Abstract
SETD8 (also known as SET8, PR-SET7, or KMT5A (lysine methyltransferase 5A)) is the only known lysine methyltransferase that catalyzes monomethylation of histone H4 lysine 20 (H4K20). In addition to H4K20, SETD8 monomethylates non-histone substrates such as the tumor suppressor p53 and proliferating cell nuclear antigen (PCNA). Because of its role in regulating diverse biological processes, SETD8 has been pursued as a potential therapeutic target. We recently reported the first substrate-competitive SETD8 inhibitor, UNC0379 (1), which is selective for SETD8 over 15 other methyltransferases. We characterized this inhibitor in a battery of biochemical and biophysical assays. Here we describe our comprehensive structure-activity relationship (SAR) studies of this chemical series. In addition to 2- and 4-substituents, we extensively explored 6- and 7-substituents of the quinazoline scaffold. These SAR studies led to the discovery of several new compounds, which displayed similar potencies as compound 1, and interesting SAR trends.
Introduction
Post-translational modifications (PTMs) of histones are critical in regulating gene expression and transcription.1–3 Among a myriad of PTMs, histone lysine methylation has been recognized as a major mechanism in chromatin regulation. Histone lysine methylation typically takes place at the N-terminal tails of core histone proteins (H3, H4, H2A, and H2B) and is catalyzed by protein lysine methyltransferases (PKMTs). It is worth noting that PKMTs also methylate many non-histone substrates.4
PKMTs have been increasingly recognized as a class of potential therapeutic targets by the medicinal chemistry and drug discovery community. Consequently, a number of selective inhibitors of PKMTs have been discovered during recent years.5–31 Related to PKMTs, protein arginine methyltransferases (PRMTs) catalyze methylation of arginine residues of histone and non-histone proteins.32 A number of selective inhibitors of PRMTs have also been reported.33–35
SETD8 (also known as SET8, PR-SET7, or KMT5A (lysine methyltransferase 5A)), first characterized in 2002, is the only known PKMT that catalyzes monomethylation of histone H4 lysine 20 (H4K20).36–38 Monomethylation of H4K20 (H4K20me) and SETD8 have been implicated in the DNA damage response and cell cycle progression.38 In addition, SETD8 promotes epithelial–mesenchymal transition (EMT) by physically associating with TWIST, a master regulator of EMT.39 SETD8 also monomethylates lysine 382 (K382) of the tumor suppressor p53 and lysine 248 (K248) of proliferating cell nuclear antigen (PCNA) and plays a potential role in human carcinogenesis.40, 41
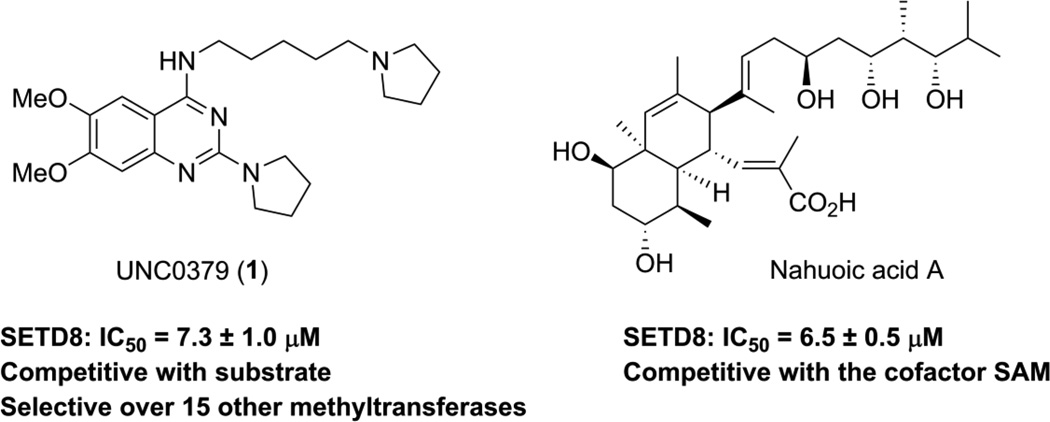
We recently reported UNC0379 (1) as the first substrate-competitive small-molecule inhibitor of SETD8 (Figure 1).42 Compound 1 is active in multiple biochemical (e.g., radioactive methyl transfer, microfluidic capillary electrophoresis) and biophysical (e.g., isothermal titration calorimetry, surface plasmon resonance) assays, and importantly, selective for SETD8 over 15 other methyltransferases.42 The only other known selective inhibitor of SETD8 is marine nature product nahuoic acid A, which is competitive with the co-factor S-adenosyl-L-methionine (SAM) (Figure 1).21 In this article, we describe our comprehensive structure-activity relationship (SAR) studies of the quinazoline template represented by compound 1, which resulted in the discovery of interesting SAR trends and novel analogs with similar potencies as compound 1.
Figure 1.
Structures of the known selective inhibitors of SETD8.
Results and discussion
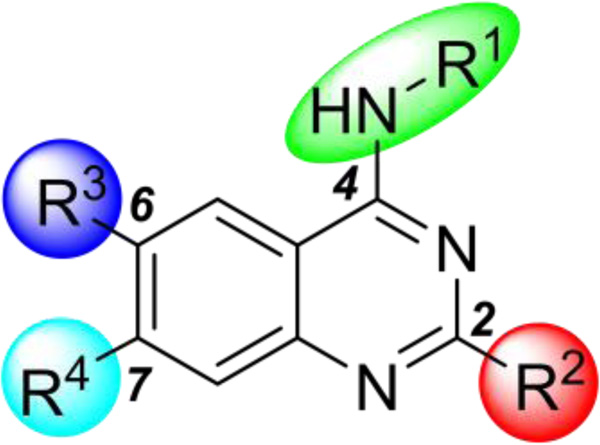
Our strategy for studying SAR of the UNC0379 series was to extensively explore the 2-, 4-, 6-, and 7-substituents (Figure 2). We previously reported initial SAR results of the 2- and 4-substituents.42 In these new studies, we investigated not only additional 2- and 4-substituents, but also 6- and 7-substituents, the two regions that were not previously explored for SETD8.
Figure 2.
The four highlighted regions were explored for studying SAR of the UNC0379 series.
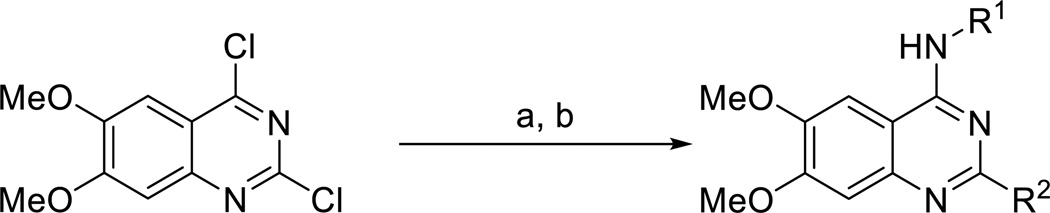
We first explored the 4-amino moiety of the quinazoline scaffold. Compounds 1 – 15 (Scheme 1 and Table 1) were synthesized from commercially available 2,4-dichloro-6,7-dimethoxyquinazoline and corresponding amines in good yields. As previously reported,6 we displaced the 4-chloro group with the first set of amines at room temperature. The 2-chloro group was substituted by the second set of amines under microwave heating conditions, yielding the desired 2,4-diamino-6,7-dimethoxyquinazolines (Scheme 1).
Scheme 1.
Typical synthesis of 2,4-diamino-6,7-dimethoxyquinazolines.a
aReagents and conditions: (a) R1 amines, THF, N,N-diisopropylethylamine, room temperature; (b) R2 amines, n-BuOH, DIPEA, microwave, heat.
Table 1.
SAR of the 4-Amino Moiety.
 | ||
|---|---|---|
| Compound | R1 Group | SETD8 IC50 (µM)a |
| 1 |  |
7.3 ± 1.0b |
| 2 |  |
7.9 ± 0.8b |
| 3 | 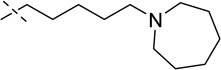 |
7.9 ± 1.2 |
| 4 | 7.9 ± 1.4b | |
| 5 | 26 ± 5 | |
| 6 | 40 ± 1 | |
| 7 | 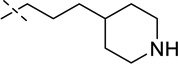 |
40 ± 6 |
| 8 | 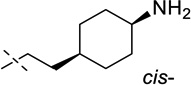 |
43 ± 5 |
| 9 | 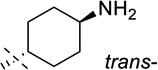 |
> 250 |
| 10 | 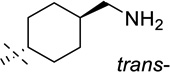 |
> 250 |
| 11 | 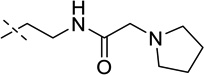 |
63 ± 19 |
| 12 | 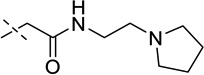 |
> 250 |
| 13 | 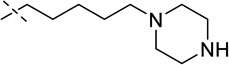 |
32 ± 8b |
| 14 | 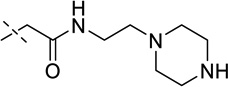 |
> 250 |
| 15 | 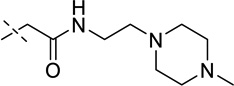 |
> 250 |
IC50 determination experiments were performed in duplicate.
IC50 value was reported previously.42
The ring size of the terminal cyclic amino group did not have significant impact on SETD8 potency (Table 1). Pyrrolidine (compound 1), piperidine (compound 2), and azepane (compound 3) resulted in similar potencies. The replacement of the cyclic amino group with an acyclic amino group such as dimethyl amine group (compound 4) didn’t lead to a significant potency change either. However, increasing the length of the side chain from 5 to 6 carbons (compound 4 versus compound 5) resulted in a decrease in potency. We previously reported that decreasing the length of the side chain also led to a decrease in potency.42 Interestingly, replacing the dimethyl amine group with the primary amino group slightly decreased potency (compound 5 versus compound 6). We next explored various conformation-constrained analogs and found that these compounds (7 – 10) were not as potent as compound 1. In addition to exploring the length of the side chain, we also attempted to replace the straight 5-carbon chain with several amide-containing linkers and found that the amide 11 was significantly less potent than compound 1 and the amide 12 was completely inactive. We previously reported that compound 13 was weakly active against SETD8.42 Consistent with the result of the amide 12, the amides 14 and 15 did not display any activity against SETD8. Taken together, these results suggest that: (1) the terminal pyrrolidine group can be modified without potency loss; and (2) the 5-carbon linker is optimal. In addition, we previously demonstrated that the basicity of the pyrrolidine nitrogen was important for maintaining potency for SETD8.42
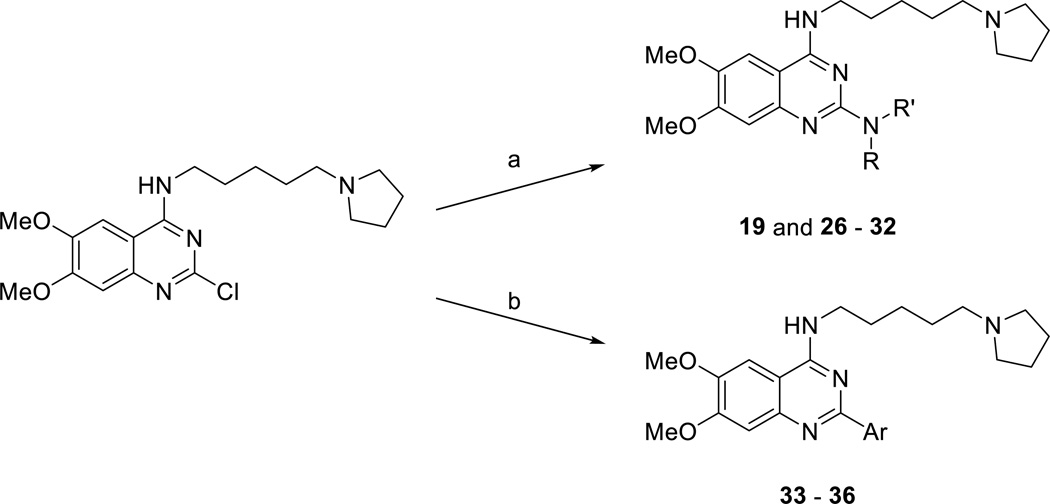
We next investigated various 2-substituents at the quinazoline core (Scheme 2 and Table 2). Synthesis of compounds 16 – 18 was described previously.42 Compounds 20 – 25 were prepared according to the synthetic route illustrated in Scheme 1. Buchwald–Hartwig amination reaction conditions43 were used to synthesize compounds 19 and 26 – 32 from the known intermediate 2-chloro-6,7-dimethoxy-N-(5-(pyrrolidin-1-yl)pentyl)quinazolin-4-amine42 and commercially available amines (Scheme 2). Compounds 19 and 26 – 32 could not be generated using standard nucleophilic conditions (Scheme 1) in good yields. A Suzuki coupling reaction44 was used to prepare compounds 33 – 36 from the same intermediate and commercially available aromatic boronic acids (Scheme 2).
Scheme 2.
Synthesis of compounds 19 and 26 – 36.a
aReagents and conditions: (a) Amines, Pd(OAc)2, (+)-BINAP, Cs2CO3, THF, microwave, heat; (b) Aromatic boronic acid, Pd(PPh3)4, K2CO3, dioxane/water, microwave, heat. Ar, aromatic ring.
Table 2.
SAR of the 2-Substituted Group.
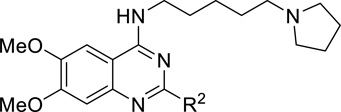 | ||
|---|---|---|
| Compound | R2 Group | SETD8 IC50 (µM)a |
| 1 | 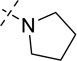 |
7.3 ± 1.0b |
| 16 |  |
94 ± 18b |
| 17 | 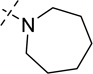 |
> 250b |
| 18 | 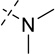 |
9.2 ± 1.2b |
| 19 | 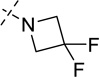 |
110 ± 4 |
| 20 | 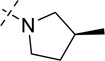 |
15 ± 3 |
| 21 |  |
37 ± 9b |
| 22 | 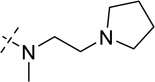 |
> 250 |
| 23 | 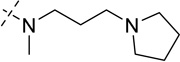 |
> 250 |
| 24 | 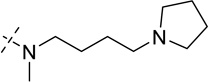 |
> 250 |
| 25 | 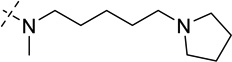 |
> 250 |
| 26 | 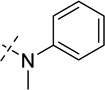 |
> 250 |
| 27 | 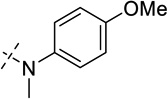 |
> 250 |
| 28 | 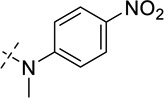 |
> 250 |
| 29 | 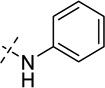 |
> 250 |
| 30 | 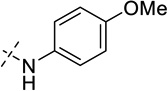 |
> 250 |
| 31 | 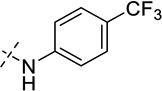 |
> 250 |
| 32 | 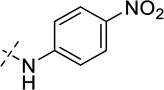 |
> 250 |
| 33 | 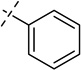 |
> 250 |
| 34 | 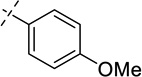 |
> 250 |
| 35 | 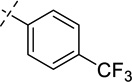 |
> 250 |
| 36 | 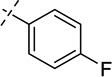 |
> 250 |
IC50 determination experiments were performed in duplicate.
IC50 value was reported previously.42
As shown in Table 2, replacing the pyrrolidine (compound 1) with either the piperidine (compound 16) or azepane (compound 17) resulted in a significant loss of the potency, suggesting that a larger group is disfavored. On the other hand, the replacement of the pyrrolidine (compound 1) with the dimethyl amine group (compound 18) didn’t lead to a significant potency change. These results were reported previously.42 Interestingly, the 3,3-difluoroazetidine (compound 19) resulted in a large loss of potency, possibly due to the increase of the compound’s polarity. We attempted to synthesize a simple azetidine analog, but found that the target molecule was unstable and could not be isolated. Adding a methyl substituent to the pyrrolidine (compound 20) led to about two-fold drop in potency. We previously reported that compound 21 displayed some activity for SETD8.42 Based on this result, we attempted to introduce a pyrrolidine with a 2 – 5-carbon linker and were disappointed to find that these compounds (22 – 25) did not exhibit any activity for SETD8. We also explored unsubstituted and substituted N-methylanilines and anilines. As shown in Table 2, unsubstituted N-methylaniline (compound 26) and N-methylanilines with either an electron-donating group (methoxy, compound 27) or an electron-withdrawing group (nitro, compound 28) at the para-position were inactive. Similarly, unsubstituted aniline (compound 29) and anilines with either an electron-donating group (compound 30) or an electron-withdrawing group (compounds 31 and 32) at the para-position did not display any activity. Lastly, we attempted to replace the pyrrolidine with a phenyl or substituted phenyl group containing an electron-donating or electron-withdrawing group at the para-position, but found that none of these compounds (33 – 36) were active against SETD8. Taken together, these results suggest that the SAR at 2-substituent is very tight. The vast majority of the modifications we made led to a significant or complete loss of potency for SETD8.
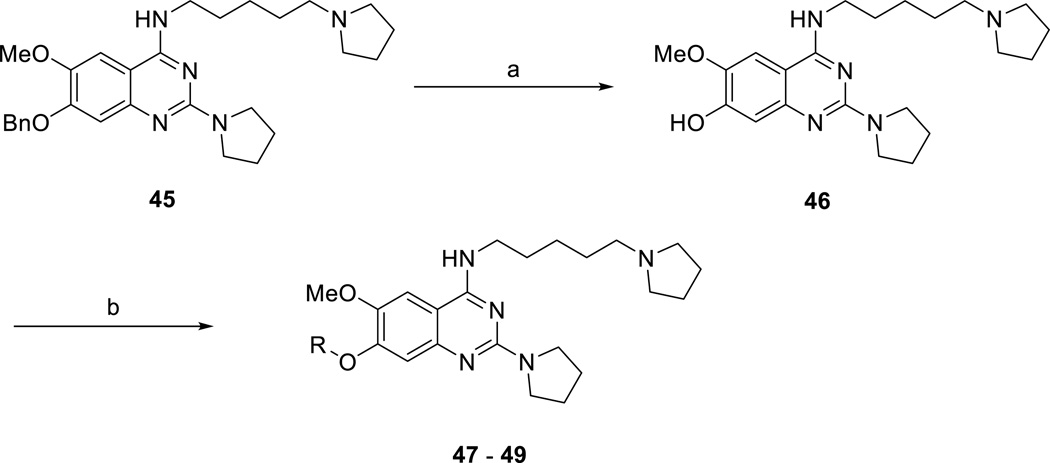
We also extensively explored the 6- and 7-substituents (Scheme 3 and Table 3), which were not studied previously. Compounds 37 – 45 were prepared from different 6,7-substituted 2,4-dichloroquinazolines (synthesis of these intermediates is detailed in the supplementary information) according to the synthetic route illustrated in Scheme 1. Synthesis of compounds 46 – 49 is outlined in Scheme 3. Briefly, debenzylation of compound 45 via hydrogenation produced compound 46. Nucleophilic substitution reactions between the phenol 46 and various alkyl bromides afforded compounds 47 – 50.
Scheme 3.
Synthesis of compounds 46 – 49.a
aReagents and conditions: (a) H2, Pd/C, EtOH, room temperature; (b) RBr, K2CO3, DMF, room temperature.
Table 3.
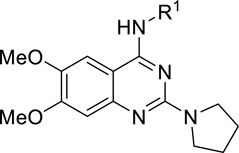
SAR of the 6- and 7-Substituted Groups.
 | |||
|---|---|---|---|
| Compound | R3 Group | R4 Group | SETD8 IC50 (µM)a |
| 1 | MeO- | MeO- | 7.3 ± 1.0b |
| 37 | H- | H- | > 250 |
| 38 | H- | MeO- | > 250 |
| 39 | MeO- | H- | 52 ± 11 |
| 40 | EtO- | MeO- | 9.5 ± 0.9 |
| 41 | i-PrO- | MeO- | 61 ± 12 |
| 42 | Cl- | MeO- | 46 ± 11 |
| 43 | MeO- | EtO- | 11 ± 1 |
| 44 | MeO- | i-PrO- | 48 ± 10 |
| 45 | MeO- | BnO- | 130 ± 44 |
| 46 | MeO- | HO- | > 250 |
| 47 | MeO- | 8.0 ± 0.8 | |
| 48 | MeO- | 40 ± 12 | |
| 49 | MeO- | 24 ± 3 | |
| 50 | MeO- | 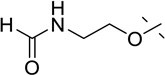 |
8.7 ± 0.4 |
IC50 determination experiments were performed in duplicate.
IC50 value was reported previously.42
We found that the removal of both methoxy groups (compound 37) or 6-methoxy group (compound 38) completely abolished activity (Table 3). Interestingly, the removal of the 7-methoxy group (compound 39) led to about 7-fold loss in potency, but it retained some activity against SETD8. These results suggest that the 6-methoxy group may be more important for maintaining SETD8 potency compared with the 7-methoxy group.
We next investigated several 6-substituents while holding the 7-methoxy group constant and found that the replacement of the 6-methoxy group with the 6-ethoxy group did not result in a significant change in potency (compound 1 versus compound 40). On the other hand, the 6-isopropoxy group (compound 41) and 6-chloro group (compound 42) led to about 8-fold and 60-fold loss in potency, respectively, suggesting that a larger group or a less electron-donating group is disfavored at this position.
Lastly, we explored various 7-substituents while holding the 6-methoxy group constant. Slightly increasing the size of the 7-methoxy group to the 7-ethoxy group (compound 43) did not significantly change potency. On the other hand, the larger 7-isopropoxy group (compound 44) and 7-benzyloxy group (compound 45) led to more than 6-fold and 17-fold potency drops, respectively. Interestingly, the 7-hydroxy group (compound 46) completely abolished activity against SETD8. We next studied whether a linear chain could be tolerated at the 7-position. We were pleased to find that compound 47, which contains the 7-methoxyethyloxy group, was as potent as compound 1. However, the 7-methoxypropoxy group (compound 48) and 7-hydroxypropoxy group (compound 49) led to a significant decrease in potency. Interestingly, compound 50, which contains the 7-formylaminoethyloxy group, retained the same potency as compounds 1 and 47. Taken together, these results suggest that the 7-position is amenable to modifications and there may be an opportunity to create more potent inhibitors of SETD8 by further exploring this region.
Conclusions
We comprehensively studied SAR of the quinazoline scaffold represented by UNC0379 (compound 1), which was recently discovered as the first substrate-competitive inhibitor of SETD8. We found a number of interesting SAR trends. They include that: (1) at 4-position, the terminal pyrrolidine group can be modified without potency loss and the 5-carbon linker is optimal; (2) at 2-position, modifications are generally not tolerated and pyrrolidine and dimethylamino groups are optimal; (3) at 6-position, the methoxy and ethoxy groups are preferred and a larger group or a less electron-donating group is disfavored; and (4) at 7-position, modifications can be well tolerated and further exploration of this region may result in more potent SETD8 inhibitors. These SAR studies also led to the discovery of several novel compounds (3, 40, 43, 47 and 50), which exhibited similar potencies as compound 1. During the revision of this paper, several novel inhibitors of SETD8 have been reported.45
Supplementary Material
Acknowledgements
The research described here was supported by the grant R01GM103893 from the U.S. National Institute of General Medical Sciences of the National Institutes of Health. Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canada Foundation for Innovation, Eli Lilly Canada, GlaxoSmithKline, the Ontario Ministry of Economic Development and Innovation, the Novartis Research Foundation, Pfizer, AbbVie, Takeda, Janssen, Boehringer Ingelheim, Bayer and the Wellcome Trust.
Footnotes
This article is part of the MedChemComm “Epigenetics” themed issue.
Electronic supplementary information (ESI) available: synthetic procedures, characterization data, and biochemical assays. See DOI: 10.1039/b000000x/
References
- 1.Copeland RA, Solomon ME, Richon VM. Nat Rev Drug Discov. 2009;8:724–732. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- 2.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Nat Rev Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 3.Helin K, Dhanak D. Nature. 2013;502:480–488. doi: 10.1038/nature12751. [DOI] [PubMed] [Google Scholar]
- 4.Clarke SG. Trends in Biochemical Sciences. 2013;38:243–252. doi: 10.1016/j.tibs.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kubicek S, O'Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, Kelly TA, Jenuwein T. Molecular cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 6.Liu F, Chen X, Allali-Hassani A, Quinn AM, Wasney GA, Dong A, Barsyte D, Kozieradzki I, Senisterra G, Chau I, Siarheyeva A, Kireev DB, Jadhav A, Herold JM, Frye SV, Arrowsmith CH, Brown PJ, Simeonov A, Vedadi M, Jin J. Journal of medicinal chemistry. 2009;52:7950–7953. doi: 10.1021/jm901543m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang Y, Ganesh T, Horton JR, Spannhoff A, Liu J, Sun A, Zhang X, Bedford MT, Shinkai Y, Snyder JP, Cheng X. J Mol Biol. 2010;400:1–7. doi: 10.1016/j.jmb.2010.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu F, Chen X, Allali-Hassani A, Quinn AM, Wigle TJ, Wasney GA, Dong A, Senisterra G, Chau I, Siarheyeva A, Norris JL, Kireev DB, Jadhav A, Herold JM, Janzen WP, Arrowsmith CH, Frye SV, Brown PJ, Simeonov A, Vedadi M, Jin J. Journal of medicinal chemistry. 2010;53:5844–5857. doi: 10.1021/jm100478y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vedadi M, Barsyte-Lovejoy D, Liu F, Rival-Gervier S, Allali-Hassani A, Labrie V, Wigle TJ, DiMaggio PA, Wasney GA, Siarheyeva A, Dong A, Tempel W, Wang S-C, Chen X, Chau I, Mangano T, Huang X-P, Simpson CD, Pattenden SG, Norris JL, Kireev DB, Tripathy A, Edwards A, Roth BL, Janzen WP, Garcia BA, Petronis A, Ellis J, Brown PJ, Frye SV, Arrowsmith CH, Jin J. Nat Chem Biol. 2011;7:566–574. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu F, Barsyte-Lovejoy D, Allali-Hassani A, He Y, Herold JM, Chen X, Yates CM, Frye SV, Brown PJ, Huang J, Vedadi M, Arrowsmith CH, Jin J. Journal of medicinal chemistry. 2011;54:6139–6150. doi: 10.1021/jm200903z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferguson AD, Larsen NA, Howard T, Pollard H, Green I, Grande C, Cheung T, Garcia-Arenas R, Cowen S, Wu J, Godin R, Chen H, Keen N. Structure. 2011;19:1262–1273. doi: 10.1016/j.str.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 12.Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, Johnston LD, Scott MP, Smith JJ, Xiao Y, Jin L, Kuntz KW, Chesworth R, Moyer MP, Bernt KM, Tseng JC, Kung AL, Armstrong SA, Copeland RA, Richon VM, Pollock RM. Cancer cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao Y, Chen P, Diao J, Cheng G, Deng L, Anglin JL, Prasad BVV, Song Y. J Am Chem Soc. 2011;133:16746–16749. doi: 10.1021/ja206312b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan Y, Wang Q, Paulk J, Kubicek S, Kemp MM, Adams DJ, Shamji AF, Wagner BK, Schreiber SL. ACS chemical biology. 2012;7:1152–1157. doi: 10.1021/cb300139y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, Sacks JD, Raimondi A, Majer CR, Song J, Scott MP, Jin L, Smith JJ, Olhava EJ, Chesworth R, Moyer MP, Richon VM, Copeland RA, Keilhack H, Pollock RM, Kuntz KW. Nat Chem Biol. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 16.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Iii AD, Diaz E, Lafrance LV, Mellinger M, Duquenne C, Tian X, Kruger RG, McHugh CF, Brandt M, Miller WH, Dhanak D, Verma SK, Tummino PJ, Creasy CL. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 17.Verma SK, Tian X, LaFrance LV, Duquenne C, Suarez DP, Newlander KA, Romeril SP, Burgess JL, Grant SW, Brackley JA, Graves AP, Scherzer DA, Shu A, Thompson C, Ott HM, Aller GSV, Machutta CA, Diaz E, Jiang Y, Johnson NW, Knight SD, Kruger RG, McCabe MT, Dhanak D, Tummino PJ, Creasy CL, Miller WH. ACS Med Chem Lett. 2012;3:1091–1096. doi: 10.1021/ml3003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng W, Ibáñez G, Wu H, Blum G, Zeng H, Dong A, Li F, Hajian T, Allali-Hassani A, Amaya MF, Siarheyeva A, Yu W, Brown PJ, Schapira M, Vedadi M, Min J, Luo M. J Am Chem Soc. 2012;134:18004–18014. doi: 10.1021/ja307060p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi W, Chan H, Teng L, Li L, Chuai S, Zhang R, Zeng J, Li M, Fan H, Lin Y, Gu J, Ardayfio O, Zhang J-H, Yan X, Fang J, Mi Y, Zhang M, Zhou T, Feng G, Chen Z, Li G, Yang T, Zhao K, Liu X, Yu Z, Lu CX, Atadja P, Li E. Proc Natl Acad Sci U S A. 2012;109:21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu W, Chory EJ, Wernimont AK, Tempel W, Scopton A, Federation A, Marineau JJ, Qi J, Barsyte-Lovejoy D, Yi J, Marcellus R, Iacob RE, Engen JR, Griffin C, Aman A, Wienholds E, Li F, Pineda J, Estiu G, Shatseva T, Hajian T, Al-awar R, Dick JE, Vedadi M, Brown PJ, Arrowsmith CH, Bradner JE, Schapira M. Nat Commun. 2012;3:1288. doi: 10.1038/ncomms2304. [DOI] [PubMed] [Google Scholar]
- 21.Williams DE, Dalisay DS, Li F, Amphlett J, Maneerat W, Chavez MA, Wang YA, Matainaho T, Yu W, Brown PJ, Arrowsmith CH, Vedadi M, Andersen RJ. Organic letters. 2013;15:414–417. doi: 10.1021/ol303416k. [DOI] [PubMed] [Google Scholar]
- 22.Anglin JL, Deng L, Yao Y, Cai G, Liu Z, Jiang H, Cheng G, Chen P, Dong S, Song Y. Journal of medicinal chemistry. 2012;55:8066–8074. doi: 10.1021/jm300917h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konze KD, Ma A, Li F, Barsyte-Lovejoy D, Parton T, Macnevin CJ, Liu F, Gao C, Huang XP, Kuznetsova E, Rougie M, Jiang A, Pattenden SG, Norris JL, James LI, Roth BL, Brown PJ, Frye SV, Arrowsmith CH, Hahn KM, Wang GG, Vedadi M, Jin J. ACS chemical biology. 2013;8:1324–1334. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knutson SK, Warholic NM, Wigle TJ, Klaus CR, Allain CJ, Raimondi A, Porter Scott M, Chesworth R, Moyer MP, Copeland RA, Richon VM, Pollock RM, Kuntz KW, Keilhack H. Proc Natl Acad Sci U S A. 2013;110:7922–7927. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beguelin W, Popovic R, Teater M, Jiang Y, Bunting KL, Rosen M, Shen H, Yang SN, Wang L, Ezponda T, Martinez-Garcia E, Zhang H, Zheng Y, Verma SK, McCabe MT, Ott HM, Van Aller GS, Kruger RG, Liu Y, McHugh CF, Scott DW, Chung YR, Kelleher N, Shaknovich R, Creasy CL, Gascoyne RD, Wong KK, Cerchietti L, Levine RL, Abdel-Wahab O, Licht JD, Elemento O, Melnick AM. Cancer cell. 2013;23:677–692. doi: 10.1016/j.ccr.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu F, Barsyte-Lovejoy D, Li F, Xiong Y, Korboukh V, Huang XP, Allali-Hassani A, Janzen WP, Roth BL, Frye SV, Arrowsmith CH, Brown PJ, Vedadi M, Jin J. Journal of medicinal chemistry. 2013;56:8931–8942. doi: 10.1021/jm401480r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sweis RF, Pliushchev M, Brown PJ, Guo J, Li FL, Maag D, Petros AM, Soni NB, Tse C, Vedadi M, Michaelides MR, Chiang GG, Pappano WN. Acs Medicinal Chemistry Letters. 2014;5:205–209. doi: 10.1021/ml400496h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garapaty-Rao S, Nasveschuk C, Gagnon A, Chan EY, Sandy P, Busby J, Balasubramanian S, Campbell R, Zhao F, Bergeron L, Audia JE, Albrecht BK, Harmange JC, Cummings R, Trojer P. Chemistry & biology. 2013;20:1329–1339. doi: 10.1016/j.chembiol.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 29.Daigle SR, Olhava EJ, Therkelsen CA, Basavapathruni A, Jin L, Boriack-Sjodin PA, Allain CJ, Klaus CR, Raimondi A, Scott MP, Waters NJ, Chesworth R, Moyer MP, Copeland RA, Richon VM, Pollock RM. Blood. 2013;122:1017–1025. doi: 10.1182/blood-2013-04-497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nasveschuk CG, Gagnon A, Garapaty-Rao S, Balasubramanian S, Campbell R, Lee C, Zhao F, Bergeron L, Cummings R, Trojer P, Audia JE, Albrecht BK, Harmange J-CP. ACS Medicinal Chemistry Letters. 2014;5:378–383. doi: 10.1021/ml400494b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He Y, Korboukh I, Jin J, Huang J. Acta biochimica et biophysica Sinica. 2012;44:70–79. doi: 10.1093/abbs/gmr109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bedford MT, Richard S. Molecular cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 33.Siarheyeva A, Senisterra G, Allali-Hassani A, Dong A, Dobrovetsky E, Wasney Gregory A, Chau I, Marcellus R, Hajian T, Liu F, Korboukh I, Smil D, Bolshan Y, Min J, Wu H, Zeng H, Loppnau P, Poda G, Griffin C, Aman A, Brown PJ, Jin J, Al-awar R, Arrowsmith CH, Schapira M, Vedadi M. Structure. 2012;20:1425–1435. doi: 10.1016/j.str.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Liu F, Li F, Ma A, Dobrovetsky E, Dong A, Gao C, Korboukh I, Liu J, Smil D, Brown PJ, Frye SV, Arrowsmith CH, Schapira M, Vedadi M, Jin J. Journal of medicinal chemistry. 2013;56:2110–2124. doi: 10.1021/jm3018332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yost JM, Korboukh I, Liu F, Gao C, Jin J. Current Chemical Genomics. 2011;5:72–84. doi: 10.2174/1875397301005010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, Lis JT, Allis CD, Reinberg D. Molecular cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 37.Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Simon JA, Zhang Y. Current biology : CB. 2002;12:1086–1099. doi: 10.1016/s0960-9822(02)00924-7. [DOI] [PubMed] [Google Scholar]
- 38.Beck DB, Oda H, Shen SS, Reinberg D. Genes & development. 2012;26:325–337. doi: 10.1101/gad.177444.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang F, Sun L, Li Q, Han X, Lei L, Zhang H, Shang Y. The EMBO journal. 2012;31:110–123. doi: 10.1038/emboj.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, Dutta S, Appella E, Gozani O. Molecular cell. 2007;27:636–646. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takawa M, Cho H-S, Hayami S, Toyokawa G, Kogure M, Yamane Y, Iwai Y, Maejima K, Ueda K, Masuda A, Dohmae N, Field HI, Tsunoda T, Kobayashi T, Akasu T, Sugiyama M, Ohnuma S-i, Atomi Y, Ponder BAJ, Nakamura Y, Hamamoto R. Cancer Research. 2012;72:3217–3227. doi: 10.1158/0008-5472.CAN-11-3701. [DOI] [PubMed] [Google Scholar]
- 42.Ma A, Yu W, Li F, Bleich RM, Herold JM, Butler KV, Norris JL, Korboukh V, Tripathy A, Janzen WP, Arrowsmith CH, Frye SV, Vedadi M, Brown PJ, Jin J. Journal of medicinal chemistry. 2014;57:6822–6833. doi: 10.1021/jm500871s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolfe JP, Buchwald SL. Organic Syntheses. 2004;10:423–430. [Google Scholar]
- 44.Miyaura N, Suzuki A. Chemical Reviews. 1995;95:2457–2483. [Google Scholar]
- 45.Blum G, Ibáñez G, Rao X, Shum D, Radu C, Djaballah H, Rice JC, Luo M. ACS chemical biology. 2014 doi: 10.1021/cb500515r. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.