Abstract
Messenger RNA plays a pivotal role in regulating cellular activities. The expression dynamics of specific mRNA contains substantial information on the intracellular milieu. Unlike the imaging of stationary mRNAs, real-time intracellular imaging of the dynamics of mRNA expression is of great value for investigating mRNA biology and exploring specific cellular cascades. In addition to advanced imaging methods, timely extracellular stimulation is another key factor in regulating the mRNA expression repertoire. The integration of effective stimulation and imaging into a single robust system would significantly improve stimulation efficiency and imaging accuracy, producing fewer unwanted artifacts. In this study, we developed a multifunctional nanocomplex to enable self-activating and spatiotemporal imaging of the dynamics of mRNA sequential expression during the neural stem cell differentiation process. This nanocomplex showed improved enzymatic stability, fast recognition kinetics, and high specificity. With a mechanism regulated by endogenous cell machinery, this nanocomplex realized the successive stimulating motif release and the dynamic imaging of chronological mRNA expression during neural stem cell differentiation without the use of transgenetic manipulation. The dynamic imaging montage of mRNA expression ultimately facilitated genetic heterogeneity analysis. In vivo lateral ventricle injection of this nanocomplex enabled endogenous neural stem cell activation and labeling at their specific differentiation stages. This nanocomplex is highly amenable as an alternative tool to explore the dynamics of intricate mRNA activities in various physiological and pathological conditions.
Keywords: mRNA, imaging, gold nanoparticle, drug delivery, neural stem cell
Messenger RNA (mRNA) is one of the key factors that regulate cellular functions and is actively involved in numerous activities in the intracellular milieu.1 The dynamic alteration of specific mRNA at a particular time or location in cells usually implies significant changes of cellular condition.2 Aberrant mRNA dynamics is associated with developmental and pathological abnormalities, such as interrupted embryonic development3 and cellular apoptosis.4 The dynamics of mRNA expression is a blueprint of cancer progress from benign to malignant stages.5 The mRNA expression signature of a given neuron reflects its developmental history, activity, and interaction with other cells and the environment.6 Therefore, intracellular imaging of the dynamics of mRNA expression is of great value for better investigating mRNA biology and exploring specific cellular cascades.
Real-time imaging of mRNA dynamics has recently drawn much attention,7,8 as mRNAs have intricate dynamics,9 which is evidenced by the delicately timed and spatially controlled processes in cells. The understanding of intracellular mRNA generation, progression, and mutual interaction was mainly dependent either on fluorescence in situ hybridization (FISH) of fixed cells or on biochemical separation of subcellular components followed by PCR amplification. Those methods, however, present only the static situation of cells at the terminating time point, when samples were prepared.9 In contrast, live cell dynamic imaging provides spatiotemporal mRNA profiling, which reveals extensive information on cell progress.
Recently, several methods have been developed for live cell mRNA imaging, such as molecular beacons10−15 and nanoflares.16,17 Some probes enabled concurrent imaging of multiple mRNAs in living cells to improve detection accuracy.16,18,19 Various real-time mRNA imaging probes have been developed,19,20 most of which, however, are capable of imaging only the stationary mRNA expression at a single time point in living cells. In addition, those imaging probes were designed to reveal the prevalence of pre-existing mRNAs in cells,20 while the pivotal dynamics of mRNA sequential expression during specific cellular events, such as differentiation or apoptosis, remains largely elusive. Moreover, mRNA dynamic expression is usually initiated by extracellular stimuli.21,22 Such environmental cues usually result in rather low stimulation efficiency and a short half-life in media unless used in high concentration, which adversely exerts unexpected cytotoxicity.23 In addition, the challenge of precise time coordination between stimulating factor function and imaging probe introduction may act as another barrier for real-time mRNA dynamic imaging.
To achieve the goal of dynamic imaging of mRNA sequential expression, the multifunctional probe should be easily delivered into cells, stable upon entry into cells, efficiently and homogeneously distributed in the cytosol, and specific and sensitive to its target. Ideally, it should incorporate a stimulating motif undergoing self-cleavage through endogenous cellular machinery in cells. In this study, we designed a multifunctional nanocomplex (fNC) meeting all the above criteria to enable self-activating and spatiotemporal imaging of the dynamics of mRNA sequential expression in differentiating neuron stem cells.
Results and Discussion
Design of Multifunctional Nanocomplexes
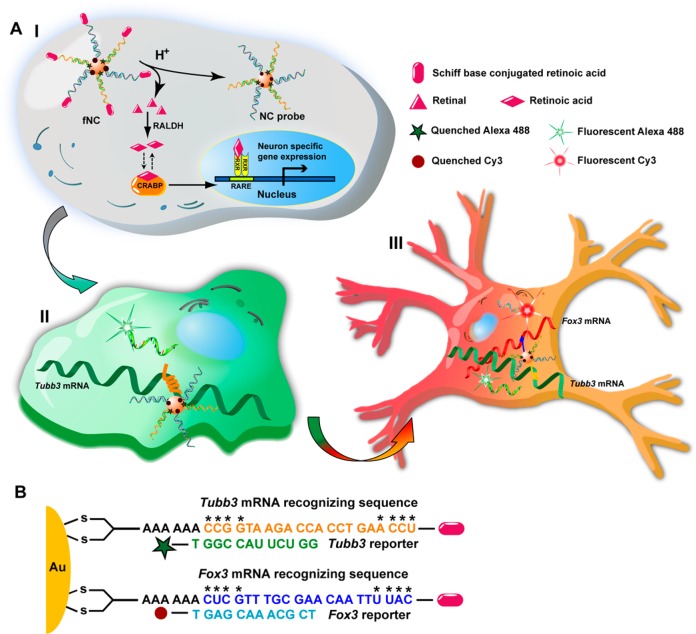
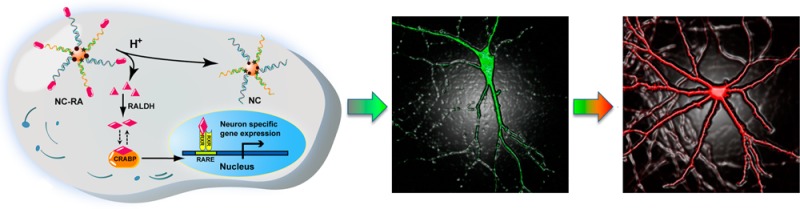
The nanocomplex consists of a cellular machinery initiated system for successive gene expression stimulation and imaging. To stimulate gene expression, nature has provided us an instrumental guidance: in cells, the metabolism of a visual pigment, in which a small molecule binds to a pocket protein, is reversibly regulated by balanced Schiff base formation and hydrolysis through endogenous subcellular machinery.24 Inspired by this well-established, naturally occurring mechanism, we designed a nanocomplex with the release of a stimulating motif controlled by Schiff base formation and hydrolysis. To construct the mRNA imaging probe, we used gold nanoparticles (AuNPs), a widely used potent delivery vehicle,25,26 as the core functionalized with a dense layer of dithiol group terminated recognizing DNA oligonucleotides hybridized with different fluorophore-capped reporter sequences. The fluorophores were initially quenched by a wide-spectrum AuNP nanoquencher (Figure 1A). Unlike molecular beacons, which require transfection agents for delivery and are susceptible to enzyme degradation, oligo-modified AuNPs enable self-delivery and have high stability to resist enzyme cleavage.27 The termini of recognizing oligonucleotides of AuNP are tethered with stimulating motifs via the labile Schiff base to enable self-activation of the fNC inside cells (Figure 1A and B). The fNC is stable at neutral pH, whereas upon cellular uptake, the Schiff base (imine) linkage is hydrolyzed to yield small molecules (retinals) and oligo-modified AuNP imaging probes. In mammalian cells, an enzyme, aldehyde dehydrogenase (RALDH), is responsible for efficient conversion of retinal into retinoic acid (RA), which specifically binds with cellular RA binding protein (CRABP) to activate the downstream gene transcription process for neuron-specific gene expression (Figure 1A).28 Briefly, the low-pH microenvironment triggered the Schiff base conjugated retinal release from gold nanoparticles, and the released retinal was converted into RA by RALDH. The biologically active RA ultimately bound with CRABP to initiate neural stem cell differentiation. During the subsequent cell progression period, the mRNA targets will be chronologically expressed in the cytosol, where they will compete with the fluorophore-tagged reporter sequences and hybridize with recognizing oligonucleotides on AuNP by forming long and stable duplexes. This sequence-specific competitive binding causes the release of a reporter sequence from fNC to generate fluorescent signal proportional to the mRNA target expression level (Figure 1A). The fNC has high colloidal stability in the cytosol,26 endonuclease resistance,27 and stimulating motif self-cleavage capacity. Importantly, this nanocomplex system demonstrates, to the best of our knowledge, the first example of a dynamic imaging study of mRNA sequential expression using robust nanomaterials without transgenetic manipulation.
Figure 1.
(A) Schematic illustration of a multifunctional nanocomplex (fNC) as an integrated system for tandem activation and imaging of the dynamics of mRNA sequential expression during the process of neural stem cell differentiation. Upon cellular uptake of the nanocomplex, the release of stimulating motif (RA) was controlled by Schiff base formation and hydrolysis through an endogenous mechinary. The released RA efficiently activated the subsequent signaling pathway to stimulate specific gene expression. In the meanwhile, the NC probe underwent sequence-specific recognition and spatiotemporal imaging of sequential mRNA expression during the process of neural stem cell differentiation. RALDH: retinaldehyde dehydrogenase; CRABP: cellular RA binding protein; RARE: RA responsive element; RXR: retinoid X receptor. (B) Design of the recognizing and reporter oligonucleotides for Tubb3 and Fox 3 mRNA imaging. Oligonucleotides marked with * indicate methylation.
Preparation of Nanocomplexes with Multifunctions
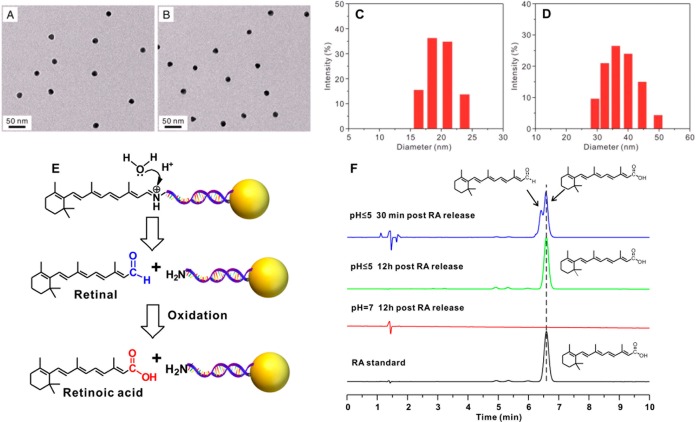
To demonstrate the function and efficacy of this fNC for self-activating and imaging the dynamics of mRNA expression, we applied it, as a proof of concept, to image the dynamics of mRNA sequential expression during neural stem cell (NSC) differentiation (Figure 1B). The differentiation of NSCs plays a central role not only in the developing mammalian nervous system but also in the adult nervous system of all mammalian organisms.29 Transplanted NSCs are capable of ameliorating damage or pathological disorders by their differentiation into neural subtypes in the central nervous system.30 During NSC differentiation stimulated by an external modulator,21 the genetic repertoire undergoes remarkable changes, where genes are chronologically regulated at specific developmental stages.31 In addition, the proliferation of NSCs undergoing differentiation is in a relatively static condition, which facilitates imaging of the differentiation process in a fairly restricted environment without interference from the potential cellular division.21 Thus, real-time spatiotemporal imaging of the dynamics of mRNA sequential expression can provide insightful information on the biochemical basics of stem cell biology. To visualize the dynamics of mRNA expression by fNCs, we chose two representative neuron-specific mRNA pairs, Tubb3 and Fox3, which are sequentially expressed during NSC differentiation from progenitor cells to neurons. Tubb3 has been suggested to be one of the earliest markers of signal neuronal commitment,32 while Fox3 shows dominance in neurons at a later stage.33 Retinoic acid, commonly used to differentiate stem cell populations into neurons,22 was chosen as the stimulating motif and incorporated in the nanocomplexes. Since mRNA has secondary and tertiary structures, the designed DNA should be complementary to the outward folding side of an mRNA, which is ready to be approached by other molecules. In addition, the reporter DNA oligos are relatively shorter than the entire complementary sequence of recognizing DNA. This design leads to a lower Tm value and thermodynamic energy, so that the reporter DNA is likely to be replaced by the complementary sequence of the target mRNA, leading to more stable hybridization. The recognizing DNA was terminated at the 3′ end by an alkanedithiol group to facilitate highly stable anchoring on the AuNP surface, and the 5′ end was flanked with primary amine to react with aldehyde from a retinoic acid analogue, retinal, to form a pH-labile Schiff base.34 The DNA sequence consists of a 6-base-pair (bp) adenine (A6) spacer and a 21-bp fragment complementary to the corresponding nucleotide sequence (Tubb3:228–248; Fox3:1349–1369) in mRNAs. The recognizing DNA was additionally methylated on selective nucleotides to enhance stability and binding affinity. The reporter sequence with a 5′ end fluorophore tag (Tubb3: Alexa 488; Fox3: Cy3) (Table S1) is complementary to the 12 bp of the corresponding recognizing sequence. The 15 nm AuNP was used as the core. The multifunctional nanocomplex is monodispersed (Figure 2A and B) and highly stable (Figure 2C and D). The UV/vis spectrum of fNC shows a 4 nm red shift of the surface plasmon resonance (SPR) band compared with that of naked AuNPs (Figure S1). This suggests the change of the dielectric constant of the surrounding environment of the AuNP after dense oligonucleotide functionalization. In contrast, the width of the SPR remains unchanged, indicating the monodispersed condition of the fNC. This result is consistent with the TEM imaging (Figure 2A and B). Dynamic light scattering (DLS) measurements (Figure 2C and D) show the colloidal fNC with a diameter of 38 ± 1.6 nm, and the size of the original AuNP was 19 ± 0.6 nm, along with particle surface charge shifting from −15 ± 1.8 mV to −27 ± 2.1 mV, confirming the successful modification with DNA oligonucleotides on the AuNP surface. Retinal conjugation did not dramatically alter the surface charge (−24 ± 2.0 mV). The dense layer of methylated ssDNA oligonucleotides provides additional stability for AuNPs. On average, the fNC was calculated to carry 22 ± 1 Tubb3-recognizing DNA oligonucleotides tagged with Alexa 488 and 23 ± 2 Fox3-recognizing DNA oligonucleotides tagged with Cy3 (Figure S2A and B). The RA moieties tethered on the termini of oligonucleotides were 35 ± 3 molecules in each particle estimated by the HPLC method (Figure S2C). The calculated quenching efficiency of the gold nanoparticles was 92% for Alexa 488 and 87% for Cy3 (Figure S3).
Figure 2.
Nanocomplex morphology characterization and retinoic acid release. TEM images of naked gold nanoparticles (A) and multifunctional nanocomplexes (B). Dynamic light scattering (DLS) analysis: (C) naked gold nanoparticles in pure water; (D) multifunctional nanocomplexes in PBS. (E) Principle of Schiff base mediated stimulating motif (retinal) self-cleavage and oxidization under different physiological pH values. (F) HPLC chromatogram of corresponding released retinal or retinoic acid under different pH values and time points. The duplicate peak from the 30 min RA release chromatogram is attributed to the mixture of retinal and retinoic acid in solution.
Low Concentration of Stimulating Motif Released from fNC Activates NSC Differentiation
The fNC was integrated with a stimulating motif function via Schiff base. The fNC is stable at neutral pH (pH = 7) such that no free RA is detected, whereas at lower pH (pH < 5) the Schiff base is hydrolyzed into retinal, followed by oxidation into a functional RA molecule (Figure 2E and F).28 In our study, the calculated fNC attaching RA applied for NSC stimulation was about 15 nM, which is much lower than the commonly used 1 μM concentration of free RA in media. Immunostaining results confirmed the identical NSC differentiation ability by these two conditions, but 15 nM free RA dissolved in culture medium did not show observable NSC differentiation (Figure S4). Another report showed consistent results that spatially positioned and concentration-controlled RA delivery into NSCs by nanoformulation offered 2 orders of magnitude higher efficiency than free soluble RA to induce an NSC proneurogenic niche in the mouse subventricular zone.23 This indicates that the nanoparticle delivery platform23,35−37 provides additional benefits to deliver and protect RA in cells to stimulate NSC differentiation. Moreover, this fNC takes advantage of endogenous subcellular machinery to spontaneously trigger RA release for NSC differentiation and simultaneously delivers the imaging probe, which avoids the need for complicated time coordination to image the dynamics of mRNA sequential expression during stem cell differentiation.
Specificity and Stability of fNC as an Imaging Probe
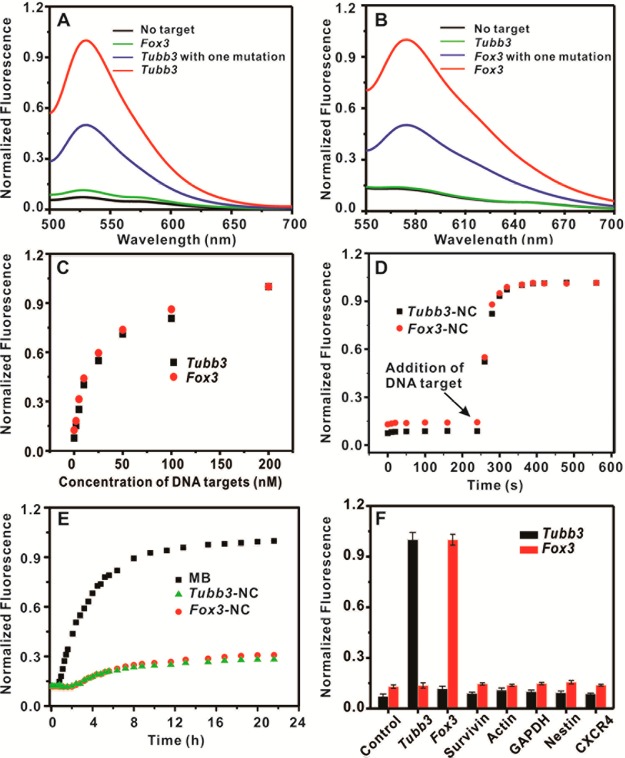
The binding specificity of fNC was first evaluated by adding a completely matched DNA target in fNC solution. To demonstrate the sequence specificity, a single mismatch (mutation) was introduced in the same DNA target. To exclude the possibility of cross-reaction between two targets on a fNC, the cross hybridization of the fNC with an individual target was also evaluated. The results showed that the fNC responded with a significant signal (over 7-fold change) in the presence of a completely matched DNA target for both Tubb3 and Fox3, while a single mutation in the DNA target resulted in 2.5 times lower signal response in both DNA targets (Figure 3A and B). This indicates that fNC is able to differentiate between the completely matched complementary target and mismatched target. In addition, there is no potential cross-reaction between two DNA targets as evidenced by signal magnitude to the background level after cross hybridization. Figure 3C shows the fluorescence intensity of fNC increases with increasing concentration of DNA target from 0 to 200 nM. This indicates that the fluorescence intensity is highly associated with the concentration of the DNA target and could represent the target sequence content for sensitive determination of gene expression level. The in vitro detection limit of fNC to Tubb3 and Fox3 was 1.5 nM and 1.2 nM, respectively. Next, we assessed the hybridization kinetics of fNC to DNA targets and the stability of fNC upon nuclease exposure. Results showed that fNC responded to the target sequence in less than 20 s (Figure 3D). This extremely short time delay should not interfere with real-time imaging data analysis in cells. Moreover, the fNC was resistant to nuclease degradation, whereas the equivalent molecular beacon, widely used for subcellular imaging,10 underwent degradation over long-term incubation with DNase I (Figure 3E). The off-target gene detection was evaluated by incubating various DNA sequences with fNCs. Figure 3F shows high specificity of fNCs to Tubb3 and Fox3 among other off-target genes. As for the stability of the nanocomplex, we introduced this dithiol modification to increase the stability. For the DNA duplex dissociation, the data shown in Figure 3C demonstrated that the nanocomplex is sensitive to spiked complementary DNA oligos, and the hybridization rate is very fast (Figure 3D). However, if mismatched DNA oligo was mixed with the nanocomplex, the nanocomplex remained stable at spiked DNA oligo concentrations as high as 200 nM (Figure 3F). Hence, the nanocomplex presented high stability in various conditions. Collectively, our results show that fNC is a robust probe for Tubb3 and Fox3 mRNA pair detection.
Figure 3.
In vitro characterization of multifunctional nanocomplexes. (A) Fluorescence spectra of fNCs in the presence of no target (black line), Tubb3 target (red line), cross-talk Fox3 target (green line), or Tubb3 target with one mismatch (blue line). (B) Fluorescence spectra of fNCs in the presence of no target (black line), Fox3 target (red line), cross-talk Tubb3 target (green line), or Fox3 target with one mismatch (blue line). (C) Dynamic response of nanocomplexes to corresponding DNA targets with various concentrations of 0, 2, 5, 10, 25, 50, 100, and 200 nM. (D) Hybridization kinetics of fNCs associated with the complementary targets. The fNCs (1 nM) were mixed with 200 nM of Tubb3 DNA target (black square) or Fox3 DNA target (red circle), respectively. (E) Enzymatic stability study of fNCs (red and green) and molecular beacons (black). fNCs (1 nM) and molecular beacons (25 nM) were treated with DNase I (50 U/L). (F) Specificity of the nanocomplex over several DNA targets. The nanocomplex was mixed with Tubb3 (black) or Fox3 (red) complementary targets and other noncomplementary targets (targets of surviving, β-actin, GAPDH, Nestin, and CXCR4) measured with a 490 nm excitation wavelength for Tubb3 and 550 nm for Fox3 (n = 3).
Cytotoxicity of fNCs to Neural Stem Cells
Next, we evaluated the possible cytotoxicity of fNCs to neural stem cells by conducting an MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay to evaluate cell proliferation. No detectable cytotoxicity to NSCs at concentrations of 0.1–1 nM over 48 h incubation was found (Figure S5), suggesting that fNCs are biocompatible for imaging of intracellular NSC gene expression.
Evaluation of fNCs as mRNA Imaging Probes in Highly Differentiated NSCs
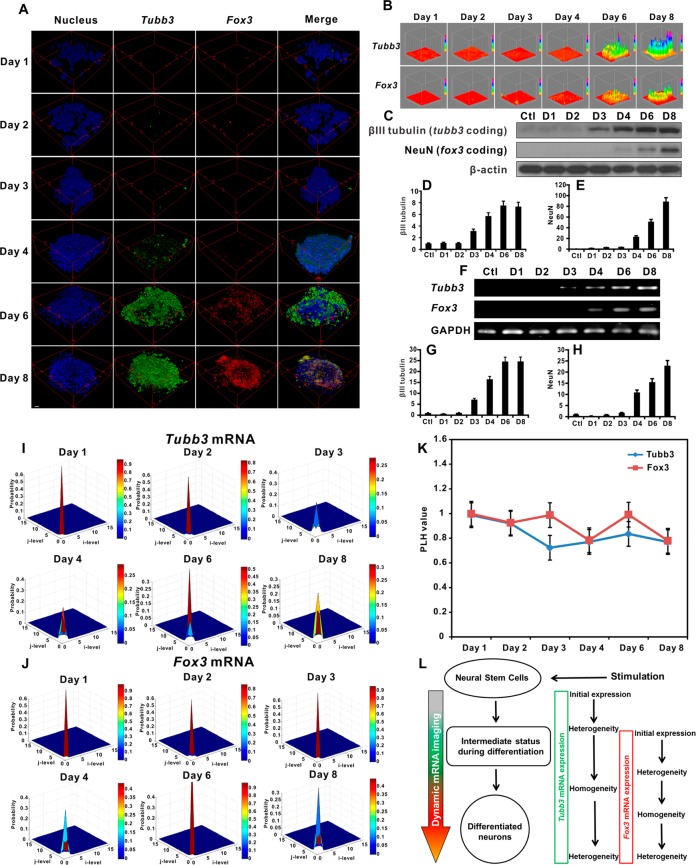
We then investigated the fNC’s ability to detect intracellular mRNA in differentiated NSCs. NSCs were first stimulated by free RA (1 μM) to differentiate into neural subtype cells over 8 days before incubation with fNCs. Figure 4A shows the intracellular Tubb3 mRNA (green) and Fox3 mRNA (red) distribution in single cells detected by fNCs after 4 h incubation. At day 8 after RA stimulation, NSCs are partially differentiated into neurons, in which βIII tubulin (encoded by Tubb3) and NeuN (encoded by Fox3) proteins are coexpressed in cells.38 This is consistent with our results in Figure 4A, showing the co-localization of Tubb3 and Fox3 mRNAs in the cytoplasm. In addition to the static imaging of mRNA expression, we then studied the kinetics of fNCs on detecting Tubb3 and Fox3 mRNAs in a well-differentiated NSC colony under identical conditions. Results (Figures 4B and S6) showed dominant Tubb3 and Fox3 mRNA expression levels at 4 h incubation with fNCs, and fluorescence intensity became almost saturated after 6 h.39 In contrast, in the initial 3 h, minimal fluorescent signal could be detected, probably due to the relatively slow kinetics of fNC endocytosis and the endo/lysosome escape progress. The dark field imaging results (Figure S7) also confirmed this endocytosis progress. In addition, it was reported that the spherical DNA oligos on the gold nanoparticle surface undergo 3D structural rearrangement during cellular uptake. This spatial structure change supports the significant endocytosis and efficient intracellular release through a lipid-raft-dependent, caveolae-mediated pathway.40 The mRNA imaging observed with fNCs in differentiated NSCs validated the time-dependent trend of endocytosis of engineered gold nanoparticles in cells26 and confirmed the efficient mRNA detection by fNCs in NSCs.
Figure 4.
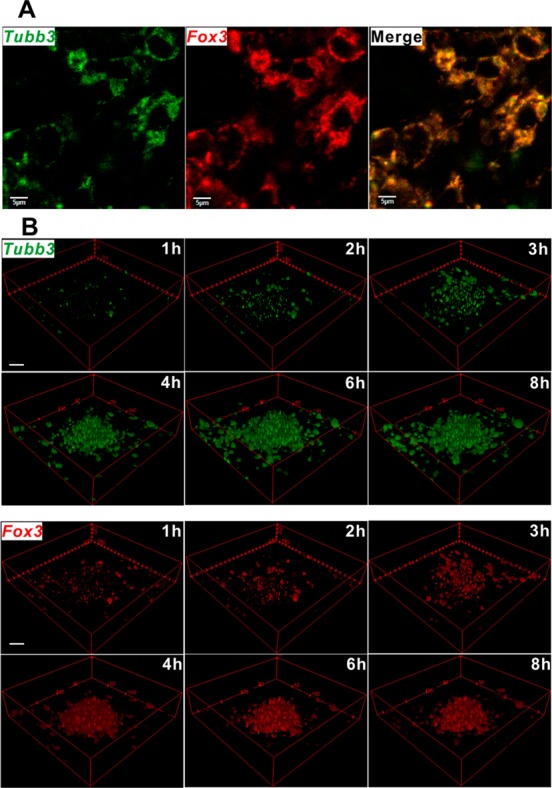
(A) Fluorescence imaging of Tubb3 (green color) and Fox3 (red color) mRNAs by fNCs at day 8 in differentiated NSCs stimulated by free RA in advance. Scale = 5 μm. (B) Fluorescence recovery kinetics of fNCs in detecting Tubb3 (green color) and Fox3 (red color) mRNAs at day 8 in differentiated NSCs stimulated by free RA in advance. The images were reconstructed into 3D format without compromising the originality for better illustration. Scale = 20 μm.
Self-Activating and Spatiotemporal Imaging Dynamics of mRNA Sequential Expression during NSC Differentiation
We applied fNCs to spatiotemporally image the dynamics of mRNA sequential expression during NSC differentiation from neural stem cells to neurons in the self-activating condition. Results in Figure 5A and B show that fNCs could efficiently stimulate NSC differentiation followed by sensitive and specific imaging of the dynamics of Tubb3 and Fox3 mRNA sequential expressions, respectively. From the imaging result, we observed that Tubb3 mRNA expression is about 2 days earlier than Fox3 mRNA expression, and both peak at day 8. This imaging result is in line with the inherent functions of Tubb3, which encodes βIII tubulin protein, an early marker of signal neuronal commitment,32 and Fox3, encoding NeuN, which is expressed in neurons.33 The imaging results were further validated by Western blots for corresponding coding protein expressions (Figure 5C–E) and PCR analysis for mRNA levels (Figure 5F–H). Our data also demonstrated the enhanced differentiation stimulation efficiency of RA delivered by nanoscale vector.23 In contrast, fNCs without the RA motif can hardly stimulate NSC differentiation, nor can they image any mRNA expression upon incubation over 8 days (Figure S8).
Figure 5.
(A) Self-activating and spatiotemporal imaging of Tubb3 and Fox3 mRNA sequential expressions during NSC differentiation in living cells. Green: Tubb3 mRNA; red: Fox3 mRNA; blue: nuclei stained with Hoechst 33342. Scale = 20 μm. (B) Hot maps of Tubb3 and Fox3 mRNA sequential expression. (C–E) Western blot analysis of Tubb3 coding βIII tubulin and Fox3 coding NeuN protein expression levels at different NSC differentiation stages. (F–H) PCR analysis of Tubb3 and Fox3 mRNA expression levels at different NSC differentiation stages. (I) Texture feature heterogeneity analysis of Tubb3 mRNA expression imaging. (J) Texture feature heterogeneity analysis of Fox3 mRNA expression imaging. (K) Photolithographic hologram (PLH) value of mRNA heterogeneity analysis of Tubb3 and Fox3 expression (n = 5). (L) Mechanistic insights of successive mRNA expression initiation and dynamic imaging during neural stem cell differentiation and associated texture feature pattern progress of the heterogeneity of mRNA expression.
Texture Feature Analysis of the Dynamics of mRNA Expression Network
Dynamic gene expression imaging provides advantages over conventional static single-point imaging to provide a continuous overview of gene expression dynamics and facilitates analysis of gene expression progress in real time. To correlate the mRNA expression pattern network, we employed a texture feature analysis method41 to investigate the gene expression pattern obtained from fNC imaging during NSC differentiation (Figure 5I,J). In the pictorial data, the texture feature represents important information on the statistic distribution of gray-scale variation in a group of defined pixels.41 In our study, the texture feature analysis is able to specifically identify the spatial relationship of a single pixel with the adjacent pixels in the two-dimensional direction and generally describes the “roughness” and distribution pattern of the gene expression condition in a single colony. For both Tubb3 and Fox3 mRNAs, the homogeneous pattern (the overall uniform gene expression level of a cell colony or cluster derived from the almost identical gene expression level of individual cells in a specific time point) on the first day is attributed to the stem-like condition of NSCs, indicating neither gene was expressed at that stage. However, Tubb3 expression became less homogeneous at day 2 due to the early onset expression of mRNA. The homogeneity increased gradually in the following 2 days, as more nascent NSCs initiated the onset of Tubb3 expression. Heterogeneous (the overall nonuniform gene expression level of a cell colony or cluster derived from the distinctive gene expression level of individual cells at a specific time point) colonial cell differentiation at day 4 and after led to the decline of Tubb3 heterogeneity (Figure 5K and L). Fox3 mRNA showed a similar dynamic expression texture pattern but delayed for 2–3 days (Figure 5K and L). These imaging and texture pattern analysis results are in high agreement with observed growth patterns during stem cell differentiation.42
In Vivo Self-Activation and Imaging of Endogenous Neural Stem Cells by Nanocomplex
We stereotactically injected the fNC directly into the lateral ventricle of the mouse brain to investigate the in vivo behavior of the nanocomplex. Figure 6 shows the parallel comparison of endogenous NSC activation and labeling effects post fNC injection. fNC without RA could not extensively activate NSC differentiation, resulting in a sporadic imaging signal distribution (Figure 6A). In contrast, fNC led to strong fluorescent signals for both Tubb3 and Fox3 mRNAs (Figure 6B). The area adjacent to the lateral ventricle showed little, if any, detectable fluorescent signal (Figure S9). This negligible fluorescent signal may come from the tissue background noise or the diffusion of fNCs taken up by mature neurons. Overall, our results clearly demonstrate that (1) the synthesized nanocomplex is capable of activating endogenous NSC by the cellular machinery triggered RA release, followed by efficient mRNA imaging; (2) the attachment of RA on the distal end of nanoparticles is critical to regulate NSC differentiation ability (Figure 6A). Importantly, the required RA concentration is much lower (15 nM) than the commonly used 1 μM to regulate NSC differentiation in animals. The in vivo experiment confirmed the high efficiency of nanocomplexes for simultaneous gene regulation and imaging. Unfortunately, we could not perform noninvasive real-time imaging in vivo due to the limitation of microscopic light penetration depth to the lateral ventricle in the brain. New techniques implanting microfibers into the deep compartment of the mouse brain would help the spatiotemporal imaging of the mRNA expression dynamics of endogenous differentiating neural stem cells in the presence of the fNCs.
Figure 6.
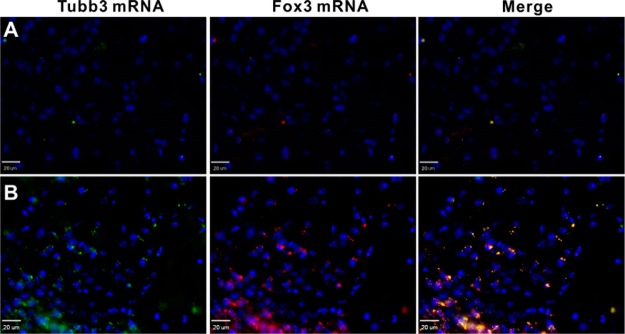
In vivo self-activation and imaging of endogenous neural stem cells by fNCs. (A) Images of nanocomplexes without RA-treated mouse brain. (B) Images of nanocomplexes with treated mouse brain. Blue: nucleus; green: Tubb3 coding mRNA expression; red: Fox3 coding mRNA expression. Scale bar = 20 μm.
Conclusion
In summary, we have developed a multifunctional nanocomplex integrating stimulating motif self-releasing capacity and dynamic mRNA expression imaging function. Our results demonstrated the high stability of fNCs to enzymatic degradation, the high specificity to target genes, and potent self-cleavage RA delivery to stimulate NSC differentiation. Moreover, fNCs enabled the spatiotemporal imaging of the dynamics of mRNA sequential expression in the complicated cascades of differentiating NSCs in a self-activating manner. In comparison to static mRNA imaging techniques at a single time point, fNCs provide a temporal tool for evaluating mRNA expression that can enhance our understanding of genetic regulation processes. Importantly, this conceptual fNC platform can be easily reprogrammed by amending corresponding oligonucleotide sequences and stimulating motifs to detect other sophisticated biological events, such as embryo development and cancer progression. We envision that this robust multifunctional nanocomplex would provide an alternative opportunity to explore dynamics of complicated biological events in living cells. An inherent limitation of this nanocomplex design is that it cannot image the downregulation of interested genes during neural stem cell differentiation, even though it demonstrated superior imaging capacity of gene upregulation in this report.
Materials and Methods
1. Synthesis of Nanocomplexes
Citrate-modified gold colloidial nanoparticles (15 ± 0.7 nm) were synthesized using published procedures.43 Two dithiolated DNA oligonucleotides were mixed separately with corresponding reporter strands at a molar ratio of 1:1.2 in PBS buffer, heated to 75 °C, and maintained for 30 min, then slowly cooled to room temperature and stored in the dark to allow hybridization. These preformed DNA duplexes were mixed together and added to a gold colloid solution (3 nM) at final concentrations of 540 nM of Alexa 488–Tubb3 DNA duplex and 560 nM of Cy3–Fox3 DNA duplex, then shaken overnight. After 12 h, phosphate buffer saline (10×; pH = 7.4) was added to the mixture to bring the NaCl concentration gradually to 0.3 M during a period of 8 h. Then the resulting nanocomplexes were centrifuged at 12 000 rpm for 15 min three times and were redispersed in PBS buffer. Particles were sterilized using a 0.2 μm acetate syringe filter (GE Healthcare). (PBS buffer components used in this paper: 10 mM phosphate, pH 7.4, 138 mM NaCl, 2.7 mM KCl, and 2 mM MgCl2.)
2. Synthesis of Multifunctional Nanocomplexes with Retinal
The resulting nanocomplexes of 0.05 nmol in 1× PBS (pH 7.4) were mixed with 10 nmol of retinal in DMSO and shaken overnight in the dark. Then the resulting fNCs were purified using centrifugation (12 000 rpm, 15 min, three times) and were redispersed in PBS buffer.
3. DNA Loading Determination
The concentration of fNCs was determined by using UV–vis absorption spectroscopy (λ = 524 nm, ε = 3.64 × 108 M–1 cm–1).44 DNA oligonucleotides were set free from fNCs by dissolving their gold backbones in a 0.1 M KCN solution. The concentration of DNA duplexes was determined by fluorescence measurements and comparing to a standard curve (Figure S2). The loading number of DNA oligonucleotides was calculated by dividing the concentration of dye-containing oligonucleotides by the concentration of gold colloidal nanoparticles.
4. Hybridization Experiment
For multiplex analysis detection, the nanocomplex of 1 nM in PBS buffer was treated with the Tubb3 targets (single-base mismatched or complementary DNA targets) or Fox3 targets (single-base mismatched or complementary DNA targets) at various concentrations of 0, 2, 5, 10, 25, 50, 100, and 200 nM. After incubation for 1 h at 37 °C, the fluorescence was observed at appropriate excitation wavelengths. The fluorescence of Alexa 488 was excited at 490 nm and measured at 530 nm (range from 500 to 700 nm), and the fluorescence of Cy3 was excited at 550 nm and measured at 570 nm (range from 550 to 700 nm). The nanocomplex had a linear range from 0 to 25 nM, and the limit of detection was calculated by the derived calibration curve, as defined by IUPAC. All experiments were repeated at least three times.
5. Specificity Experiments
The completely complementary DNA targets for designed nanocomplexes and other cancer marker targets as controls were spiked in 1 mL of buffer containing 1 nM of nanocomplex, while the DNA targets were 200 nM. All experiments were repeated at least three times.
6. Nuclease Assay
Nanocomplexes were diluted to a concentration of 1 nM in buffer (10 mM PBS, pH 7.4, 2 mM MgCl2, 1 mM CaCl2, and 50 mg/L bovine serum albumin). After allowing the samples to equilibrate for 10 min, DNase I (Amplified grade, Invitrogen) with a final concentration of 50 U/L was added immediately before reading. Molecular beacons were tested in an analogous manner at a concentration of 25 nM. The fluorescence of these samples was monitored for 24 h and was collected at predetermined intervals during this time period.
7. Cytotoxicity of fNCs
The cytotoxicity of fNCs to neural stem cells was evaluated by the MTT assay and cell proliferation curve. In the MTT assay, 100 μL of complete culture medium containing 5000 NSCs was cultured in a 96-well plate for 24 h before experiments. Various concentrations of fNCs (with or without RA) were incubated with cells for 48 h. The medium was replaced with 100 μL of fresh media containing 10 μL of MTT solution (5 mg/mL), followed by incubation for another 4 h. The media was then removed, and 100 μL of dimethyl sulfoxide (DMSO) was added into each well to dissolve the formed formazan crystals. The absorbance at 570 nm was recorded using a microplate reader. The absorbance from the control cells was set as 100% cell viability (n = 6). For the cell proliferation curve, 0.25 million cells were initially cultured in a 24-well plate in complete medium with various concentrations of fNCs (without RA). The medium was changed every other day until day 8. The cell number in each well was counted by cytometer every day (n = 4).
8. Characterization of Nanocomplexes Using TEM, UV Spectra, and DLS
Transmission electron microscopy (TEM) measurements were carried out on a JEOL 200X operated at 200 kV. UV–vis spectra were measured using a Shimadzu UV1701. Dynamic light scattering analysis was performed using a Zetasizer Nano ZS (Malvern Instruments Ltd., UK).
9. In Vitro RA Release of fNCs
The prepared fNC (5 nM final concentration, diluted from a 40 nM stock solution in corresponding buffer into a final volume of 200 μL) was incubated in either PBS buffer (pH = 7) or sodium acetate buffer (pH = 5) for a predetermined time (30 min or 12 h), followed by ultracentrifugation at 14 000 rpm for 30 min to precipitate DNA oligo attached gold nanoparticles. The supernatants containing released RA or derivatives were harvested for HPLC analysis. For the HPLC analysis, we used a gradient mobile phase with water/acetonitrile (20%/80%–10%/90%) over 12 min at 360 nm UV detection wavelength.
10. Quenching Effect of AuNPs of Fluorophores Conjugated on Oligos
In each experiment, different recognizing DNA-modified gold nanoparticles were added gradually to 50 nM of corresponding reporter in PBS buffer. The fluorescence signal was scanned by a fluorescence spectrometer. The quenching effect was calculated as the following equation: QE = quenched fluorescence/initial fluorescence × 100%. The quenched fluorescence represents the remaining fluorescence from the initial recorded fluorescence signal minus the fluorescence after addition of DNA-Au NPs. When 1 nM of DNA-Au NPs was used, the calculated quenching efficiency of gold nanoparticles was 92% for Alexa 488 and 87% for Cy3. This quenching effect difference of AuNPs is attributed to the various absorption spectral overlap of gold nanoparticle with distinct emission spectra of different fluorophores.
11. Real-Time Imaging and Kinetics of fNCs without RA on Tubb3 and Fox3 mRNAs in Differentiated NSCs at Day 8
Mouse neuroectodermal neural stem cells (NE-4C, ATCC CRL-2925) were cultured in eight-well Lab-Tech Chamber plates (NUNC, Thermo Scientific) for 24 h prior to experiment. The next day, free retinoic acid dissolved in DMSO was diluted into 1 μM working solution with complete medium. The cell culture medium was changed to the freshly prepared free-RA-containing medium for 48 h incubation with NSCs. Then, fresh medium without RA was changed every other day until day 8. On day 8, an fNC stock solution was diluted with complete medium to a 0.5 nM working solution before incubation with differentiated NSCs. The kinetics of fNC to image mRNA targets in differentiated NSCs was obtained by continuously imaging a single cell colony over 8 at 1 h intervals by laser scanning confocal microscopy (LSFM) (Olympus, FV10). Single living cell imaging results were obtained under similar conditions, while the imaging focus was set to the periphery of the cell colony with high magnification to ensure single-cell resolution. The 3D imaging results were achieved by postprocessing with Matlab software (Cambridge, MA, USA) and reconstructed with ImageJ software (NIH).
12. Self-Activating and Spatiotemporal Imaging of mRNA Sequential Expression Dynamics during NSC Differentiation
Mouse neuroectodermal neural stem cells were cultured in eight-well Lab-Tech Chamber plates (NUNC, Thermo Scientific) for 24 h prior to experiments. A fNC of 0.5 nM was used in this experiment. The stock solution of the fNC was diluted with complete medium and incubated with NSCs. The first day after incubation, 0.5 μL of Hoechst 33342 stock solution was added into each well for living cell nucleus staining 30 min prior to imaging. A marker was made on the chamber to ensure the consistently same cell colony imaging over 8 days. On the second day prior to imaging, cells were washed with HBSS solution (pH = 7.4) twice, followed by changing the fresh medium. The culture medium was changed every other day until day 8, and imaging was performed every day. Hoechst 33342 was added whenever necessary to illustrate the cell nucleus and cell colony location. For the negative control experiment, 0.5 nM of fNC without the RA motif was incubated with NSCs and imaged over 8 days using the same above protocol. The 3D imaging results were achieved by postprocessing ith Matlab software and reconstructed with ImageJ software.
13. Western Blot Study of βIII Tubulin and NeuN for Imaging Result Validation
NSCs were incubated with 0.5 nM fNCs at predetermined time points. Then, cells were washed with ice cold PBS (calcium and magnesium free) three times before being treated with RIPA buffer (Pirece) with protease inhibitor cocktail (Roche) for 10 min on ice. Cell lysates were harvested and subjected to centrifugation at 13 000 rpm at 4 °C for 10 min. The resultant supernatants containing protein were harvested, followed by 4–12% gradient SDS-PAGE gel separation. Nitrocellulose membranes were used for transferring separated protein bands. The membranes were blocked by 5% skim milk for 1 h at room temperature and then immunoblotted with anti βIII tubulin (1:1000) and anti NeuN (1:1000) primary monoclonal antibodies (Cell Signaling Technology, Inc.). HRP conjugated goat anti-rabbit secondary antibody (1:3000) was used to visualize immunoreactive bands. Anti β-actin rabbit mAb (Cell Signaling Technology, Inc.) (1:2000) was used as the housekeeping control.
14. qPCR Analysis of Tubb3 and Fox3 mRNA Expression Levels during NSC Differentiation
NSCs were incubated with 0.5 nM fNCs at predetermined time points. Total RNAs were extracted from treated NSCs using Tripure isolation reagent (Roche). RNA (1 μg) was reversely transcribed into cDANs using SuperScript First Strand Synthesis kit (Invitrogen), followed by quantitative real-time PCR analysis using a SYBR Green Master Mix kit (Applied Biosciences) on a StepOne Plus real-time PCR system (Applied Biosystems, Foster City, CA, USA). The primers used in this study are Tubb3: 5′-CTTTTCGTCTCTAGCCGCGT-3 (forward); 5′-CTCATCGCTGATGACCTCCC-3′ (reverse). Fox3: 5′-GGCAATGGTGGGACTCAAAA-3′ (forward); 5′-GGGACCCGCTCCTTCAAC-3′ (reverse). GAPDH was used as the housekeeping control: 5′-GGTCTCCTCTGACTTCAACA-3′ (forward); 5′-AGCCAAATTCGTTGTCATAC-3′ (reverse).
15. In Vivo Animal Model
All experiments were performed in accordance with NIH guidelines for the care and use of laboratory animals. Adult Balb/c mice (7–8 weeks) were anesthetized under 2% isoflurane, and the brain was placed on a stereotaxic apparatus. The following stereotaxic coordination parameters were used for the direct injection of nanocomplex (5 μL, 10 nM) into the lateral ventricle: rostral: 0.5 mm, lateral: 0.75 mm, ventral: 2.5 mm. After completing injection, the micropipette was kept for 5 min before being withdrawn. Mice were placed on a warm pad until awakened. At a predetermined time, mice were perfused using 3.7% polyformaldehyde (20 mL) before being sacrificed. Brains were dissected and sliced into 15 μm tissue slides for imaging using fluorescence microscopy.
Acknowledgments
This study was supported, in part, by the Center for Neuroscience and Regenerative Medicine (CNRM) program at Henry M. Jackson Foundation and the Intramural Research Program (IRP) of the National Institute of Biomedical Imaging and Bioengineering (NIBIB) at the National Institutes of Health (NIH).
Supporting Information Available
This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Jansen R. P. mRNA Localization: Message on the Move. Nat. Rev. Mol. Cell Biol. 2001, 2, 247–256. [DOI] [PubMed] [Google Scholar]
- Buxbaum A. R.; Wu B.; Singer R. H. Single Beta-Actin mRNA Detection in Neurons Reveals a Mechanism for Regulating Its Translatability. Science 2014, 343, 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock S. L.; Stauber M.; Prell A.; Hughes J. R.; Ish-Horowicz D.; Schmidt-Ott U. Differential Cytoplasmic mRNA Localisation Adjusts Pair-Rule Transcription Factor Activity to Cytoarchitecture in Dipteran Evolution. Development 2004, 131, 4251–4261. [DOI] [PubMed] [Google Scholar]
- Marciano P. G.; Brettschneider J.; Manduchi E.; Davis J. E.; Eastman S.; Raghupathi R.; Saatman K. E.; Speed T. P.; Stoeckert C. J. Jr.; Eberwine J. H.; McIntosh T. K. Neuron-Specific mRNA Complexity Responses During Hippocampal Apoptosis after Traumatic Brain Injury. J. Neurosci. 2004, 24, 2866–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadran S.; Remacle F.; Levine R. D. miRNA and mRNA Cancer Signatures Determined by Analysis of Expression Levels in Large Cohorts of Patients. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 19160–19165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felfly H.; Xue J.; Zambon A. C.; Muotri A.; Zhou D.; Haddad G. G. Identification of a Neuronal Gene Expression Signature: Role of Cell Cycle Arrest in Murine Neuronal Differentiation. In Vitro. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 301, R727–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.; Cui Y.; Lee L. P.; Irudayaraj J. Quantitative Imaging of Single mRNA Splice Variants in Living Cells. Nat. Nanotechnol. 2014, 9, 474–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W.; Zhang T.; Yang H.; Diao W.; Li N.; Tang B. Multiplexed Detection and Imaging of Intracellular mRNAs Using a Four-Color Nanoprobe. Anal. Chem. 2013, 85, 10581–10588. [DOI] [PubMed] [Google Scholar]
- Tyagi S. Imaging Intracellular RNA Distribution and Dynamics in Living Cells. Nat. Methods 2009, 6, 331–338. [DOI] [PubMed] [Google Scholar]
- Qiu L.; Wu C.; You M.; Han D.; Chen T.; Zhu G.; Jiang J.; Yu R.; Tan W. A Targeted, Self-Delivered, and Photocontrolled Molecular Beacon for mRNA Detection in Living Cells. J. Am. Chem. Soc. 2013, 135, 12952–12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhlanga M. M.; Vargas D. Y.; Fung C. W.; Kramer F. R.; Tyagi S. tRNA-Linked Molecular Beacons for Imaging mRNAs in the Cytoplasm of Living Cells. Nucleic Acids Res. 2005, 33, 1902–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. J.; Martinez K.; Lin H.; Tan W. Hybrid Molecular Probe for Nucleic Acid Analysis in Biological Samples. J. Am. Chem. Soc. 2006, 128, 9986–9987. [DOI] [PubMed] [Google Scholar]
- Wang F.; Wang Z.; Hida N.; Kiesewetter D. O.; Ma Y.; Yang K.; Rong P.; Liang J.; Tian J.; Niu G.; Chen X. A Cyclic HSV1-TK Reporter for Real-Time PET Imaging of Apoptosis. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, 5165–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao G.; Gao Y.; Li N.; Yu Z.; Zhuo L.; Tang B. Simultaneous Detection of Intracellular Tumor mRNA With Bi-Color Imaging Based on a Gold Nanoparticle/Molecular Beacon. Chemistry 2011, 17, 11210–11215. [DOI] [PubMed] [Google Scholar]
- Kam Y.; Rubinstein A.; Nissan A.; Halle D.; Yavin E. Detection of Endogenous K-Ras mRNA in Living Cells at a Single Base Resolution by a PNA Molecular Beacon. Mol. Pharmaceutics 2012, 9, 685–693. [DOI] [PubMed] [Google Scholar]
- Prigodich A. E.; Randeria P. S.; Briley W. E.; Kim N. J.; Daniel W. L.; Giljohann D. A.; Mirkin C. A. Multiplexed Nanoflares: mRNA Detection in Live Cells. Anal. Chem. 2012, 84, 2062–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seferos D. S.; Giljohann D. A.; Hill H. D.; Prigodich A. E.; Mirkin C. A. Nano-Flares: Probes for Transfection and mRNA Detection in Living Cells. J. Am. Chem. Soc. 2007, 129, 15477–15479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N.; Chang C.; Pan W.; Tang B. A Multicolor Nanoprobe for Detection and Imaging of Tumor-Related mRNAs in Living Cells. Angew. Chem., Int. Ed. 2012, 51, 7426–7430. [DOI] [PubMed] [Google Scholar]
- Chen T.; Wu C. S.; Jimenez E.; Zhu Z.; Dajac J. G.; You M.; Han D.; Zhang X.; Tan W. DNA Micelle Flares for Intracellular mRNA Imaging and Gene Therapy. Angew. Chem., Int. Ed. 2013, 52, 2012–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.; Chen T.; Han D.; You M.; Peng L.; Cansiz S.; Zhu G.; Li C.; Xiong X.; Jimenez E.; Yang C. J.; Tan W. Engineering of Switchable Aptamer Micelle Flares for Molecular Imaging in Living Cells. ACS Nano 2013, 7, 5724–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amamoto R.; Arlotta P. Development-Inspired Reprogramming of the Mammalian Central Nervous System. Science 2014, 343, 1239882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S.; Lie D. C.; DeCicco K. L.; Shi Y.; DeLuca L. M.; Gage F. H.; Evans R. M. Retinoic Acid Is Required Early during Adult Neurogenesis in the Dentate Gyrus. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 3902–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos T.; Ferreira R.; Maia J.; Agasse F.; Xapelli S.; Cortes L.; Braganca J.; Malva J. O.; Ferreira L.; Bernardino L. Polymeric Nanoparticles to Control the Differentiation of Neural Stem Cells in the Subventricular Zone of the Brain. ACS Nano 2012, 6, 10463–10474. [DOI] [PubMed] [Google Scholar]
- Wang W.; Nossoni Z.; Berbasova T.; Watson C. T.; Yapici I.; Lee K. S.; Vasileiou C.; Geiger J. H.; Borhan B. Tuning the Electronic Absorption of Protein-Embedded All-Trans-Retinal. Science 2012, 338, 1340–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z. J.; Tang R.; Yeh Y. C.; Miranda O. R.; Rotello V. M.; Vachet R. W. Determination of the Intracellular Stability of Gold Nanoparticle Monolayers Using Mass Spectrometry. Anal. Chem. 2012, 84, 4321–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykman L. A.; Khlebtsov N. G. Uptake of Engineered Gold Nanoparticles into Mammalian Cells. Chem. Rev. 2014, 114, 1258–12588. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Wang Z.; Liu D.; Yan X.; Wang F.; Niu G.; Yang M.; Chen X. Biomimetic RNA-Silencing Nanocomplexes: Overcoming Multidrug Resistance in Cancer Cells. Angew. Chem., Int. Ed. 2014, 53, 1997–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duester G. Retinoic Acid Synthesis and Signaling during Early Organogenesis. Cell 2008, 134, 921–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage F. H. Mammalian Neural Stem Cells. Science 2000, 287, 1433–1438. [DOI] [PubMed] [Google Scholar]
- Cummings B. J.; Uchida N.; Tamaki S. J.; Salazar D. L.; Hooshmand M.; Summers R.; Gage F. H.; Anderson A. J. Human Neural Stem Cells Differentiate and Promote Locomotor Recovery in Spinal Cord Injured Mice. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 14069–14074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke D. L.; Johansson C. B.; Wilbertz J.; Veress B.; Nilsson E.; Karlstrom H.; Lendahl U.; Frisen J. Generalized Potential of Adult Neural Stem Cells. Science 2000, 288, 1660–1663. [DOI] [PubMed] [Google Scholar]
- Dimos J. T.; Rodolfa K. T.; Niakan K. K.; Weisenthal L. M.; Mitsumoto H.; Chung W.; Croft G. F.; Saphier G.; Leibel R.; Goland R.; Wichterle H.; Henderson C. E.; Eggan K. Induced Pluripotent Stem Cells Generated from Patients with ALS Can Be Differentiated into Motor Neurons. Science 2008, 321, 1218–1221. [DOI] [PubMed] [Google Scholar]
- Ding S.; Wu T. Y.; Brinker A.; Peters E. C.; Hur W.; Gray N. S.; Schultz P. G. Synthetic Small Molecules that Control Stem Cell Fate. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 7632–7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes E. H.; Jencks W. P. On the Mechanism of Schiff Base Formation and Hydrolysis. J. Am. Chem. Soc. 1962, 84, 823–837. [Google Scholar]
- Wang Z.; Huang P.; Bhirde A.; Jin A.; Ma Y.; Niu G.; Neamati N.; Chen X. A Nanoscale Graphene Oxide Peptide Biosensor for Real-Time Specific Biomarker Detection on the Cell Surface. Chem. Commun. 2012, 48, 9768–9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Chui W. K.; Ho P. C. Design of A Multifunctional PLGA Nanoparticulate Drug Delivery System: Evaluation of Its Physicochemical Properties and Anticancer Activity to Malignant Cancer Cells. Pharm. Res. 2009, 26, 1162–1171. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Zhang X.; Huang P.; Zhao W.; Liu D.; Nie L.; Yue X.; Wang S.; Ma Y.; Kiesewetter D.; Niu G.; Chen X. Dual-Factor Triggered Fluorogenic Nanoprobe for Ultrahigh Contrast and Subdiffraction Fluorescence Imaging. Biomaterials 2013, 34, 6194–6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga B. V.; Hadinger N.; Gocza E.; Dulberg V.; Demeter K.; Madarasz E.; Herberth B. Generation of Diverse Neuronal Subtypes in Cloned Populations of Stem-Like Cells. BMC Dev. Biol. 2008, 8, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chithrani B. D.; Ghazani A. A.; Chan W. C. Determining the Size and Shape Dependence of Gold Nanoparticle Uptake into Mammalian Cells. Nano Lett. 2006, 6, 662–668. [DOI] [PubMed] [Google Scholar]
- Choi C. H.; Hao L.; Narayan S. P.; Auyeung E.; Mirkin C. A. Mechanism for the Endocytosis of Spherical Nucleic Acid Nanoparticle Conjugates. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 7625–7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralick R. M.; Shanmugam K.; Dinstein I. H. Textural Features for Image Classification. IEEE Trans. Syst., Man, Cybern. 1973, SMC-3, 610–621. [Google Scholar]
- Hu B. Y.; Weick J. P.; Yu J.; Ma L. X.; Zhang X. Q.; Thomson J. A.; Zhang S. C. Neural Differentiation of Human Induced Pluripotent Stem Cells Follows Developmental Principles But with Variable Potency. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 4335–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabar K. C.; Freeman R. G.; Hommer M. B.; Natan M. J. Preparation and Characterization of Au Colloid Monolayers. Anal. Chem. 1995, 67, 735–743. [Google Scholar]
- Prigodich A. E.; Lee O. S.; Daniel W. L.; Seferos D. S.; Schatz G. C.; Mirkin C. A. Tailoring DNA Structure to Increase Target Hybridization Kinetics on Surfaces. J. Am. Chem. Soc. 2010, 132, 10638–10641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.