Abstract
Monitoring of hepatitis B virus (HBV) DNA in serum by molecular methods has become the standard for assessment of the replicative activity of HBV. Several molecular assays for the detection and quantification of HBV DNA have been described. However, they usually lack automated sample preparation. Moreover, those assays, which are based on PCR, are limited by a short dynamic range (2 to 3 log units). In the present study, the use of RealArt HBV LC PCR Reagents in conjunction with automated extraction on the COBAS AMPLIPREP analyzer was evaluated. Members of an HBV proficiency program panel were tested; linearity, interassay, and intra-assay variations were determined. The performance of the assay in a routine clinical laboratory was evaluated with a total of 117 clinical specimens. When members of the HBV proficiency program panel were tested by the new molecular assay, the results were found to be within ±0.5 log unit of the results obtained by reference laboratories. Determination of linearity resulted in a quasilinear curve over more than 6 log units. The interassay variation of the RealArt HBV LC PCR Reagents by use of the automated sample preparation protocol ranged from 16 to 73%, and the intra-assay variation ranged from 9 to 40%. When clinical samples were tested by the new assay with the automated sample preparation protocol and the results were compared with those obtained by the COBAS AMPLICOR HBV MONITOR Test with manual sample preparation, the results for 76% of all samples with positive results by both tests were found to be within ±0.5 log unit and the results for another 18% were found to be within between 0.5 and 1.0 log unit. In conclusion, the real-time PCR assay with automated sample preparation proved to be suitable for the routine molecular laboratory and required less hands-on time.
Assays for quantification of hepatitis B virus (HBV) DNA in serum have been shown to be useful for pretreatment evaluation, clinical staging, monitoring of antiviral therapy, detection of the emergence of drug resistance, and detection of relapses after the discontinuation of antiviral therapy (1, 5).
Several commercially available assays for quantification of HBV DNA have been brought onto the market and have been found to be useful for the routine diagnostic laboratory (2, 6, 8, 9). Recently, real-time PCR has been introduced. This new technique combines amplification and detection of amplification products in the same closed vessel, thus reducing the analytical turnaround time as well as the risk of contamination (3, 7). To improve assay performance, ready-to-use reagents have been introduced (RealArt HBV LC PCR Reagents; Artus GmbH, Hamburg, Germany). They have been optimized for use on the LightCycler instrument (Roche Applied Science, Penzberg, Germany).
For automated sample preparation, a new automated device, the COBAS AMPLIPREP analyzer (Roche Molecular Systems Inc., Branchburg, N.J.), has recently been developed (4, 10). Sample preparation protocols with this instrument were initially dependent on the use of specific capture probes, thus limiting applications to extraction of hepatitis C virus RNA and human immunodeficiency virus RNA. Recently, a new kit for preparation of total nucleic acids from serum and plasma has been introduced. Following extraction, eluted nucleic acids can be used with any nucleic acid amplification technology. Furthermore, the sample preparation protocol allows addition of an internal control or quantitative standard into each sample. In this way the assay can compensate for loss of the target and the presence of trace amounts of potential inhibitors in the consecutive amplification step.
In the present study, we established a new molecular assay for quantitative detection of HBV DNA in human serum. The assay consisted of automated sample preparation on the COBAS AMPLIPREP instrument and real-time PCR with the RealArt HBV LC Reagents. Members of an HBV proficiency program panel were tested; linearity was determined with a dilution series of a high-titer sample. Both interassay and intra-assay variations were analyzed. The clinical performance of the new assay in the routine diagnostic laboratory was evaluated with routine clinical samples, and the results were compared with those obtained by the COBAS AMPLICOR HBV MONITOR Test (Roche).
MATERIALS AND METHODS
Molecular assays.
Tests with the COBAS AMPLIPREP Total Nucleic Acid Isolation kit (Roche) for isolation of nucleic acids, tests with the RealArt HBV LC PCR Reagents (Artus), and the COBAS AMPLICOR HBV MONITOR Test (Roche) were performed according to the instructions of the manufacturers.
Automated sample preparation protocol.
For automated sample preparation, isolation of HBV DNA was done with the COBAS AMPLIPREP Total Nucleic Acid Isolation kit (Roche) on the COBAS AMPLIPREP analyzer. A processing volume of 200 μl was chosen because of the limited sample amounts available in the routine clinical laboratory; i.e., a minimum of 300 μl of serum was added to bar code-labeled tubes. Prior to the start of DNA extraction, 1,800 μl of an internal control provided by Artus was added to 360 μl of an internal control-quantitation standard diluent. This volume was sufficient for extraction of 24 samples. Adequate amounts of this mixture were automatically introduced into each sample by the instrument. Purified nucleic acids were eluted at 80°C with 75 μl of specimen diluent.
Manual sample preparation protocol.
Manual isolation of HBV DNA was done according to the instructions of the manufacturer provided in the COBAS AMPLICOR HBV MONITOR Test package insert.
Amplification and detection by the new molecular assay.
The RealArt HBV LC PCR Reagents constitute a ready-to use system for PCR amplification and the detection of HBV DNA on the LightCycler instrument. The RealArt HBV LC PCR Reagents master mixture contains reagents and enzymes for amplification of a 120-bp region of the HBV genome and for parallel detection of the specific amplification products. In addition, the RealArt HBV LC PCR Reagents contain a heterologous internal control for identification of possible PCR inhibition. For quantitation, plasmids constructed and linearized in vitro are used as standards. The standards are included in the kit and calibrated against an original stock solution of the World Health Organization International Standard for HBV DNA (http://www.nibsc.ac.uk/catalog/standards/ifu/97-746ifu.pdf). Five standard samples diluted 10-fold were used to generate the standard curve.
Study design.
All experiments were done in an International Standards Organization (ISO9001, 2000)-certified laboratory, the Molecular Diagnostics Laboratory, Institute of Hygiene.
In the first step, samples from the members of the Quality Control for Molecular Diagnostics Hepatitis B Virus Proficiency Program Panel 2002 (http://www.qcmd.org) were tested with the RealArt HBV LC PCR Reagents by using the automated sample preparation protocol of the COBAS AMPLIPREP Total Nucleic Acid Isolation kit. The samples in this panel contained different concentrations of HBV types A and D.
In the second step, the linearity of the RealArt HBV LC PCR Reagents was determined by use of the automated sample preparation protocol. A routine clinical serum sample, which contained 1.6 × 108 HBV DNA copies/ml, as determined by the COBAS AMPLICOR HBV MONITOR Test, was taken. A dilution series (0.5 log steps; i.e., 1:3.16 dilutions) was prepared by using HBV-negative human serum. Each dilution was analyzed three times, and the mean HBV DNA titer of each sample was determined.
In the third step, the interassay variation of the RealArt HBV LC PCR Reagents with the automated sample preparation protocol on the COBAS AMPLIPREP instrument was determined. Eight samples, which contained different amounts of HBV DNA ranging from 2.5 × 102 to 1.5 × 108 IU/ml, were aliquoted and tested one time on each of five different days.
The intra-assay variation of the new assay was determined in the fourth step. Four routine clinical samples with different amounts of HBV DNA ranging from 2.8 × 102 to 1.2 × 107 IU/ml were analyzed five times in one run.
In the fifth step, the performance of the RealArt HBV LC PCR Reagents with the automated sample preparation protocol on the COBAS AMPLIPREP instrument was evaluated in a routine diagnostic laboratory. A total of 117 clinical serum samples were tested. Another aliquot of each of the samples had been tested earlier by the COBAS AMPLICOR HBV MONITOR Test in the same routine diagnostic laboratory.
RESULTS
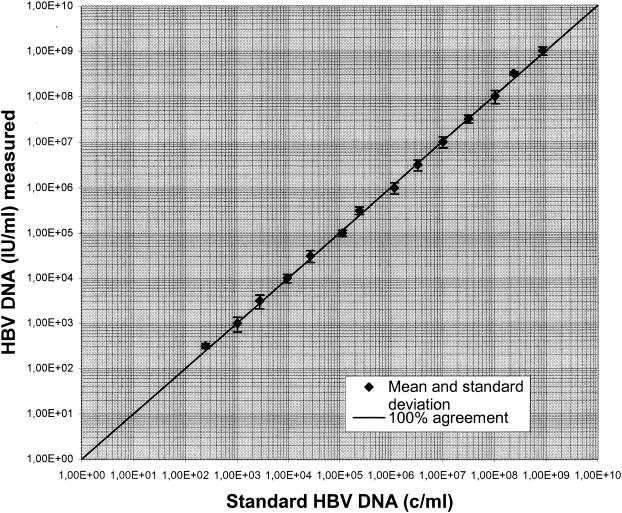
When the samples from among the members of an HBV proficiency program panel were tested, the results were found to be within ±0.5 log unit of those obtained by reference laboratories (Table 1). Linearity was tested with a dilution series of a high-titer routine clinical sample. A quasilinear curve was observed with up to 1.0 × 109 IU of HBV DNA/ml. HBV DNA was inconsistently detected in dilutions containing less than 2.5 × 102 IU of HBV DNA/ml (Fig. 1).
TABLE 1.
Results obtained by the new molecular assay in comparison with those obtained by reference laboratories with samples from the Quality Control for Molecular Diagnostics Hepatitis B Virus Proficiency Program Panel 2002a
| Sample no. | Results obtained by the new molecular assay (IU of HBV DNA/ml) | Results obtained by reference laboratories (no. of HBV DNA copies/ml) | Log unit difference |
|---|---|---|---|
| 1 | 1.6 × 106 | 2.7 × 106 | 0.23 |
| 2 | 1.5 × 104 | 3.3 × 104 | 0.34 |
| 3 | 2.3 × 105 | 1.8 × 105 | −0.12 |
| 4 | 6.0 × 104 | 1.8 × 105 | 0.46 |
| 5 | 2.4 × 104 | 3.9 × 104 | 0.21 |
| 6 | 1.1 × 104 | 3.3 × 104 | 0.48 |
| 7 | 8.8 × 102 | 9.0 × 102 | 0.01 |
| 8 | Negative | Negative | Negative |
One replicate of each sample was tested.
FIG. 1.
Linearity of the results for a 0.5-log-unit dilution series of a high-titer routine clinical serum obtained by the new molecular assay. c, number of HBV DNA copies.
For determination of interassay variation, the mean serum HBV DNA titers ranged from 2.5 × 102 to 1.5 × 108 IU of HBV DNA/ml when samples were tested one time on each of 5 days. Coefficients of variation were found to be between 16 and 73% (Table 2). Intra-assay variation was determined by testing four routine clinical samples with titers ranging from 2.8 × 102 to 1.2 × 107 IU/ml five times in one run. Coefficients of variation were found to be between 9 and 40% (Table 3).
TABLE 2.
Results of interassay testinga
| Sample no. | IU of HBV DNA/ml detected
|
Coefficient of variation (%) | |
|---|---|---|---|
| Mean | Standard deviation | ||
| 1 | 2.5 × 102 | 9.5 × 101 | 38 |
| 2 | 8.1 × 103 | 3.6 × 103 | 45 |
| 3 | 1.4 × 104 | 4.4 × 103 | 32 |
| 4 | 1.7 × 104 | 4.6 × 103 | 27 |
| 5 | 8.8 × 105 | 1.4 × 105 | 16 |
| 6 | 6.7 × 106 | 1.5 × 106 | 22 |
| 7 | 1.5 × 107 | 1.1 × 107 | 73 |
| 8 | 1.5 × 108 | 6.8 × 107 | 47 |
One replicate of each sample was tested on each of 5 days.
TABLE 3.
Results of intra-assay testinga
| Sample no. | IU of HBV DNA/ml detected
|
Coefficient of variation (%) | |
|---|---|---|---|
| Mean | Standard deviation | ||
| 1 | 2.8 × 102 | 8.5 × 101 | 31 |
| 2 | 3.2 × 105 | 1.3 × 105 | 40 |
| 3 | 1.8 × 106 | 3.7 × 105 | 21 |
| 5 | 1.2 × 107 | 1.1 × 106 | 9 |
Five replicates were tested in one run.
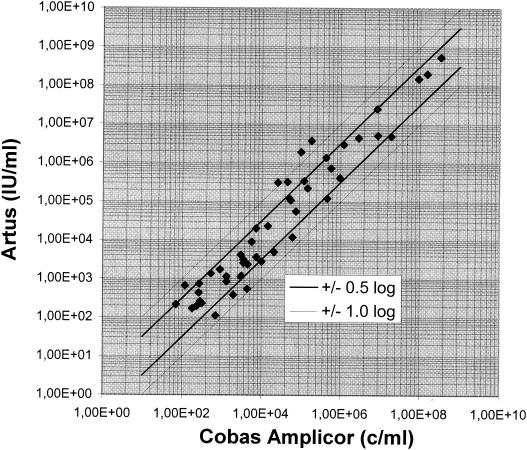
When 117 clinical samples were tested with the RealArt HBV LC PCR Reagents and the automated sample preparation protocol, all samples with positive results (n = 49) were also found to be positive by the COBAS AMPLICOR HBV MONITOR Test, and the results for 37 samples were found to be within ±0.5 log unit by both tests. Among the remaining 12 samples, the results for 9 samples were found to be within 0.5 and 1.0 log unit and the results for the other 3 samples were found to be within 1.0 and 1.3 log units (Fig. 2). Among the 68 negative samples, the result for 1 sample was found to be invalid due to a low internal standard rate with the RealArt HBV LC PCR Reagents and the automated sample preparation protocol.
FIG. 2.
Correlation between viral load measurements for 49 HBV DNA-positive serum samples by the new molecular assay (with the results expressed in international units per milliliter) and the COBAS AMPLICOR HBV MONITOR Test (with the results expressed as the number of copies [c] per milliliter).
During the whole study, quantification standards, which are included in the RealArt HBV LC PCR Reagents kit, were diluted 1:10 in negative serum, extracted, and used as positive controls. The results for all of them were found to be within ±0.5 log unit of the value expected for the diluted standard.
Automated HBV DNA extraction with the COBAS AMPLIPREP instrument was completed within 150 min for extraction of 24 samples. This included 10 min to set up the COBAS AMPLIPREP instrument. After preparation of the master mixture, aliquoting into capillaries, and addition of extracted samples, amplification and detection on the LightCycler instrument were completed within 1 h.
DISCUSSION
Several commercially available assays for quantification of HBV DNA have been brought onto the market and have been found to be useful for the routine diagnostic laboratory (2, 6, 8, 9). Those assays, which are based on PCR, show very limited dynamic ranges (2 to 3 log units). Therefore, a large number of routine samples contain HBV DNA in amounts that exceed the upper limit of detection and must be diluted and retested, which leads to unnecessary additional costs. In contrast to conventional PCR, real-time PCR shows a more extended range. The RealArt HBV LC PCR Reagents kit is a ready-to-use system for quantification of HBV DNA by real-time PCR on the LightCycler instrument. In addition, the kit includes a second heterologous amplification system that identifies possible PCR inhibition. In the present study, we established a new molecular assay for the quantitative detection of HBV DNA in human serum which consisted of automated sample preparation on the COBAS AMPLIPREP instrument and real-time PCR with the RealArt HBV LC Reagents.
At present, sample preparation is considered the major weakness in molecular assays. The COBAS AMPLIPREP instrument uses sample volumes between 100 and 850 μl. According to the limited sample volumes available in the routine diagnostic laboratory, an input volume of 200 μl was chosen. Prior to the start of the fully automated extraction procedure, an adequate amount of internal standard was manually introduced into the reagents cassette. Amplification may fail because of interference from PCR inhibitors. Therefore, it is obligatory to incorporate an internal control into every molecular assay used in the routine diagnostic laboratory. By the new molecular assay, the internal control could be recovered from 67 of 68 HBV DNA-negative clinical samples.
Members of an HBV proficiency program panel were tested by the new molecular assay. The results that reference laboratories obtain with the members of the panel are expressed as the number of HBV DNA copies per milliliter. Directive 98/79/EC of the European Parliament and of the European Council (http://europa.eu.int/documents/eur-lex/index_en.htm) requires that the manufacturers of new diagnostic molecular assays use an international standard for quantitation or standards calibrated against the international standard. Therefore, the results obtained by the new assay are expressed as international units of HBV DNA per milliliter. At present, however, no general conversion factor is available, and there is thus a need for clarification. Despite the different units (number of HBV DNA copies per milliliter versus international units of HBV DNA per milliliter), all results were found to be within ±0.5 log unit of the results obtained by reference laboratories, which may indicate close correlation between the two different sets of units.
The linear range of the RealArt HBV LC PCR Reagents with the automated sample preparation protocol was determined by analysis of dilutions of a serum sample with a high titer of HBV DNA. The new molecular assay revealed sufficient linearity up to 1.0 × 109 IU/ml. In contrast to the COBAS AMPLICOR HBV MONITOR Test, accurate results for samples with HBV DNA levels above 2.0 × 105 IU/ml could be obtained without preparation of appropriate dilutions prior to sample preparation.
In this study, the interassay variation of the RealArt HBV LC PCR Reagents with the automated sample preparation protocol ranged from 16 to 73% and the intra-assay variation ranged from 9 to 40%. These results are in concordance with those reported for other molecular assays based on PCR amplification (6).
When the RealArt HBV LC PCR Reagents with the automated sample preparation protocol were evaluated with clinical samples in a routine diagnostic laboratory and the results were compared with those obtained by the COBAS AMPLICOR HBV MONITOR Test, the results for 37 of 49 samples with positive results were found to be within ±0.5 log unit. The results for 9 of the remaining 12 samples were found to be between 0.5 and 1.0 log unit, and those for the other 3 samples were found to be within between 1.0 and 1.3 log units. Again, this shows the good correlation between the results expressed as international units of HBV DNA per milliliter obtained with the RealArt HBV LC PCR Reagents and those expressed as the number of HBV DNA copies per milliliter obtained by the COBAS AMPLICOR HBV MONITOR Test.
The new assay with automated sample preparation showed good overall functionality and user-friendliness. The automated sample preparation protocol saved hands-on time. It must, however, be taken into consideration that aliquots of the extracted samples must be pipetted manually into the capillaries, which contain the master mixtures. Nevertheless, because of the significantly lower number of manipulations required, the probability that false-positive results will be obtained because of contamination may be less.
In conclusion, the new assay with automated nucleic acid purification on the COBAS AMPLIPREP instrument proved to be a step forward in meeting the requirements of the routine diagnostic laboratory. Compared to conventional assays, the new assay took less time to complete and required less hands-on work.
Acknowledgments
This work was supported in part by a grant from the Austrian-Hungarian Scientific and Education Cooperation Action Program and Artus GmbH.
REFERENCES
- 1.Berger, A, W. Preiser, and H. W. Doerr. 2001. The role of viral load determination for the management of human immunodeficiency virus, hepatitis B virus and hepatitis C virus infection. J. Clin. Virol. 20:23-30. [DOI] [PubMed] [Google Scholar]
- 2.Hendricks, D. A., B. J. Stowe, B. S. Hoo, J. Kolberg, B. D. Irvine, P. D. Neuwald, M. S. Urdea, and R. P. Perrillo. 1995. Quantitation of HBV DNA in human serum using a branched DNA (bDNA) signal amplification assay. Am. J. Clin. Pathol. 104:537-546. [DOI] [PubMed] [Google Scholar]
- 3.Ho, S. K., W. C. Yam, E. T. Leung, L. P. Wong, J. K. Leung, K. N. Lai, and T. M. Chan. 2003. Rapid quantification of hepatitis B virus DNA by real-time PCR using fluorescent hybridization probes. J. Med. Microbiol. 52:397-402. [DOI] [PubMed] [Google Scholar]
- 4.Jungkind, D. 2001. Automation of laboratory testing for infectious diseases using the polymerase chain reaction—our past, our present, our future. J. Clin. Virol. 20:1-6. [DOI] [PubMed] [Google Scholar]
- 5.Kessler, H. H., S. Preininger, E. Stelzl, E. Daghofer, B. I. Santner, E. Marth, H. Lackner, and R. E. Stauber. 2000. Identification of different states of hepatitis B virus infection with a quantitative PCR assay. Clin. Diagn. Lab. Immunol. 7:298-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kessler, H. H., E. Stelzl, E. Daghofer, B. I. Santner, E. Marth, H. Lackner, and R. E. Stauber. 2000. Semiautomated quantification of hepatitis B virus DNA in a routine diagnostic laboratory. Clin. Diagn. Lab. Immunol. 7:853-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohmoto, M., M. Enomoto, Y. Yano, S. Otani, S. Minamitani, A. Tamori, D. Habu, T. Takeda, S. Shiomi, and S. Seki. 2003. Detection of serum hepatitis B virus DNA by real-time quantitative polymerase chain reaction (TaqMan PCR) during lamivudine treatment: comparison with three other assays. Hepatol. Res. 26:125-133. [DOI] [PubMed] [Google Scholar]
- 8.Marin, I. J., M. Poljak, K. Seme, J. Meglic-Volkar, M. Maticic, G. Lesnicar, and V. Brinovec. 2001. Comparative evaluation of semiautomated COBAS AMPLICOR hepatitis B virus (HBV) MONITOR test and manual microwell plate-based AMPLICOR HBV MONITOR test. J. Clin. Microbiol. 39:758-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noborg, U., A. Gusdal, E. K. Pisa, A. Hedrum, and M. Lindh. 1999. Automated quantitative analysis of hepatitis B virus DNA by using the Cobas Amplicor HBV Monitor test. J. Clin. Microbiol. 37:2793-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stelzl, E., A. Kormann-Klement, J. Haas, E. Daghofer, B. I. Santner, E. Marth, and H. H. Kessler. 2002. Evaluation of an automated sample preparation protocol for quantitative detection of hepatitis C virus RNA. J. Clin. Microbiol. 40:1447-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]