Abstract
Stimulated macrophages generate potent anti-microbial reactive oxygen and nitrogen species within their phagosomes. Previous studies have shown that the E3 ubiquitin ligase natural killer lytic-associated molecule (NKLAM) is a macrophage phagosomal protein that plays a role in macrophage anti-bacterial activity. In vivo, NKLAM-knockout (KO) mice produce less nitric oxide (NO) upon exposure to lipopolysaccharide (LPS) than wild type (WT) mice. In vitro, we found that NO production and inducible nitric oxide synthase (iNOS) protein were diminished in LPS-stimulated KO bone marrow-derived and splenic macrophages. Additionally, LPS-stimulated NKLAM-KO macrophages displayed defects in STAT1 tyrosine phosphorylation and production of interferon beta (IFNβ). The JAK/STAT pathway is critical for the production of IFNβ, which augments iNOS protein expression in mice. iNOS protein expression is also regulated by the transcription factor NFκB, thus we investigated whether NKLAM influences NFκB function. LPS-stimulated NKLAM-KO macrophages showed evidence of delayed nuclear translocation of the NFκB subunit p65. This was associated with a reduction in p65/DNA colocalization. The defect in p65 translocation was independent of IKBα degradation. NKLAM-KO macrophages also expressed less p65 and showed evidence of defective p65 phosphorylation at serine 536. Importantly, LPS-stimulated NKLAM-KO macrophages have diminished NFκB transcriptional activity as assessed by transfection of a luciferase reporter plasmid. Collectively, our data implicate NKLAM as a novel modulator of macrophage iNOS expression.
Keywords: Innate immunity, iNOS, Macrophage, NKLAM, NFκB, Nitric oxide, p65
INTRODUCTION
Macrophages are a first line of defense against pathogens and serve as a critical bridge between the innate and adaptive arms of the immune system. Macrophages employ several bactericidal mechanisms to eradicate foreign pathogens. One such mechanism is the generation of nitric oxide (NO) from L-arginine via the expression and activation of inducible nitric oxide synthase (iNOS; NOS2). iNOS is a dimeric protein that is expressed in several immune cells including macrophages, dendritic cells, and neutrophils (Cruz, et al., 2001, Ratajczak-Wrona, et al., 2013). The production of NO by iNOS in response to infection is a critical defense mechanism. Studies have shown that iNOS-knockout mice are unable to mount an efficient immune response (Alayan, et al., 2006). Stimulation with microbial products such as lipopolysaccharide (LPS), lipoteichoic acid or pro-inflammatory cytokines induces the transcriptional upregulation of iNOS protein (Zhang, et al., 2013). Previous studies have shown that treatment with exogenous IFNβ enhances iNOS protein expression and NO production (Jacobs and Ignarro, 2001, Yao, et al., 2001). Once activated, iNOS is capable to producing large amounts of NO. Nitric oxide can combine with the simultaneously produced super oxide anion resulting in peroxynitrite, a potent bactericidal free radical that induces oxidative damage to proteins, lipids, and nucleic acids (Habib and Ali, 2011, Tecder-Unal, et al., 2008). The mouse iNOS promoter contains several transcription factor binding sites such as interferon-stimulated response elements (ISRE), IFNγ activation sites, Oct-1 and NFκB binding sites (Korhonen, et al., 2005, Xie, et al., 1993).
The regulation of iNOS transcription by transcription factor NFκB has been well studied. There are two NFκB consensus binding sites in the iNOS promoter that are critical for the regulation of iNOS transcription (Kim, et al., 1997). In the canonical NFκB pathway, NFκB is sequestered in the cytoplasm by its inhibitor IKBα. Upon stimulation with bacterial products such as LPS, IKBα is rapidly phosphorylated, ubiquitinated and degraded in the 26S proteasome. The binding of IKBα to NFκB subunit p65 masks a nuclear localization signal in p65. Once IKBα is degraded, NFκB is freed and translocates to the nucleus where it transcriptionally regulates a large number of immunologically important genes (Zerfaoui, et al., 2008). NFκB p65 is subject to several forms of post-translational modifications including phosphorylation, acetylation, and ubiquitination (Hoesel and Schmid, 2013). These modifications affect the nuclear translocation, DNA-binding and transcriptional activity of NFκB (Perkins, 2006). According to the NFκB barcode hypothesis, it is the unique combination of these modifications that direct transcription in a gene-specific pattern (Moreno, et al., 2010).
Natural killer lytic-associated molecule (NKLAM) is a membrane-bound E3 ubiquitin ligase and a member of the RING1-in between RING-RING2 (RBR) family of proteins. Three cysteine-rich regions in the N-terminus are critical for the ubiquitin ligase activity of NKLAM (Fortier and Kornbluth, 2006). NKLAM was originally discovered in our laboratory as a protein that co-localized with granzyme B in natural killer (NK) cytolytic granule membranes (Kozlowski, et al., 1999). In addition to NK cells, NKLAM is expressed in other mononuclear cells such as monocytes and macrophages (Kozlowski, et al., 1999, Portis, et al., 2000). NKLAM is weakly expressed under resting conditions but is upregulated by cytokines (e.g. IFNγ) and bacterial products such as LPS (Lawrence and Kornbluth, 2012). NK cells from NKLAM-deficient (KO) mice have diminished anti-tumor cytolytic activity (Hoover, et al., 2009). In vivo, NKLAM plays a role in controlling tumor metastasis (Hoover, et al., 2012). Recent studies from our laboratory have also demonstrated a role for NKLAM in macrophage bacterial killing. We found that macrophages (bone marrow-derived and peritoneal) from NKLAM-KO mice are significantly defective in killing Escherichia coli. Despite its localization to phagosomes, NKLAM does not regulate bacteria uptake or phagosome acidification (Lawrence and Kornbluth, 2012). Thus, NKLAM is likely involved in regulating some other macrophage killing mechanism within the phagosome. One such mechanism is iNOS protein expression and NO production. Results presented here indicate that NKLAM as a positive regulator of iNOS protein expression, and does so, at least in part, by modulating the activity of the transcription factor NFκB.
Materials and Methods
Macrophage cultures
All experiments on mice were approved by the Institutional Animal Care and Use Committee at Saint Louis University. Wild type (WT) C57BL/6 and corresponding age-matched NKLAM-KO mice were used in all studies. For isolation of bone marrow, euthanized mice were sprayed with 70% ethanol and the femurs and tibias were dissected. The bones were flushed with DMEM and the collected marrow was resuspended in BM20 media (DMEM supplemented with 20% fetal bovine serum (FBS), 20% L929-cell conditioned media, 2 mM L-glutamine, 100 U/mL penicillin, 100 U/mL streptomycin, and 1 mM sodium pyruvate). The bone marrow cells were cultured for 7 days in non-tissue culture petri dishes with a partial media change on day 3. For splenic macrophage isolation, spleens were harvested from healthy mice and the red blood cells were removed by Lympholyte-M (Cedarlane) density gradient centrifugation. The mononuclear cell layer was washed, resuspended in RPMI 1640 supplemented with 10% FBS and plated in tissue culture plastic dishes for 7 days. Cells obtained using this method are predominantly mature F4/80+, CD11b+ macrophages (Van Ginderachter, et al., 2006).
Nitric oxide measurement
Adherent bone marrow-derived macrophages (BMDM) were incubated with various concentrations of LPS for times indicated. Culture supernatants were assayed for nitrite using the Greiss reaction. For the in vivo studies, mice were intraperitoneally injected with sterile PBS or 25 µg of LPS and blood was obtained at 1, 2 and 6 h post-injection. The blood was centrifuged at 4000 × g for 4 min and the total concentration of nitrite present in the plasma was determined using the Griess reaction following the plasma preparation protocol of Moshage et al. (Moshage, et al., 1995). Briefly, plasma samples were diluted and deproteinized with the addition of 1/20th volume of zinc sulfate for a final concentration of 15 g/L. The samples were centrifuged and the resulting supernatants were assayed for total nitrite using a commercial kit according the manufacturer’s protocol (Cayman Chemical Company).
Macrophage transfection and luciferase activity measurement
Adherent BMDM were harvested from dishes with 1.5 mM EDTA, washed once with cold PBS, and then resuspended in DMEM plus 10% FBS. Wild type and NKLAM-KO macrophages (4 × 106) were mixed with 3 µg of the luciferase reporter plasmid pNFκB-luc (Clontech) and suspended in nucleofection solution T. Cells were nucleofected using program T-20 with the nucleofector I (Amaxa Biosystems). The nucleofected macrophages were resuspended in DMEM plus 10% FBS, transferred to 12-well plates and stimulated with 100 ng/ml LPS for the times indicated. At the desired time, the cells were collected, lysed in 1× luciferase reporter lysis buffer and snap frozen at −80°C to aid in cell disruption. The total firefly luciferase activity was measured using the Promega Luciferase Assay System (Promega). Protein concentrations were determined using bicinchoninic acid (BCA) protein assay reagents (Pierce). Transfection efficiency was assessed by flow cytometry after nucleofection of cells with the green fluorescence protein (GFP) reporter plasmid pmaxGFP (Amaxa).
Immunoblotting
Whole cell protein lysates were separated using SDS-PAGE and then transferred to PVDF membrane. Membranes were blocked with 1% (wt/vol) BSA in Tris-buffered saline plus 0.1% Tween-20 (TBS-T) and then incubated in primary antibody with rocking overnight at 4°C. The antibodies for iNOS (BD Transduction Laboratories), NFκB p65 (Santa Cruz Biotechnologies), IKBα (Cell Signaling), phosphop65 Ser536 (Cell Signaling), STAT1 (Cell Signaling), poly ADP ribose polymerase (PARP; Cell Signaling), and phospho-STAT1 (701) (Cell Signaling) were used at 1:1000. Anti β-actin antibody (Sigma Aldrich) was used at 1:4000. After three washes in TBS-T, the membranes were probed with HRP-conjugated secondary antibodies and the proteins were visualized with Bio-Rad Immun-Star Western C chemiluminescence kit. Images were captured and analyzed using a Bio-Rad Chemidoc XRS+ imager (BioRad).
Preparation of cytosolic and nuclear fractions
WT and NKLAM-KO BMDM were treated with 100 ng/ml LPS for 15, 30 or 60 min. Cytoplasmic and nuclear protein fractions were isolated using NE-PER nuclear and cytoplasmic extraction reagents (Thermo Scientific). Equal protein amounts were immunoblotted for NFκB p65. The membranes with nuclear and cytoplasmic cell extracts were reprobed PARP or beta actin, respectively, to demonstrate purity of the fractions and equivalent protein loading.
Immunofluorescence
Adherent BMDM were plated on acid-cleaned 18 mm glass coverslips and stimulated with 100 ng/ml LPS. The monolayers were fixed and permeabilized by the addition of chilled (−20°C) methanol:acetone (1:1) for 30 seconds. After washing in PBS, the coverslips were blocked with 3% BSA in PBS for 60 min at room temperature. The coverslips were placed in a humidified chamber and incubated with 1:100 anti-NFκB p65 or anti-phospho-p65 (Ser536) overnight at 4°C. The coverslips were washed 3 × 10 min in PBS at room temperature and incubated with Alexa Fluor 594 chicken anti-rabbit IgG (Life Technologies) for 60 min at room temperature. The wash was repeated and the coverslips were incubated with 1 µg/ml 4',6-diamidino-2-phenylindole (DAPI) for 10 min. The coverslips were washed in PBS a final time and mounted in polyvinyl alcohol mounting media. Images were collected with a Leica DMI 4000B microscope. All image processing was performed with ImageJ (NIH, Bethesda, MD) or Photoshop. All images were processed identically.
Quantitative real-time PCR
Unstimulated and LPS-stimulated adherent macrophages (4 × 105) in 12-well plates were rinsed quickly with ice-cold PBS, and then 350 µL of Qiagen RLT buffer was added per well and the plates were snap frozen at −80°C. RNA was prepared with the RNeasy Mini Kit (Qiagen) and cDNA was made using TaqMan Reverse Transcription Gene Expression Assay kit (Applied Biosystems). Quantitative PCR analysis of iNOS, 18S rRNA, and IFNβ was performed using qPCR primers purchased from Integrated DNA Technologies. Samples were run on an ABI 7500 Real-Time PCR System (Applied Biosystems) using iTaq Sybr green supermix (Bio-Rad). The change in Ct (ΔCt) was calculated with 18S rRNA as a housekeeping gene. iNOS data are presented as 2−/(ΔCt) (Livak and Schmittgen, 2001) and IFNβ are presented as 2−(ΔΔCt).
Statistical analysis
Statistical differences were assessed using a two-tailed, unpaired Student’s t-test or one-way ANOVA with Microsoft Excel software. For both tests, a p value of 0.05 or lower was considered statistically significant. Data represent the mean ± s.e.m.
RESULTS
Lack of NKLAM expression is associated with diminished nitric oxide production in response to LPS in vitro and in vivo
Our previous studies demonstrated that NKLAM-KO macrophages are inherently defective in killing E. coli (Lawrence and Kornbluth, 2012). Macrophages employ a number of bactericidal mechanisms to combat pathogens; these include phagosome acid hydrolases, phagosomal pH reduction and NO production (Flannagan, et al., 2009). We therefore examined the effect of LPS stimulation on NO production in WT and NKLAM-KO BMDM. Adherent macrophages were treated with 100 ng/ml LPS for 18 h and supernatants were assayed for nitrite concentration. NKLAM-KO macrophages showed evidence of defective nitric oxide production. At 18 h, NKLAM-KO macrophages had accumulated less than half the nitrite levels of WT macrophages (Fig. 1A). To support these initial in vitro studies, we injected 25 µg of LPS or sterile PBS into the peritoneal cavity of WT and NKLAM-KO mice. Blood was collected at 1, 2 and 6 h post-injection and total plasma nitrite levels were measured using the Griess reaction after conversion of nitrate to nitrite. As shown in Fig. 1B, the plasma nitrite levels in LPS-injected WT mice were significantly increased at 1, 2 and 6 h post-injection, compared to PBS-injected WT mice. In contrast, the plasma nitrite levels in NKLAM-KO mice were not significantly elevated above PBS-injected control mice until 6 h post-injection, and did not reach the levels observed in control WT mice. These results suggest that NKLAM plays a positive role in regulating the production of NO in response to LPS stimulation in vitro and in vivo.
Figure 1. Nitric oxide production in WT and NKLAM-KO BMDM.
A) WT (black column) and NKLAM-KO (white column) BMDM were treated with 100 ng/ml LPS for 18 h. The culture supernatants were assayed for nitrite using the Griess reaction. Data represent the mean ± s.e.m. of three experiments; *p ≤ 0.05, compared to LPS-stimulated WT BMDM. B) LPS (25 µg) or sterile PBS was injected into the peritoneal cavity of WT and NKLAM-KO mice. Blood was obtained at the times indicated and the concentration of total plasma nitrite was determined using the Griess assay after conversion of nitrate to nitrite; * p ≤ 0.05, compared to WT mice injected with sterile PBS; # p ≤ 0.05, compared to NKLAM-KO mice injected with sterile PBS.
Lack of NKLAM expression is associated with diminished iNOS mRNA and protein expression in response to LPS
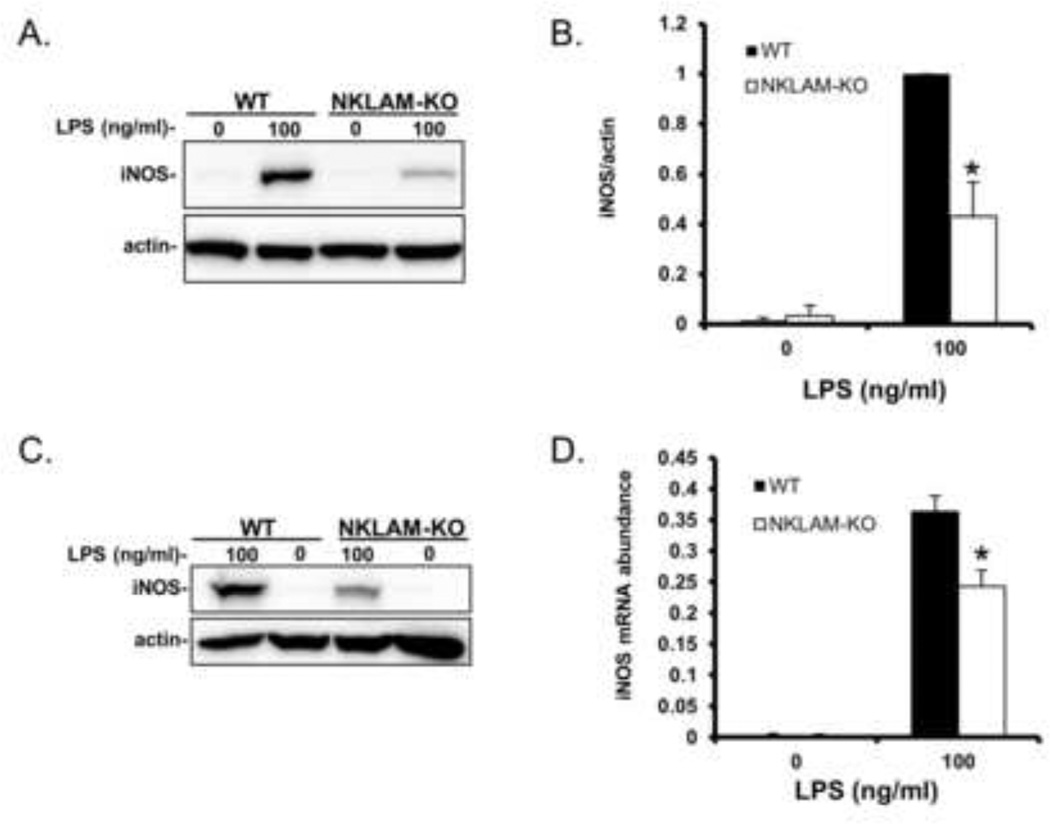
Based upon our results that demonstrate defective NO production in NKLAM-KO macrophages, we next examined the expression of iNOS protein in LPS-stimulated macrophages. As shown in Fig. 2A and B, LPS-treated WT BMDM expressed greater than twice the amount of iNOS protein as compared to NKLAM-KO BMDM. Additionally, we found that splenic macrophages respond similarly to LPS with respect to iNOS expression. iNOS expression was higher in LPS-treated (100 ng/ml) WT splenic macrophages than in NKLAM-KO splenic macrophages (Fig. 2C). We also found that iNOS mRNA was significantly greater in WT BMDM than in NKLAM-KO BMDM after 2 h of LPS stimulation (Fig. 2D).
Figure 2. iNOS expression in WT and NKLAM-KO BMDM stimulated with LPS.
A) WT and NKLAM-KO BMDM were treated with 100 ng/ml LPS for18 h and whole cell lysates were made and immunoblotted for iNOS. B) Densitometric analysis for Fig. 2A; * p ≤ 0.05, n = 3. C) Splenic macrophages isolated from WT and NKLAM-KO mice were untreated or treated with 100 ng/ml LPS for 18 h and whole cell lysates were immunoblotted for iNOS protein. For all immunoblots, beta actin was used as a loading control. D) WT and NKLAM-KO BMDM were stimulated with 100 ng/ml LPS for 2 h. iNOS mRNA expression was determined by quantitative real-time PCR and the data are expressed as 2−ΔCt; * p ≤ 0.03, n = 4.
NKLAM-KO macrophages have defective interferon beta production in response to LPS stimulation
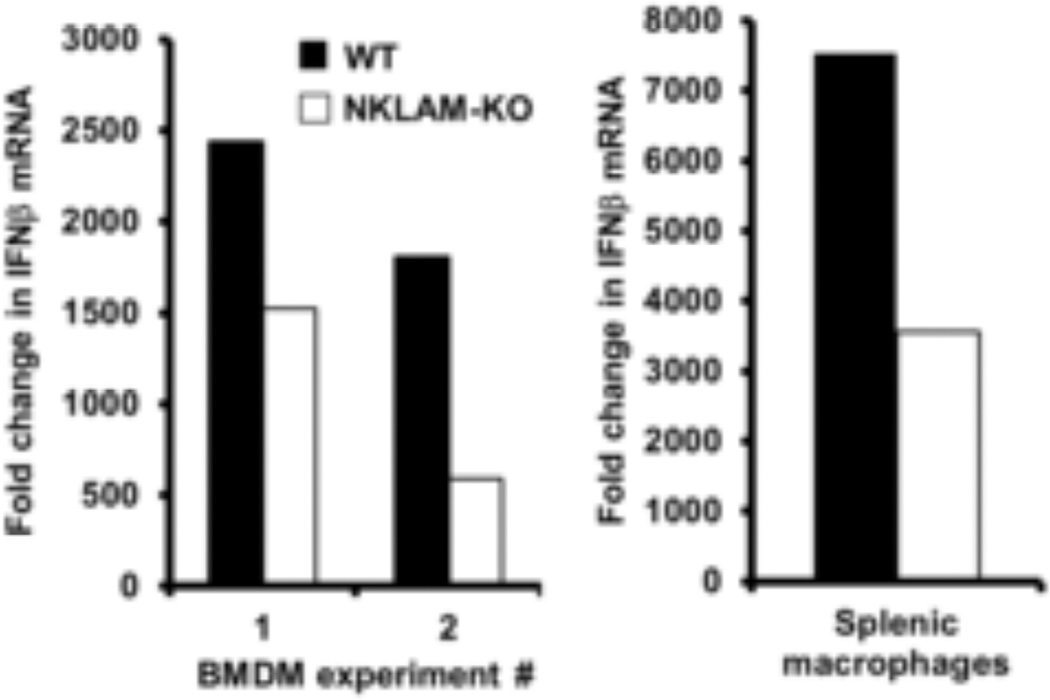
Murine macrophages secrete IFNβ in response to LPS stimulation and IFNβ augments the expression of iNOS (Gao, et al., 1998). For our next set of experiments, we stimulated WT and NKLAM-KO BMDM and spleen macrophages with 100 ng/ml LPS and quantitated the levels of IFNβ mRNA by real time PCR. As shown in 3 independent experiments we found that IFNβ mRNA was 38–67% less in NKLAM-KO BMDM than in WT BMDM stimulated with LPS. Supportively, IFNβ mRNA from LPS-stimulated splenic macrophages from NKLAM-KO mice was ~50% less compared to WT splenic macrophages (Fig. 3).
Figure 3. Interferon beta mRNA expression in WT and NKLAM-KO splenic macrophages and BMDM following LPS stimulation.
WT (black column) and NKLAM-KO (white column) BMDM and splenic macrophages were treated with 100 ng/ml LPS for 2 h. Total RNA was isolated and IFNβ mRNA expression was determined by quantitative real-time PCR. Three individual experiments are presented. Graphs depict the fold change (2−ΔΔCt) in IFNβ mRNA levels at 2 h LPS.
STAT1 phosphorylation at tyrosine 701 is defective in LPS-stimulated NKLAM-KO macrophages
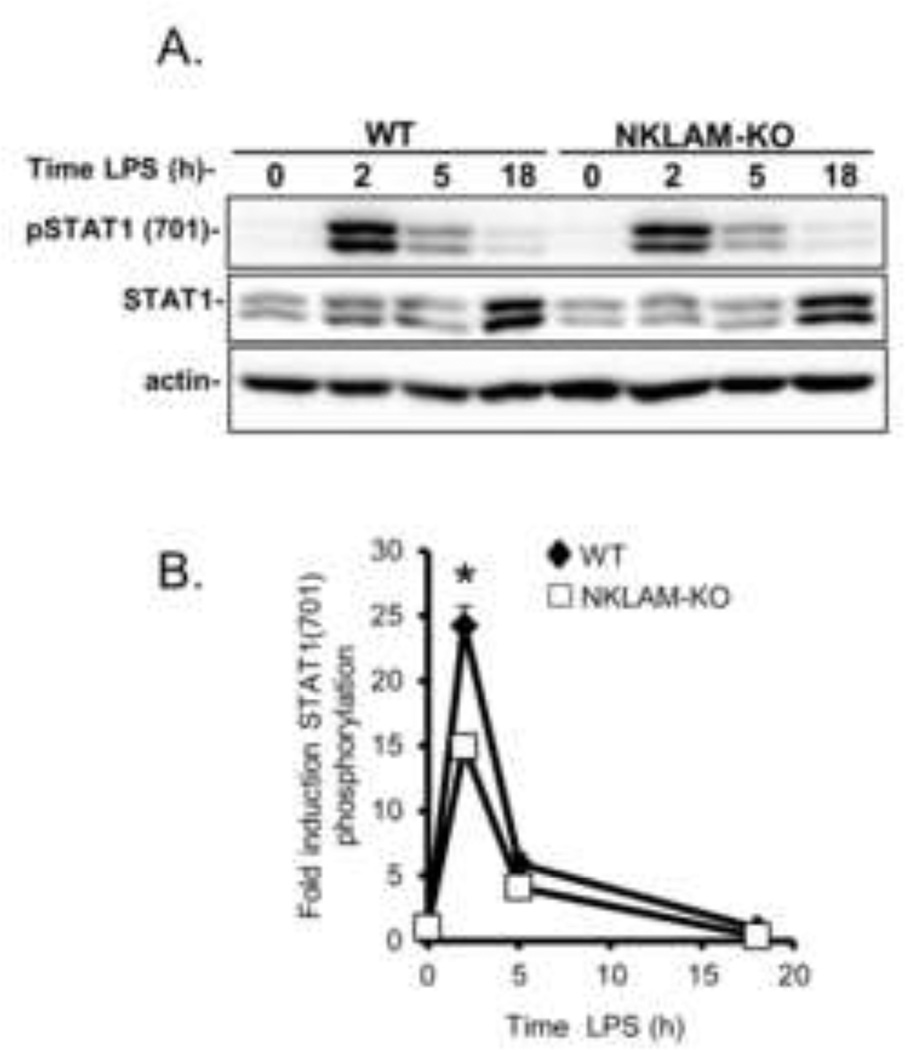
Interferons regulate the induction of target genes via the JAK/STAT pathway. Following ligation of cell surface receptors, JAK phosphorylates STAT (signal transducer and activator of transcription) proteins that dimerize before entering the nucleus and initiate target gene transcription (Rauch, et al., 2013). In murine macrophages, STAT1 is phosphorylated on tyrosine 701 in response to LPS stimulation (Jacobs and Ignarro, 2001, Rhee, et al., 2003). For our next set of experiments we explored the effect of NKLAM expression on STAT1 phosphorylation in response to LPS. WT and NKLAM-KO BMDM were stimulated with 100 ng/ml LPS for the times indicated. Whole cell lysates were prepared and immunoblotted for phospho-STAT1 (701) and total STAT1. As shown in Fig. 4A and B, LPS stimulation for 2 h induced maximal STAT1 phosphorylation at tyrosine 701. Although the kinetics of STAT1 phosphorylation at tyrosine 701 was similar, there was significantly less induction of STAT1 phosphorylation by LPS in NKLAM-KO BMDM than in WT macrophages. The levels of total STAT1 were comparable in NKLAM-KO and WT macrophages. These results suggest that NKLAM plays a role in the signal transduction pathway that regulates LPS-induced STAT1 phosphorylation.
Figure 4. STAT1 phosphorylation induction by LPS in WT and NKLAM-KO BMDM.
WT and NKLAM-KO BMDM were treated with 100 ng/ml LPS for the times indicated. A) Whole cell lysates were prepared and total STAT1 and phospho-STAT1 (701) were detected by immunoblot. Immunoblots are representative of 3 independent experiments. B) Quantitation by densitometric analysis of the fold increase in phospho-STAT1 (701) relative to untreated cells. Data represent the mean ± s.e.m. of three experiments; * p < 0.05, comparing phosphorylation of STAT1 in WT and NKLAM-KO BMDM after 2 h of LPS stimulation.
LPS-induced NFκB p65 nuclear translocation is delayed in NKLAM-KO macrophages
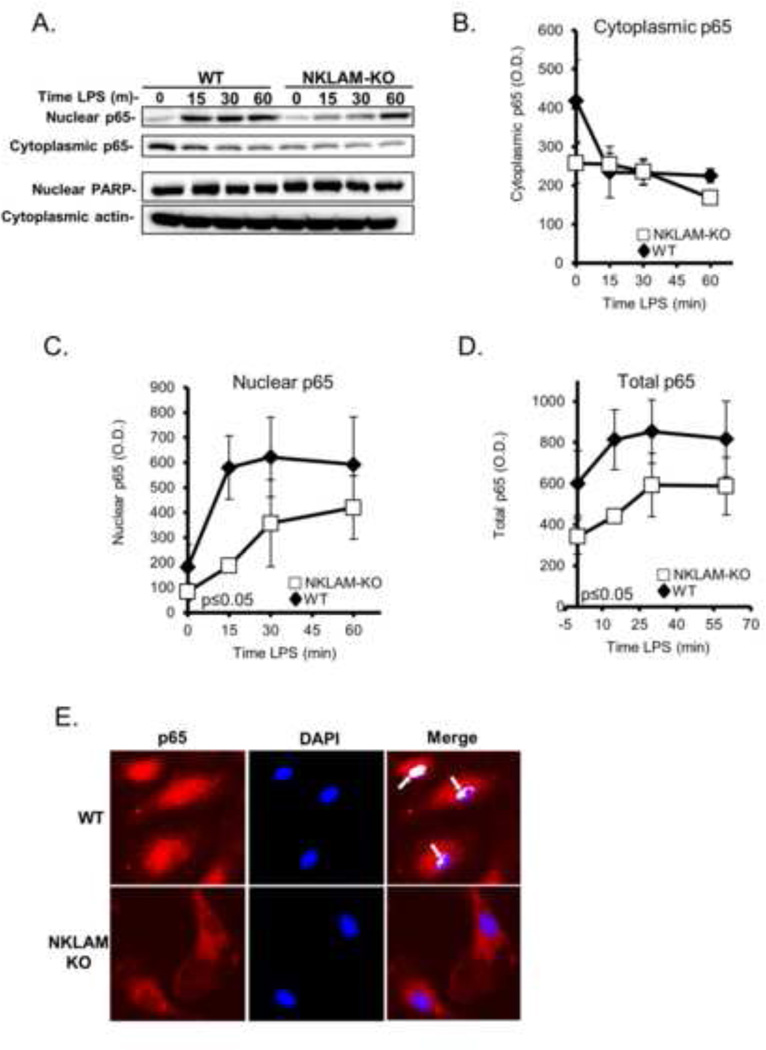
NFκB regulates nearly 250 different genes, including iNOS (Dhruv, et al., 2013). In macrophages, stimulation with LPS induces the translocation of NFκB p65 subunit from the cytoplasm into the nucleus. NFκB translocation is regulated in part by the ubiquitination of multiple components of the Toll-like receptor (TLR) pathway that are triggered by exposure to LPS. Based on the diminished iNOS expression observed in NKLAM-KO macrophages, we investigated the possible involvement of NFκB p65 in regulating iNOS expression in WT and NKLAM-KO BMDM. Adherent macrophages were stimulated with 100 ng/ml LPS and the nuclear and cytosolic fractions were isolated. Equal amounts of protein were immunoblotted for p65. In unstimulated cells, the levels of cytosolic p65 were higher in WT macrophages than in NKLAM-KO macrophages (Fig. 5A, B). In WT BMDM, p65 cytosolic levels decreased sharply after 15 min of LPS stimulation. This decrease in cytoplasmic p65 was associated with a concomitant rise in nuclear p65 expression, consistent with translocation. Following this initial decrease, WT cytosolic p65 levels remained constant. In contrast, the cytosolic p65 levels in NKLAM-KO macrophages did not change as drastically as in WT macrophages following LPS stimulation. The kinetics of p65 expression in the nucleus was much more dynamic (Fig. 5A, C). In WT macrophages, there was a large (~3.2 fold) increase in nuclear p65 after 15 min of LPS stimulation, which remained elevated out to 60 min. The levels of nuclear p65 increased more modestly (~2 fold) in NKLAM-KO macrophages after 15 min of LPS stimulation and continued to gradually increase over time. Overall, p65 expression in the nucleus of NKLAM-KO macrophages was significantly less than in WT macrophages (Fig. 5C). Total p65 expression was also less in NKLAM-KO than WT BMDM (Fig. 5D).
Figure 5. NFκB p65 expression and nuclear translocation in WT and NKLAM-KO BMDM.
A–D) WT and NKLAM-KO BMDM were treated with 100 ng/ml LPS for the times indicated. Isolated cytosolic and nuclear protein fractions were immunoblotted for p65. Immunoblotting for PARP (nuclear lysates) and actin (cytoplasmic lysates) was done to show purity of the fractions and equivalent protein loading. A) Immunoblots represent one of 3 identical experiments. B–D) Densitometric analysis of p65 in cytoplasmic, nuclear and total cellular lysates from WT and NKLAM-KO macrophages treated with 100 ng/ml LPS for the times indicated. Data represent the mean ± s.e.m. of three experiments; p ≤ 0.05, one-way ANOVA, comparing response of WT and NKLAM-KO BMDM. E) Monolayers of WT and NKLAM-KO BMDM were treated with 100 ng/ml LPS for 15 min and then immunostained for p65 (red). The nuclei were counterstained with DAPI (blue). Arrows denote areas of p65/DNA colocalization (white) as defined by the ImageJ Colocalization Finder plugin.
To reveal the intracellular expression pattern of p65 during LPS stimulation in WT and NKLAM-KO BMDM, we localized p65 by immunofluorescence. As translocation of p65 into the nucleus is rapid, we stimulated macrophage monolayers with LPS (100 ng/ml) for 15 min before fixation and p65 immunostaining (Fig. 5E). The nuclei were counterstained with DAPI. Analysis of LPS-stimulated macrophages showed that in WT macrophages, p65 was highly expressed in the nucleus and perinuclear area. In NKLAM-KO macrophages, there was less overall p65 and very little within the nucleus (Fig. 5E, lower left panel). Using the NIH ImageJ Colocalization Finder plugin we were able to identify specific areas of p65/DNA colocalization, denoted in white, in WT macrophages but not in NKLAM-KO macrophages (Fig. 5E, right panels). Therefore, by both immunoblotting and immunostaining, we observed that NKLAM-KO macrophages have a defect in p65 translocation into the nucleus after LPS stimulation. Presumably, diminished translocation would result in less interaction between p65 and its target DNA sequences.
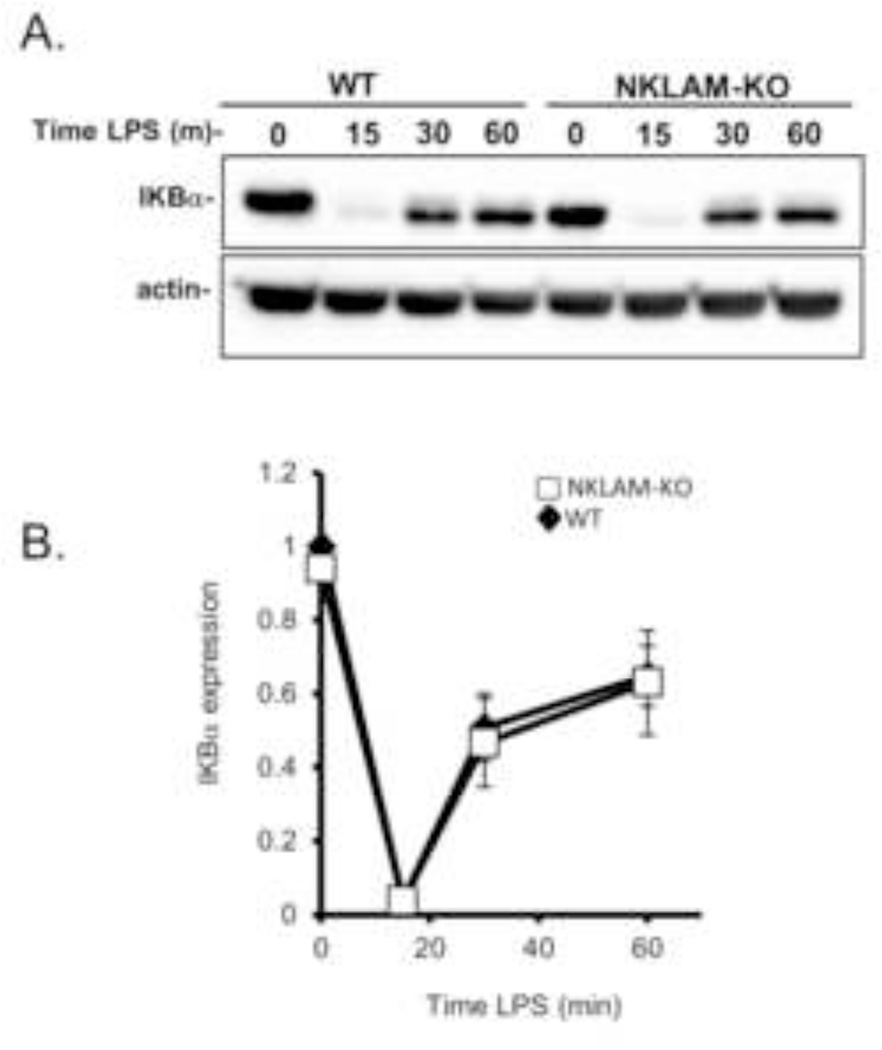
LPS-stimulated IKBα degradation is unaffected by the lack of NKLAM expression
In the canonical NFκB pathway, NFκB is sequestered in the cytoplasm by IKBα. Phosphorylation and ubiquitination of IKBα leads to its proteasomal degradation and release of NFκB which then translocates into the nucleus. The prominent role of IKBα ubiquitination in facilitating NFκB nuclear translocation coupled with our observation that p65 nuclear translocation is attenuated in NKLAM-KO macrophages led us to examine the degradation and rebound expression of IKBα during LPS stimulation. Adherent macrophages were stimulated with 100 ng/ml LPS for 15, 30 or 60 min. Cytoplasmic fractions were prepared and immunoblotted for IKBα. As shown in Fig. 6A, IKBα was nearly completely degraded after 15 min of LPS stimulation. IKBα degradation was followed by rapid rebound expression. After 30 min of LPS stimulation, the IKBα expression levels increased substantially and were ~50% of the resting IKBα levels. The kinetics and expression levels of IKBα were nearly identical in WT and NKLAM-KO BMDM, suggesting that NKLAM does not affect IKBα degradation.
Figure 6. IKBα expression kinetics in WT and NKLAM-KO macrophages stimulated with LPS.
WT and NKLAM-KO BMDM were incubated with 100 ng/ml LPS for the times indicated. A) Cytoplasmic protein fractions were isolated from each time point and immunoblotted for IKBα. Immunoblots represent one of 3 identical experiments. B) Densitometric analysis of IKBα. Data represent the mean ± s.e.m. of three experiments.
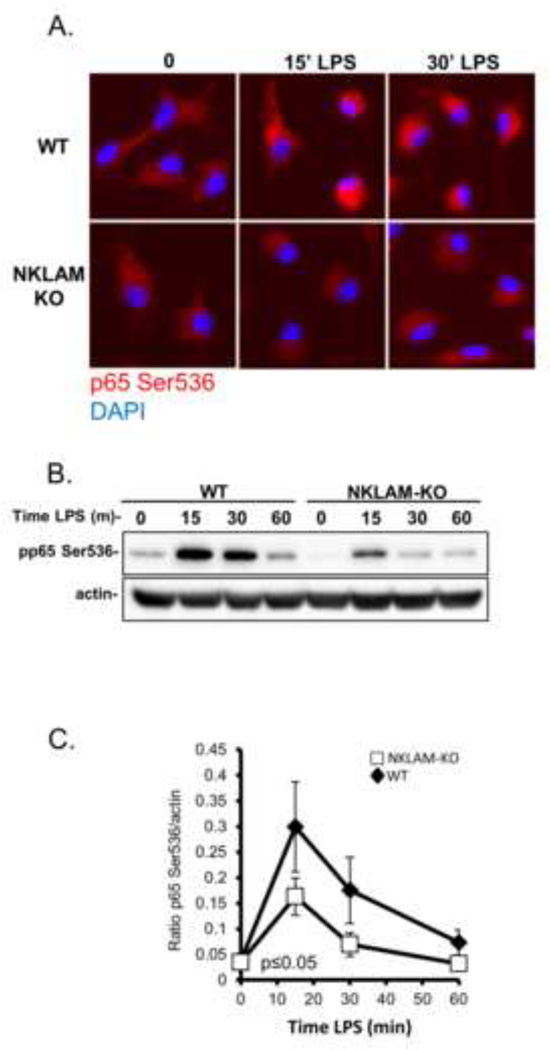
Phosphorylation of NFκB p65 at serine 536 is defective in LPS-stimulated NKLAM-KO macrophages
Post-translational modification events such as phosphorylation and acetylation are critical regulators of p65 transcriptional activity, and are believed to aid in guiding specific target gene transcription (Perkins, 2006). Phosphorylation of NFκB p65 at serine 536 has been shown to be associated with nuclear translocation and transcriptional activity (Hu, et al., 2005); thus our next set of experiments focused on the kinetics of p65 serine 536 phosphorylation in response to LPS. We stimulated WT and NKLAM-KO macrophages with 100 ng/ml LPS for 15, 30 and 60 min and examined the expression of serine 536-phosphoryated p65 by both immunofluorescence and immunoblotting. In unstimulated WT and NKLAM-KO cells, low levels of serine 536 phosphorylated p65 were seen in the cytoplasm and displayed a punctate staining pattern, consistent with previous reports (Moreno, et al., 2010). After 15 min of LPS stimulation, the intensity of serine 536 phosphorylated p65 staining increased dramatically in WT but not in NKLAM-KO macrophages (Fig. 7A). In addition, this increase in phosphorylated p65 was localized to an area close to the nucleus. In WT cells only, the increase in serine 536 phosphorylated p65 was still observable 30 min after LPS stimulation. In order to more precisely and quantitatively examine the kinetics of p65 phosphorylation, we made lysates from WT and NKLAM-KO macrophages stimulated with 100 ng/ml LPS and performed Western blot analysis. As shown in Fig. 7B, unstimulated macrophages expressed small amounts of p65 phosphorylated at serine 536. In WT macrophages, treatment with LPS induced an increase in the amount of phosphorylated p65 at 15 min that returned to baseline levels by 60 min. The kinetics of serine 536 phosphorylation of p65 in NKLAM-KO was similar to WT cells; however, the amount of phosphorylated protein was significantly less in comparison. Beta actin was used as a loading control and was similar at all time points. The lower levels of phosphorylated p65 in LPS-stimulated NKLAM-KO macrophages may account for the delay in NFκB p65 nuclear translocation observed in NKLAM-KO macrophages.
Figure 7. LPS-induced serine 536 phosphorylation of NFκB p65 in WT and NKLAM-KO BMDM.
A) WT and NKLAM-KO BMDM were stimulated with 100 ng/ml LPS for the times indicated. Phosphorylated p65 was visualized by immunostaining the monolayers with anti-phospho-p65 Ser536 antibody (red). The nuclei were counterstained with DAPI (blue). B) WT and NKLAM-KO macrophages were treated with 100 ng/ml LPS for the times indicated and cytosolic fractions were immunoblotted for p65 phosphorylated at serine 536. Beta actin was used as a loading control. C) Densitometric analysis of phosphorylated NFκB p65 induction by LPS. Data represent the mean ± s.e.m. of three experiments; *p ≤ 0.05; one-way ANOVA, comparing responses of WT and NKLAM-KO BMDM.
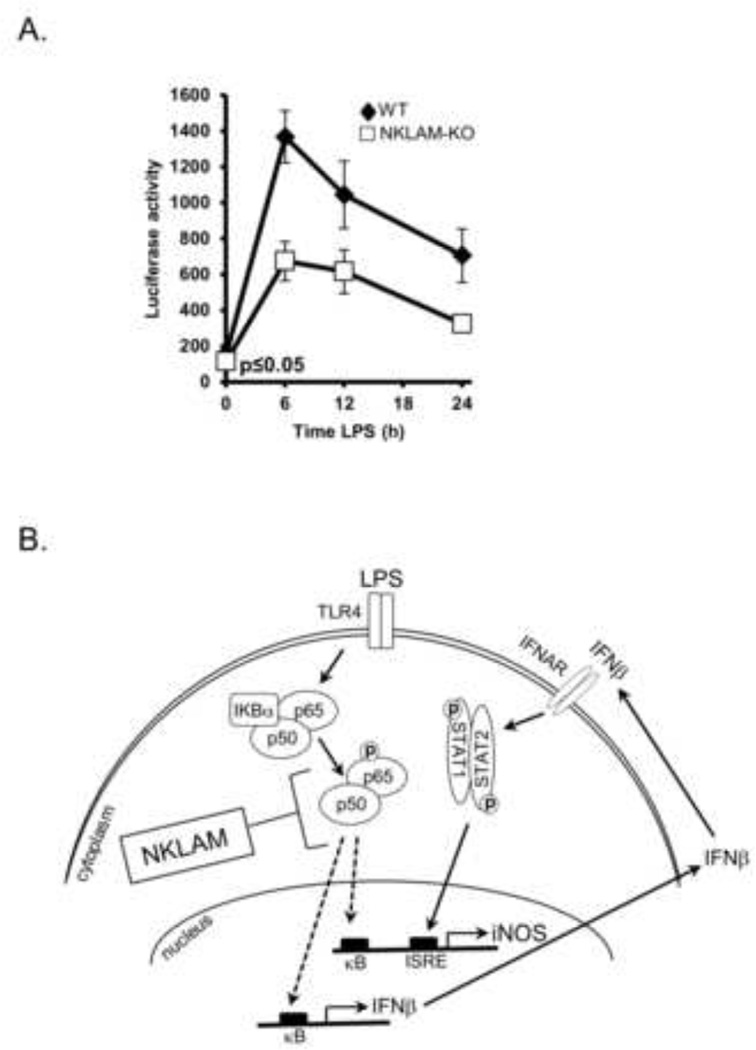
NFκB transcriptional activity is lower in NKLAM-KO than in WT BMDM
Our observation that NFκB translocation into the nucleus is delayed in NKLAM-KO macrophages prompted us to examine NFκB transcriptional activity in response to LPS stimulation. To this end, WT and NKLAM-KO BMDM were transfected by nucleofection with an NFκB firefly luciferase reporter plasmid and stimulated with 100 ng/ml LPS for 6, 12 and 24 h. NFκB proteins bind to the kappa enhancer element within the reporter plasmid, inducing luciferase transcription. The amount of luminescence is therefore a measure of NFκB transcriptional activity. Transfection efficiencies of WT and NKLAM-KO BMDM were monitored by nucleofection of the pmaxGFP plasmid. Cells were evaluated for GFP expression after 24 h by flow cytometry in a manner similar to Sims et al. (Sims, et al., 2003). Using this approach allowed us to examine the percentage of cells that are transfected and producing GFP protein. The transfection efficiency of WT and NKLAM-KO macrophages was identical, ranging from 22–24% GFP positive cells per experiment. Since the difference in nucleofection efficiency between WT and NKLAM-KO was insignificant, we normalized the relative luciferase light units per mg protein to compare WT and NKLAM-KO NFκB transcriptional activities. To ensure that the nucleofection efficiency within each group (unstimulated and LPS-stimulated) was identical, all of the nucleofected cells were aliquoted into 12-well plates from a single suspension prior to LPS stimulation. As shown in Fig. 8A, the NFκB transcriptional activity in LPS-stimulated WT cells peaked at 6 h (~7 fold compared to untreated cells), and then decreased with time. NFκB transcriptional activity from LPS-stimulated NKLAM-KO macrophages followed a similar pattern except the values were significantly lower than WT macrophages (p ≤ 0.05). These results indicate that NKLAM-KO macrophages have attenuated NFκB transcriptional activity in response to LPS stimulation.
Figure 8. NFκB p65 transcriptional activity in WT and NKLAM-KO BMDM and a model of iNOS regulation in NKLAM-KO BMDM.
A) WT and NKLAM-KO BMDM (4 × 106) were nucleofected with 3µg of the NFκB luciferase reporter plasmid pNFκB-luc and then stimulated with 100 ng/ml LPS for the times indicated. Firefly luciferase activity was measured and expressed as relative light units per mg protein. Data represent the mean ± s.e.m. of five experiments; p ≤ 0.05; one-way ANOVA. B) Proposed model of iNOS regulation in LPS-stimulated NKLAM-KO BMDM. Our data suggest that macrophages from NKLAM-KO mice have lower NFκB p65 expression, less p65 serine 536 phosphorylation (dotted circle) and delayed p65 nuclear translocation (dotted arrow). As there are defined NFκB binding sites (κB) within the promoters of iNOS and IFNβ, we propose that diminished translocation is likely to attenuate the transcription of both genes. This observation is supported by our findings of reduced levels of iNOS and IFNβ mRNA in LPS-stimulated NKLAM-KO cells compared to WT cells. Furthermore, diminished IFNβ production may account for the observed reduction in STAT1 phosphorylation which may ultimately affect STAT1-mediated iNOS upregulation through binding to the interferon-stimulated response element.
DISCUSSION
A growing body of literature has implicated members of the RBR family of ubiquitin ligases as important regulators of the immune response to foreign pathogens (de Leseleuc, et al., 2013, Manzanillo, et al., 2013). Recent studies from our laboratory demonstrated that the RBR family member NKLAM plays an important role in regulating macrophage killing of E. coli (Lawrence and Kornbluth, 2012). However, precisely how NKLAM is involved in the murine macrophage killing response has not been elucidated. This study was designed to address potential mechanisms by which NKLAM regulates the macrophage anti-bacterial response.
In this present study we show that LPS-stimulated BMDM from NKLAM-KO mice produce less NO than BMDM from WT mice (Fig.1). To support this initial in vitro observation we employed an in vivo peritonitis model to examine systemic NO production in NKLAM-KO mice. In agreement with previously reported studies, we found that i.p. injection of LPS induced a significant time-dependent increase in plasma nitrite levels (Yeh, et al., 2011). In WT mice, plasma nitrite levels were significantly elevated at 1, 2 and 6 h post-injection, and the highest levels were seen at 6h. In NKLAM-KO mice, there was a lag in NO production. The levels of plasma nitrite were not significantly elevated over PBS-injected controls until six h post-injection and never approached the levels seen in WT mice. This suggests that NKLAM may play a predominant role early in the regulation of NO production after LPS exposure in vivo.
In macrophages, iNOS is responsible for NO production in response to LPS; thus we examined the production of iNOS protein and mRNA in WT and NKLAM-KO macrophages. iNOS protein expression was both time and LPS concentration dependent and we found that overall NKLAM-KO BMDM produced less iNOS protein than WT BMDM. Transcription of iNOS was also lower in NKLAM-KO than WT BMDM after stimulation with LPS. We assessed whether differences in iNOS expression could be observed in resident splenic macrophages from WT and NKLAM-KO mice. In response to LPS stimulation, splenic macrophages from NKLAM-KO mice expressed less iNOS than WT macrophages (Fig. 2C).
In macrophages, Toll-like receptor 4 (TLR 4) stimulation by LPS induces iNOS expression. However, maximal iNOS expression is achieved through synergistic induction by inflammatory mediators such as IFNβ (Jacobs and Ignarro, 2001). Interestingly, we found less IFNβ mRNA expression after two h of LPS stimulation in NKLAM-KO spleen macrophages and BMDM than in WT macrophages, thus implicating NKLAM as a positive regulator, at least in part, of type I interferon expression. A contrasting role for RBR ubiquitin ligases as regulators of IFNβ was demonstrated by Inn et al. The authors showed that RBR family members HOIL-1 and HOIP, as part of the linear ubiquitin chain assembly complex (LUBAC), negatively regulated type I interferon production (Inn, et al., 2011). Clearly, further studies are needed to fully elucidate the multiple roles of RBR ubiquitin ligases in interferon production.
Interferon signaling is mediated by the JAK/STAT pathways. Following interferon receptor ligation, JAK phosphorylates STAT proteins, which then dimerize and enter the nucleus to initiate transcription. Ligation of TLR 4 also activates STAT phosphorylation. Rhee et al. demonstrated that STAT1 is phosphorylated on serine 727 and tyrosine 701 in macrophages stimulated with LPS (Rhee, et al., 2003). We found less STAT1 phosphorylation at tyrosine 701 in LPS-stimulated NKLAM-KO BMDM than in WT macrophages (Fig. 4). These results suggest NKLAM may play a role in the tyrosine phosphorylation state of STAT1. Due to the lack of immunologically-related substrates for NKLAM and the absence of known protein-protein interaction motifs within NKLAM, it is difficult to predict what proteins NKLAM may interact with. Studies are now in progress to determine how NKLAM may influence this phosphorylation event.
The NFκB family of transcription factors is crucial for coordinating the expression of a number of immunologically important genes. Consensus NFκB binding sites are found within the promoter regions of IFNβ (Doyle, et al., 2002) and iNOS. The central role of NFκB in iNOS and IFNβ protein expression prompted us to examine NFκB expression and translocation kinetics in WT and NKLAM-KO macrophages stimulated with LPS. We found that translocation of the NFκB p65 subunit into the nucleus was delayed in NKLAM-KO macrophages. Other studies have shown that delayed NFκB p65 translocation has a negative effect on DNA binding and transcription (Loscher, et al., 2005, Park and James, 2005). Indeed, we also found less p65/DNA colocalization in LPS-stimulated NKLAM-KO macrophages than in WT macrophages under similar conditions. One potential explanation for the observed delay in p65 translocation is that IKBα degradation or rebound expression during LPS stimulation is influenced by NKLAM. We tested this possibility by immunoblotting for IKBα in macrophages treated with LPS and found no difference in the kinetics of IKBα degradation or rebound expression between WT and NKLAM-KO macrophages. Additionally, WT and NKLAM-KO macrophages expressed similar amounts of IKBα protein. Thus, it is unlikely that NKLAM affects signaling proteins upstream of IKBα degradation. Interestingly, we observed reduced levels of NFκB p65 in unstimulated NKLAM-KO cells. In WT and NKLAM-KO macrophages stimulated with LPS, p65 expression levels increased over time. Similar LPS-stimulated p65 expression kinetics has been observed in other experimental systems (Sunday, et al., 2007). The p65 expression kinetics is nearly identical in WT and NKLAM-KO BMDM; however, total p65 expression was lower in NKLAM-KO than in WT macrophages (Fig. 5B). Thus, the lack of NKLAM expression is associated with both lower basal and LPS-stimulated p65 expression in macrophages. Other RBR family members have been shown to play a role in NFκB activation. Muller-Rischart et al. recently demonstrated that RBR family member parkin, in association with the linear ubiquitin chain assembly complex, activates NEMO (IKKγ) and thus facilitates the canonical NFκB signaling pathway (Muller-Rischart, et al., 2013). Previous studies have demonstrated that NFκB subunit p65 is a target for ubiquitin ligases, with polyubiquitination resulting in degradation (Hou, et al., 2012, Wang, et al., 2013). Further studies are needed to determine if NFκB p65 is a target for NKLAM ubiquitin ligase activity.
Phosphorylation is a key regulatory mechanism that governs the movement of NFκB subunits into the nucleus. There are at least ten phosphorylation sites in the NFκB p65 subunit (Perkins, 2006). One of the most well studied sites is serine 536. NFκB p65 subunit phosphorylation at serine 536 has been associated with p65 translocation and transcriptional activation (Hu, et al., 2005, Viatour, et al., 2005). By immunofluorescence, we observed less phospho-p65 Ser536 in NKLAM-KO macrophages stimulated with LPS than in WT macrophages (Fig. 7). The lack of phosphorylated p65 may reflect the reduced total p65 expression in NKLAM-KO macrophages both before and after LPS stimulation. We hypothesize that the lower p65 levels may reduce LPS-induced transcriptional activation of NFκB in NKLAM-KO macrophages. To test this hypothesis, we nucleofected WT and NKLAM-KO macrophages with an NFκB luciferase reporter plasmid and investigated the effect of NKLAM expression on NFκB -dependent transcriptional activity. As shown in Fig. 8A, NFκB transcriptional activity in response to LPS is significantly attenuated in NKLAM-KO macrophages.
In conclusion, we present evidence that the E3 ubiquitin ligase NKLAM is an important and positive regulator of the macrophage immune response. Our data identify NKLAM as part of an additional regulatory control point for synthesis of immunological proteins such as iNOS. Our data suggest that E3 ubiquitin ligase NKLAM modulates the transcriptional activity of NFκB. Figure 8B depicts a model of the proposed role of NKLAM in this pathway. In NKLAM-KO macrophages, NFκB p65 expression, serine 536 phosphorylation, and nuclear translocation are significantly reduced. We propose that these deficiencies significantly contribute to the observed reduction in iNOS protein via the attenuation of NFκB and IFNβ-mediated transcription of the iNOS gene. By controlling NFκB transcriptional activity, NKLAM has the potential to affect the transcriptional regulation of hundreds of immunologically important genes. Our data further strengthen the emerging concept that RBR ubiquitin ligases such as NKLAM play important roles in fundamental immune functions.
Acknowledgments
These studies were supported in part by NIH grant R56AI089758 and by a Merit Award (1 IO1BX000705) from the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development.
ABBREVIATIONS
- BMDM
Bone marrow-derived macrophage
- IFN
Interferon
- iNOS
inducible nitric oxide synthase
- KO
NKLAM-knockout
- LPS
Lipopolysaccharide
- NK
Natural killer cell
- NKLAM
Natural Killer Lytic-Associated Molecule
- NO
nitric oxide
- RBR
RING in between RING
- STAT
Signal transducer and activator of transcription
- TLR
Toll-like receptor
- WT
Wild type
- GFP
Green fluorescent protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The authors have no financial conflicts of interest
References
- Cruz MT, Duarte CB, Goncalo M, Carvalho AP, Lopes MC. LPS induction of I kappa B-alpha degradation and iNOS expression in a skin dendritic cell line is prevented by the janus kinase 2 inhibitor, Tyrphostin b42. Nitric Oxide. 2001;5:53. doi: 10.1006/niox.2000.0320. [DOI] [PubMed] [Google Scholar]
- Ratajczak-Wrona W, Jablonska E, Garley M, Jablonski J, Radziwon P, Iwaniuk A, Grubczak K. PI3K-Akt/PKB signaling pathway in neutrophils and mononuclear cells exposed to Nnitrosodimethylamine. Journal of Immunotoxicology. 2013;11:231. doi: 10.3109/1547691X.2013.826307. [DOI] [PubMed] [Google Scholar]
- Alayan J, Ivanovski S, Gemmell E, Ford P, Hamlet S, Farah CS. Deficiency of iNOS contributes to Porphyromonas gingivalis-induced tissue damage. Oral Microbiol Immunol. 2006;21:360. doi: 10.1111/j.1399-302X.2006.00304.x. [DOI] [PubMed] [Google Scholar]
- Zhang L, Liu Q, Yuan X, Wang T, Luo S, Lei H, Xia Y. Requirement of heat shock protein 70 for inducible nitric oxide synthase induction. Cell Signal. 2013;25:1310. doi: 10.1016/j.cellsig.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Jacobs AT, Ignarro LJ. Lipopolysaccharide-induced expression of interferon-beta mediates the timing of inducible nitric-oxide synthase induction in RAW 264.7 macrophages. J Biol Chem. 2001;276:47950. doi: 10.1074/jbc.M106639200. [DOI] [PubMed] [Google Scholar]
- Yao SY, Ljunggren-Rose A, Stratton CW, Mitchell WM, Sriram S. Regulation by IFN-beta of inducible nitric oxide synthase and interleukin-12/p40 in murine macrophages cultured in the presence of Chlamydia pneumoniae antigens. J Interferon Cytokine Res. 2001;21:137. doi: 10.1089/107999001750133131. [DOI] [PubMed] [Google Scholar]
- Habib S, Ali A. Biochemistry of nitric oxide. Indian J Clin Biochem. 2011;26:3. doi: 10.1007/s12291-011-0108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tecder-Unal M, Can F, Demirbilek M, Karabay G, Tufan H, Arslan H. The bactericidal and morphological effects of peroxynitrite on Helicobacter pylori. Helicobacter. 2008;13:42. doi: 10.1111/j.1523-5378.2008.00583.x. [DOI] [PubMed] [Google Scholar]
- Korhonen R, Lahti A, Kankaanranta H, Moilanen E. Nitric oxide production and signaling in inflammation. Curr Drug Targets Inflamm Allergy. 2005;4:471. doi: 10.2174/1568010054526359. [DOI] [PubMed] [Google Scholar]
- Xie QW, Whisnant R, Nathan C. Promoter of the mouse gene encoding calcium-independent nitric oxide synthase confers inducibility by interferon gamma and bacterial lipopolysaccharide. J Exp Med. 1993;177:1779. doi: 10.1084/jem.177.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Lee BS, Yi KY, Paik SG. Upstream NF-kappaB site is required for the maximal expression of mouse inducible nitric oxide synthase gene in interferon-gamma plus lipopolysaccharide-induced RAW 264.7 macrophages. Biochem Biophys Res Commun. 1997;236:655. doi: 10.1006/bbrc.1997.7031. [DOI] [PubMed] [Google Scholar]
- Zerfaoui M, Suzuki Y, Naura AS, Hans CP, Nichols C, Boulares AH. Nuclear translocation of p65 NF-kappaB is sufficient for VCAM-1, but not ICAM-1, expression in TNF-stimulated smooth muscle cells: Differential requirement for PARP-1 expression and interaction. Cell Signal. 2008;20:186. doi: 10.1016/j.cellsig.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoesel B, Schmid JA. The complexity of NF-kappaB signaling in inflammation and cancer. Mol Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins ND. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene. 2006;25:6717. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- Moreno R, Sobotzik JM, Schultz C, Schmitz ML. Specification of the NF-kappaB transcriptional response by p65 phosphorylation and TNF-induced nuclear translocation of IKK epsilon. Nucleic Acids Res. 2010;38:6029. doi: 10.1093/nar/gkq439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier JM, Kornbluth J. NK lytic-associated molecule, involved in NK cytotoxic function, is an E3 ligase. J Immunol. 2006;176:6454. doi: 10.4049/jimmunol.176.11.6454. [DOI] [PubMed] [Google Scholar]
- Kozlowski M, Schorey J, Portis T, Grigoriev V, Kornbluth J. NK lytic-associated molecule: a novel gene selectively expressed in cells with cytolytic function. J Immunol. 1999;163:1775. [PubMed] [Google Scholar]
- Portis T, Anderson J, Esposito A, Kornbluth J. Gene structure of human and mouse NKLAM, a gene associated with cellular cytotoxicity. Immunogenetics. 2000;51:546. doi: 10.1007/s002510000182. [DOI] [PubMed] [Google Scholar]
- Lawrence DW, Kornbluth J. E3 ubiquitin ligase NKLAM is a macrophage phagosome protein and plays a role in bacterial killing. Cell Immunol. 2012;279:46. doi: 10.1016/j.cellimm.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover RG, Gullickson G, Kornbluth J. Impaired NK cytolytic activity and enhanced tumor growth in NK lytic-associated molecule-deficient mice. J Immunol. 2009;183:6913. doi: 10.4049/jimmunol.0901679. [DOI] [PubMed] [Google Scholar]
- Hoover RG, Gullickson G, Kornbluth J. Natural killer lytic-associated molecule plays a role in controlling tumor dissemination and metastasis. Front Immunol. 2012;3:393. doi: 10.3389/fimmu.2012.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ginderachter JA, Meerschaut S, Liu Y, Brys L, De Groeve K, Hassanzadeh Ghassabeh G, Raes G, De Baetselier P. Peroxisome proliferator-activated receptor gamma (PPARgamma) ligands reverse CTL suppression by alternatively activated (M2) macrophages in cancer. Blood. 2006;108:525. doi: 10.1182/blood-2005-09-3777. [DOI] [PubMed] [Google Scholar]
- Moshage H, Kok B, Huizenga JR, Jansen PL. Nitrite and nitrate determinations in plasma: a critical evaluation. Clinical Chemistry. 1995;41:892. [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Flannagan RS, Cosio G, Grinstein S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nature Reviews. Microbiology. 2009;7:355. doi: 10.1038/nrmicro2128. [DOI] [PubMed] [Google Scholar]
- Gao JJ, Filla MB, Fultz MJ, Vogel SN, Russell SW, Murphy WJ. Autocrine/paracrine IFNalphabeta mediates the lipopolysaccharide-induced activation of transcription factor Stat1alpha in mouse macrophages: pivotal role of Stat1alpha in induction of the inducible nitric oxide synthase gene. J Immunol. 1998;161:4803. [PubMed] [Google Scholar]
- Rauch I, Muller M, Decker T. The regulation of inflammation by interferons and their STATs. JAKSTAT. 2013;2:1. doi: 10.4161/jkst.23820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SH, Jones BW, Toshchakov V, Vogel SN, Fenton MJ. Toll-like receptors 2 and 4 activate STAT1 serine phosphorylation by distinct mechanisms in macrophages. J Biol Chem. 2003;278:22506. doi: 10.1074/jbc.M208633200. [DOI] [PubMed] [Google Scholar]
- Dhruv HD, McDonough Winslow WS, Armstrong B, Tuncali S, Eschbacher J, Kislin K, Loftus JC, Tran NL, Berens ME. Reciprocal activation of transcription factors underlies the dichotomy between proliferation and invasion of glioma cells. PloS one. 2013;8:e72134. doi: 10.1371/journal.pone.0072134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Haseebuddin M, Young M, Colburn NH. Suppression of p65 phosphorylation coincides with inhibition of IkappaBalpha polyubiquitination and degradation. Mol Carcinog. 2005;44:274. doi: 10.1002/mc.20142. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Liss AS, Gottlieb PD. Normalization of luciferase reporter assays under conditions that alter internal controls. Biotechniques. 2003;34:938. doi: 10.2144/03345bm07. [DOI] [PubMed] [Google Scholar]
- de Leseleuc L, Orlova M, Cobat A, Girard M, Huong NT, Ba NN, Thuc NV, Truman R, Spencer JS, Adams L, Thai VH, Alcais A, Schurr E. PARK2 mediates interleukin 6 and monocyte chemoattractant protein 1 production by human macrophages. PLoS Negl Trop Dis. 2013;7:e2015. doi: 10.1371/journal.pntd.0002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, Schneider DS, Nakamura K, Shiloh MU, Cox JS. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501:512. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh C-H, Chou W, Chu C-C, So EC, Chang H-C, Wang J-J, Hsing C-H. Anticancer Agent 2-Methoxyestradiol Improves Survival in Septic Mice by Reducing the Production of Cytokines and Nitric Oxide. Shock. 2011;36:510. doi: 10.1097/SHK.0b013e318231866f. [DOI] [PubMed] [Google Scholar]
- Inn K-S, Gack MU, Tokunaga F, Shi M, Wong L-Y, Iwai K, Jung JU. Linear Ubiquitin Assembly Complex Negatively Regulates RIG-I- and TRIM25-Mediated Type I Interferon Induction. Molecular Cell. 2011;41:354. doi: 10.1016/j.molcel.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SE, Vaidya SA, O'Connell R, Dadgostar H, Dempsey PW, Wu T-T, Rao G, Sun R, Haberland ME, Modlin RL, Cheng G. IRF3 Mediates a TLR3/TLR4-Specific Antiviral Gene Program. Immunity. 2002;17:251. doi: 10.1016/s1074-7613(02)00390-4. [DOI] [PubMed] [Google Scholar]
- Loscher CE, Draper E, Leavy O, Kelleher D, Mills KH, Roche HM. Conjugated linoleic acid suppresses NF-kappa B activation and IL-12 production in dendritic cells through ERK-mediated IL-10 induction. J Immunol. 2005;175:4990. doi: 10.4049/jimmunol.175.8.4990. [DOI] [PubMed] [Google Scholar]
- Park S, James CD. ECop (EGFR-coamplified and overexpressed protein), a novel protein, regulates NF-kappaB transcriptional activity and associated apoptotic response in an IkappaBalpha-dependent manner. Oncogene. 2005;24:2495. doi: 10.1038/sj.onc.1208496. [DOI] [PubMed] [Google Scholar]
- Sunday L, Osuna C, Krause DN, Duckles SP. Age alters cerebrovascular inflammation and effects of estrogen. Am J Physiol Heart Circ Physiol. 2007;292:H2333. doi: 10.1152/ajpheart.01057.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Rischart AK, Pilsl A, Beaudette P, Patra M, Hadian K, Funke M, Peis R, Deinlein A, Schweimer C, Kuhn PH, Lichtenthaler SF, Motori E, Hrelia S, Wurst W, Trumbach D, Langer T, Krappmann D, Dittmar G, Tatzelt J, Winklhofer KF. The E3 ligase parkin maintains mitochondrial integrity by increasing linear ubiquitination of NEMO. Mol Cell. 2013;49:908. doi: 10.1016/j.molcel.2013.01.036. [DOI] [PubMed] [Google Scholar]
- Hou Y, Moreau F, Chadee K. PPARgamma is an E3 ligase that induces the degradation of NFkappaB/p65. Nat Commun. 2012;3:1300. doi: 10.1038/ncomms2270. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ren F, Wang Y, Feng Y, Wang D, Jia B, Qiu Y, Wang S, Yu J, Sung JJ, Xu J, Zeps N, Chang Z. CHIP/Stub1 functions as a tumor suppressor and represses NF-kappaB-mediated signaling in colorectal cancer. Carcinogenesis. 2013;35:983. doi: 10.1093/carcin/bgt393. [DOI] [PubMed] [Google Scholar]
- Viatour P, Merville MP, Bours V, Chariot A. Phosphorylation of NF-kappaB and IkappaB proteins: implications in cancer and inflammation. Trends Biochem Sci. 2005;30:43. doi: 10.1016/j.tibs.2004.11.009. [DOI] [PubMed] [Google Scholar]