Biomol. NMR. 2002;23:71–72. doi: 10.1023/a:1015337100759. [DOI] [PubMed] [Google Scholar]
- [42].Pond MP, Vuletich DA, Falzone CJ, Majumdar A, Lecomte JTJ. Biomol. NMR Assign. 2009;3:211–214. doi: 10.1007/s12104-009-9177-1. [DOI] [PubMed] [Google Scholar]
- [43].Pelton JG, Torchia DA, Meadow ND, Roseman S. Protein Sci. 1993;2:543–558. doi: 10.1002/pro.5560020406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. J. Biomol. NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- [45].Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- [46].Goddard TD, Kneller DG. University of California; San Francisco: 2006. [Google Scholar]
- [47].Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- [48].La Mar GN, Davis NL, Parish DW, Smith KM. J. Mol. Biol. 1983;168:887–896. doi: 10.1016/s0022-2836(83)80080-1. [DOI] [PubMed] [Google Scholar]
- [49].Lee KB, La Mar GN, Kehres LA, Fujinari EM, Smith KM, Pochapsky TC, Sligar SG. Biochemistry. 1990;29:9623–9631. doi: 10.1021/bi00493a017. [DOI] [PubMed] [Google Scholar]
- [50].La Mar GN, Satterlee JD, de Ropp JS. In: The Porphyrin Handbook. Smith KM, Kadish K, Guilard R, editors. Vol. 5. Academic Press; Burlington, MA: 2000. pp. 185–298. [Google Scholar]
- [51].Bolli A, Ciaccio C, Coletta M, Nardini M, Bolognesi M, Pesce A, Guertin M, Visca P, Ascenzi P. FEBS J. 2008;275:633–645. doi: 10.1111/j.1742-4658.2007.06223.x. [DOI] [PubMed] [Google Scholar]
- [52].Vila JA. J. Phys. Chem. B. Vol. 116. American Chemical Society; 2012. pp. 6665–6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Bertini I, Luchinat C, Parigi G, Walker FA. J. Biol. Inorg. Chem. 1999;4:515–519. doi: 10.1007/s007750050337. [DOI] [PubMed] [Google Scholar]
- [54].Boffi A, Chiancone E, Takahashi S, Rousseau DL. Biochemistry. 1997;36:4505–4509. doi: 10.1021/bi9618880. [DOI] [PubMed] [Google Scholar]
- [55].Marzocchi MP, Smulevich G. J. Raman Spect. 2003;34:725–736. [Google Scholar]
- [56].Hayashi A, Suzuki T, Shin M. Biochim. Biophys. Acta. 1973;310:309–316. doi: 10.1016/0005-2795(73)90110-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.