Abstract
Chlamydia trachomatis conjunctival samples collected over a 6-month period from individuals with clinical signs of trachoma and located in remote communities in the Australian Northern Territory were differentially characterized according to serovar and variants. The rationale was to gain an understanding of the epidemiology of an apparent increased prevalence of acute trachoma in areas thought to be less conducive to this disease. Characterization was performed through sequencing of a region of the omp1 gene spanning the four variable domains and encoding the major outer membrane protein. Nucleotide and deduced amino acid sequences were genotyped by using a BLAST similarity search and were examined by phylogenetic analyses to illustrate evolutionary relationships between the clinical and GenBank reference strains. The predominant genotype identified corresponded to that of serovar C (87.1%), followed by the genotype corresponding to serovar Ba (12.9%). All nucleotide and amino acid sequences exhibited minor levels of variation with respect to GenBank reference sequences. The omp1 nucleotide sequences of the clinical samples best aligned with those of the conjunctival C. trachomatis reference strains C/TW-3/OT and Ba/Apache-2. All clinical samples (of serovar C) exhibited four or five nucleotide changes compared with C/TW-3/OT, while all serovar Ba samples had one or two nucleotide differences from Ba/Apache-2. Phylogenetic analyses revealed close relationships between these Northern Territory chlamydial samples and the respective reference strains, although the high proportion of sequence variants suggests an evolutionarily distinct C. trachomatis population causing eye infections in Australia. Given that such genotypic information has gone unreported, these findings provide knowledge and a foundation for trachoma-associated C. trachomatis variants circulating in the Northern Territory.
Chlamydia trachomatis infections cause a wide spectrum of human diseases in multiple organ systems. Urogenital C. trachomatis infections (caused by serovars D to K) are the most common bacterial sexually transmitted diseases. They are associated in men with urethritis and occasionally epididymitis and in women with cervicitis, salpingitis, and pelvic inflammatory disease, with the potential outcome of ectopic pregnancy or tubal infertility (6, 7, 8, 23). A woman infected during pregnancy can transmit C. trachomatis intrapartum to her infant, resulting in neonatal conjunctivitis and, if untreated, nasopharyngeal colonization, which can lead to pneumonitis. Furthermore, in adults, genital serovars D to K can result in conjunctivitis, once known as “swimming pool conjunctivitis,” although the likely transmission route is from the genitalia to the eyes (12).
Besides urogenitally acquired C. trachomatis eye infections, serovars A to C can result in an array of sequelae varying from mild conjunctivitis to ocular trachoma with resultant blindness. In fact, trachoma is the leading cause of preventable blindness in the world. The pathogenesis is thought to be repeated C. trachomatis infections, resultant inflammation, secondary bacterial infection, and gradual scarring of the cornea and conjunctiva (16, 27). Hands, clothing, or flies that have had contact with infected bodily discharges readily spread C. trachomatis. Generally, the disease occurs in poor, undeveloped countries where people live in overcrowded conditions and have limited access to water and health care. Australia is the only developed country where trachoma blindness still occurs (28). Trachoma has been successfully eradicated from many regional communities in Australia, although the disease is still prevalent in areas where living conditions are overcrowded and there is inadequate personal and community hygiene, as experienced in many remote communities in the Australian Northern Territory, Western Australia, and central Australia (9, 10, 28). Incidence rates for sexually transmitted infections, including urogenital chlamydial disease, are also markedly higher in remote populations of the Northern Territory than in the rest of Australia (3).
Currently, C. trachomatis is classified into 15 different serovars based on immunogenic epitope analysis of the major outer membrane protein (MOMP) with polyclonal and monoclonal antibodies (18, 31). The MOMP is the principal immunodominant surface antigen of C. trachomatis, with antigenic determinants located across four symmetrically spaced variable domains (VDI to VDIV), which are flanked and interspaced by five constant domains (CDs) (1, 25, 32). The nucleotide sequences of these omp1 variable domains exhibit distinct variations in different serovars and have become widely used for the genotyping of C. trachomatis isolates (2, 15, 26). Typically, C. trachomatis serovars A to C are found associated with trachoma, serovars D to K are associated with urogenital infections, and serovars L1 to L3 are associated with the systemic disease lymphogranuloma venereum (26, 31). Genotypic characterization of C. trachomatis isolates not only can provide valuable insight about the serovars and their variants circulating within a given community but also can improve the understanding of their epidemiology, which may assist in strategies for improved disease control.
In this study, we report a protocol for the extraction, amplification, and sequencing of a region of the C. trachomatis omp1 gene spanning the four variable domains and the use of this protocol to examine eye swab samples collected from individuals residing in remote communities spread across geographically distinct districts of the Northern Territory. These communities were experiencing an increased prevalence of C. trachomatis eye disease, with some cases occurring in geographical areas believed to be less conducive to trachoma. The aim was to genotype a panel of C. trachomatis conjunctival samples collected in these communities for insight into their epidemiology and phylogenetic relationships with prototypic reference strains.
MATERIALS AND METHODS
Clinical samples and reference strains.
Between July and December 2002, eye swab samples (n = 37) were collected from individuals with symptoms of acute trachoma (in accordance with World Health Organization clinical diagnostic criteria for acute trachoma). Individuals were from six geographically distinct remote communities (five coastal and one inland) in the Northern Territory that were of low socioeconomic status. The communities were spread over the length and breadth of the Northern Territory, with a maximum distance of nearly 1,000 km between the most distant two, and ranged in distance from the capital, Darwin, by approximately 100 to 700 km. Communities 1 and 2 were clustered in close proximity (less than 50 km apart). Samples from these communities were initially identified as C. trachomatis positive by the COBAS Amplicor protocol (Roche Molecular Diagnostics, Pleasanton, Calif.).
The ages of the individuals tested ranged from <1 to 25 years, although most individuals were predominantly less than 13 years of age (n = 33; 89%), and the remainder were between 19 and 25 years of age. Eye swab samples were collected during annual health screening programs for school-age children (signed consent was obtained) and at local health centers as a routine step in the clinical management of conjunctivitis (no signed consent required). Samples were collected at an opportunistic time after the noted increase in cases and encompassed the wet (July) and dry (December) seasons; most samples were taken during the dry season, when school screening takes place. Nine C. trachomatis reference strains, B, D, E, F, G, I, J, K, and L2, were used as positive PCR controls. C. pneumoniae, C. psittaci and C. percori strains were used as negative PCR controls.
Nucleotide sequences of the omp1 genes derived from C. trachomatis reference strains of all 15 serovars (A to L3) were obtained from the GenBank nucleotide database (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db = Nucleotide). The reference strains were as follows (accession numbers in parentheses): A/Har-13 (J03813), A/Sa1/OT (M58938), B/Alpha-95 (U80075), B-Jali-20 (M33636), B/TW-5/OT (M17342), Ba/Apache-2 (AF063194), C/TW3 (AF202455), C/TW3/OT (M17343), C/TW-3/OT (AF352789), D/B120 (X62918), D/B185 (X62919), D/IC-Cal8 (X62920), E/Bour-1990 (X52557), E/Bour-1997 (U78763), F/IC-Cal3 (X52080), G/UW57/Cx (AF063199), H/Wash (X16007), I/UW-12 (AF063200), J/UW36/Cx (AF063202), K/UW31/Cx (AF063204), L1/440-Bu (M36533), L2/434-Bu (M14738), L3/404-Bu (X55700), and MoPn (mouse pneumonitis) (M64171). These strains were included in this study to assist in the generation of an accurate phylogenetic tree representative of sequence relationships. The majority of these sequences have been well characterized, representing prototypes of each serovar, and as such have been used in similar phylogenetic studies of chlamydial isolates (14, 15, 26). Strain MoPn was included as an out-group in the phylogenetic tree analyses in order to root the neighbor-joining trees.
DNA extraction.
Aliquots (200 μl) of COBAS Amplicor-processed eye swab samples were extracted by using automated MagNA Pure LC (Roche) with the associated DNA isolation kit I protocol. DNA was eluted in a final volume of 100 μl of MagNA Pure elution buffer (Roche).
omp1 PCR.
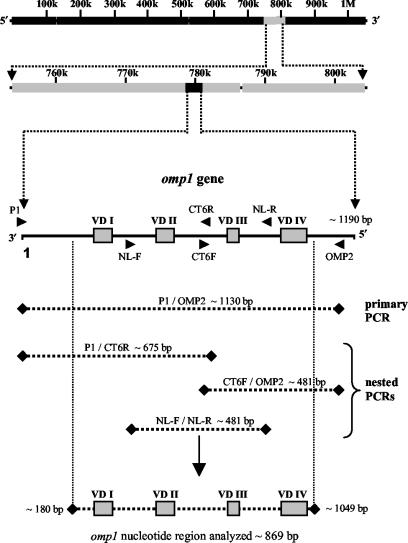
The C. trachomatis omp1 gene was initially PCR amplified by using previously described primers P1 and OMP2 (15), annealing at the omp1 start codon and approximately 1,130 bp downstream, respectively (Fig. 1). For every amplification reaction, one phosphate-buffered saline negative control was included. An aliquot of the DNA template (2 to 10 μl) was added to a PCR mixture (final reaction volume, 50 μl) composed of PCR buffer (10 mM Tris-HCl, 50 mM KCl [pH 8.3]), 2.0 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 1.0 μM each primer, and 2.5 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, Calif.). PCR amplification was carried out by using a model FTS-960 thermocycler (Corbett Research). The amplification profile included an initial polymerase activation step at 94°C for 10 min, followed by 40 cycles of denaturation (94°C for 30 s), annealing (50°C for 30 s), and extension (72°C for 2 min). An additional 7 min of extension at 72°C was included at the end of the amplification cycle. Amplification products of approximately 1.2 kb were visualized by electrophoresis through a 1% agarose gel containing ethidium bromide.
FIG. 1.
Schematic representation of the C. trachomatis genome. The omp1 gene, approximately 1.2 kb in length, was PCR amplified with various combinations of primers. Primers P1 and OMP2 were used in the primary PCR. DNA which failed to generate primary PCR products was subsequently amplified in a nested PCR with the following primer combinations: P1-CT6R, CT6F-OMP2, and NL-F-NL-R. Nucleotide sequence data from the C. trachomatis conjunctival samples was assembled into one contiguous sequence spanning the four variable domains (VDI to VDIV).
DNA samples which failed to generate a primary PCR product were subjected to amplification with additional primer sets and/or nested reamplification. The additional primer sets used were as follows: primers CT6F (5′-GCTCAATCTAAACCTAAARTMCAAG-3′) and CT6R (5′-CTTGKAYTTTAGGTTTAGATTGAGC-3′) (modified from reference 4) and primers NL-F (5′-TGGGATCGYTTTGATGTATT-3′) and NL-R (5′-CCAATGTARGGAGTGAACAT-3′) (16a). These primers were also used in mixed combinations with primers P1 and OMP2 to generate a panel of PCR products ensuring sufficient sequence overlap and fidelity (Fig. 1). Nested reamplification reactions were essentially the same as those of the P1-OMP2 primary PCR, except that only 1 to 2 μl of the primary PCR mixture was used as the template.
omp1 nucleotide sequencing.
The PCR amplicons were purified in 50 μl of sterile H2O by using a High-Pure PCR product purification kit (Roche). The amplicons were sequenced by using an optimized reduced-reaction method for dye terminator cycle sequencing. Briefly, sequencing reaction mixtures were composed of 4 μl of CEQ dye terminator cycle sequencing Quick-Start master mix (Beckman Coulter, Inc., Fullerton, Calif.), 4 μl of CEQ 2.5× sequencing reaction buffer (Beckman Coulter), 2 μl of the appropriate primer (1.6 μM stock), 100 fmol of purified amplicon, and sterile H2O to a final reaction volume of 20 μl. Sequencing reactions were carried out with 30 cycles of denaturation (96°C for 20 s), annealing (50°C for 20 s), and extension (60°C for 4 min), followed by a final incubation at 4°C. The amplicons were sequenced in both directions to ensure sufficient sequence overlap and fidelity. Postreaction cleanup of the dye-labeled sequencing products was performed with an ethanol precipitation protocol as described by Beckman Coulter. Final DNA pellets were resuspended in 40 μl of CEQ sample loading solution (Beckman Coulter), overlaid with mineral oil, and sequenced by using a CEQ 8000 capillary DNA sequencer (Beckman Coulter).
BLAST and phylogenetic analyses.
All nucleotide sequence data generated were reviewed by using CEQ 8000 sequence analysis software (Beckman Coulter) to ensure sufficient accuracy. DNA sequence data were initially submitted to the standard nucleotide-nucleotide BLAST search engine at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST) for comparative analyses with omp1 nucleotide sequences of known C. trachomatis serovars to determine consensus genotypes for the conjunctival samples. The nucleotide sequence data generated were manually assembled into complete consensus sequences, approximately 870 bp in size and spanning the four omp1 variable domains (Fig. 1), by using BLAST alignment of two sequences (http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html). This sequence length represented approximately 73% of the complete C. trachomatis omp1 gene. These sequences were realigned by using a BLAST search for reconfirmation of their determined serovars.
In order to illustrate the evolutionary relationships between the conjunctival samples and the various C. trachomatis reference strains, we used phylogenetic tree analyses. The reference strains obtained from GenBank included prototypic strains of serovars A to L3, together with strain MoPn for phylogenetic tree construction. Raw sequence data were imported into the BioEdit (version 5.0.9) program (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) (13) and saved into a FASTA file format. This file then was imported into the CLUSTAL X (version 1.83) program (ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX) (29), where phylogenetic analyses of the region spanning the four variable domains were performed (Fig. 1). Sequences generated from the conjunctival samples and obtained for both laboratory and GenBank reference strains (spanning equivalent lengths) were aligned by using the “Complete Multiple Alignment” option in CLUSTAL X. A bootstrapped neighbor-joining tree was generated by the method of Saitou and Nei (22) from 1,000 randomly generated data sets providing bootstrap confidence levels (expressed as a percentage of the replicates) for the various branch nodes to which they applied. Bootstrap values of <50% were omitted because they were considered insignificant.
Nucleotide sequence accession numbers.
Accession numbers for the nucleotide sequences spanning the four variable domains of the 31 PCR-amplified C. trachomatis conjunctival samples were obtained from GenBank and are listed in Table 1.
TABLE 1.
GenBank accession numbers for C. trachomatis conjunctival swab samplesa
| Serovar | No. of samples with same omp1 sequenceb | Sample | GenBank accession no. |
|---|---|---|---|
| Ba | 2 | 3537416 | AY378286 |
| 3537534 | AY378284 | ||
| 2 | 3969853 | AY378285 | |
| 5165682 | AY378287 | ||
| C | 1 | 4027779 | AY380092 |
| 26 | 3681739 | AY380093 | |
| 3681952 | AY380094 | ||
| 3681970 | AY380095 | ||
| 3938625 | AY380096 | ||
| 3958651 | AY380097 | ||
| 4080214 | AY380098 | ||
| 4080233 | AY380099 | ||
| 4118607 | AY380100 | ||
| 418671 | AY380101 | ||
| 4837384 | AY380102 | ||
| 4953801 | AY380103 | ||
| 5631112 | AY380104 | ||
| 5644791 | AY380105 | ||
| 5644793 | AY380106 | ||
| 5644848 | AY380107 | ||
| 5677568 | AY380108 | ||
| CH 4312 | AY380109 | ||
| CH 4313 | AY380110 | ||
| CH 4314 | AY380111 | ||
| CH 4316 | AY380112 | ||
| CH 4464 | AY380113 | ||
| CH 4543 | AY380114 | ||
| CH 4545 | AY380115 | ||
| SE 4572 | AY380116 | ||
| SE 4766 | AY380117 | ||
| SE 4767 | AY380118 |
Nucleotide sequence analyses of the region spanning VDI to VDIV demonstrated that 4 specimens corresponded to serovar Ba and 27 corresponded to serovar C (26 of the latter exhibited identical sequences).
Number of C. trachomatis conjunctival swab samples having the same omp1 nucleotide sequence (i.e., representative of the same variant).
RESULTS
omp1 PCR.
DNA extracted from all nine C. trachomatis laboratory reference strains was successfully amplified by the P1-OMP2 primary PCR, generating products of approximately 1.2 kb. DNA from C. pneumoniae, C. psittaci, and C. percori control strains was PCR negative. From the 37 COBAS Amplicor C. trachomatis-positive samples examined, 31 were successfully amplified by either P1-OMP2 primary PCR, NL-F-NL-R internal PCR, and/or P1-CT6R and CT6F-OMP2 seminested PCRs (Fig. 1). Six of the clinical samples repeatedly failed to generate PCR products, despite initial positive test results in the COBAS Amplicor. These six PCR-negative samples were retested by the COBAS Amplicor and demonstrated to be C. trachomatis positive, confirming that they were not false-positive results initially.
omp1 nucleotide and MOMP sequence analyses.
The nucleotide and deduced amino acid sequences spanning the omp1 gene and MOMP variable domains (VDI to VDIV) were analyzed in this study. Nucleotide sequence analyses of the 31 PCR-positive conjunctival samples by a BLAST similarity search demonstrated that the most prevalent genotype was that corresponding to C. trachomatis serovar C (n = 27; 87.1%), followed by serovar Ba (n = 4; 12.9%) (Table 2). A comparison of the omp1 nucleotide sequences of these samples revealed that 26 of the 27 serovar C types were identical, while for serovar Ba, 2 were of one sequence and 2 were of another sequence (one nucleotide difference). Interestingly, all of the sequences generated from these clinical samples exhibited notable variations relative to their GenBank reference counterparts (Table 2). The omp1 nucleotide sequences aligned closest with the C. trachomatis GenBank reference strains Ba/Apache-2 (AF063194) and C/TW-3/OT (AF352789), both of which were derived from conjunctival sources. The four serovar Ba conjunctival samples exhibited one nucleotide difference from Ba/Apache-2 at omp1 position 511 (common to serovar A isolates), with two of the sequences showing an additional nucleotide change at position 662. These mutations resulted in amino acid changes at positions 171 and 221 (Table 2). The 27 serovar C clinical samples exhibited four nucleotide differences from C/TW-3/OT (positions 569 and 571 [common to serovar A and B isolates], 972, and 1003 [common to serovar A isolates]), with resultant amino acid changes at positions 190, 191, and 335 (Table 2). One serovar C sample (4027779) showed an additional nucleotide change at position 697 (common to serovar B isolates), with a resultant amino acid change at position 233 (Table 2).
TABLE 2.
Nucleotide and amino acid changes within the omp1 sequence in conjunctival samples collected from remote communities in the Australian Northern Territory
| Serovar
|
Nucleotide
|
Amino acid
|
||||||
|---|---|---|---|---|---|---|---|---|
| Type | %a | No.b | No. of changes | Changec | omp1 positiond | No. of changes | Changec | MOMP positione |
| Ba | 12.9 | 2 | 1 | A → G | 511 (VDII) | 1 | S → G | 171 |
| 2 | 2 | A → G | 511 (VDII) | 2 | S → G | 171 | ||
| C → T | 662 (CD) | P → L | 221 | |||||
| C | 87.1 | 26 | 4 | T → C | 569 (CD) | 3 | I → T | 190 |
| A → G | 571 (CD) | N → D | 191 | |||||
| G → A | 972 (VDIV) | Silent | ||||||
| G → T | 1003 (VDIV) | A → S | 335 | |||||
| 1 | 5 | T → C | 569 (CD) | 4 | I → T | 190 | ||
| A → G | 571 (CD) | N → D | 191 | |||||
| A → G | 697 (CD) | N → D | 233 | |||||
| G → A | 972 (VDIV) | Silent | ||||||
| G → T | 1003 (VDIV) | A → S | 335 | |||||
Percentage of 31 C. trachomatis PCR-amplified conjunctival samples with the indicated serovar.
Number of C. trachomatis samples having the same omp1 sequence changes as those indicated.
Nucleotide and amino acid changes from the most phylogenetically related reference strain (i.e., Ba/Apache-2 and C/TW-3/OT).
Nucleotide position from the start of the C. trachomatis omp1 gene at which the change was located.
Amino acid position from the start of the MOMP at which the deduced change has occurred.
A comparison of the C. trachomatis serovars detected in the different communities revealed that all of the clinical samples of serovar Ba were from the same inland community (Table 3). In contrast, the majority of the samples of serovar C (n = 26; 96.3%) were from coastal communities. The remaining serovar C sample was from the inland community (Table 3).
TABLE 3.
C. trachomatis serovar distribution in six remote Northern Territory communities
| Communitya | No. of samples with the following serovar:
|
|||
|---|---|---|---|---|
| Ba | C | NDb | Total | |
| 1 | 1 | 1 | ||
| 2 | 7 | 1 | 8 | |
| 3 | 2 | 2 | ||
| 4 | 15 | 4 | 19 | |
| 5 | 1 | 1 | ||
| 6 | 4 | 1 | 1 | 6 |
| Total | 4 | 27 | 6 | 37 |
Communities 1 to 5 were located at different coastal regions, while community 6 was situated inland.
Clinical samples which did not generate PCR products and for which the serovar therefore was not determined (ND), despite a positive result in the COBAS Amplicor protocol.
Phylogenetic analyses of the omp1 gene and MOMP.
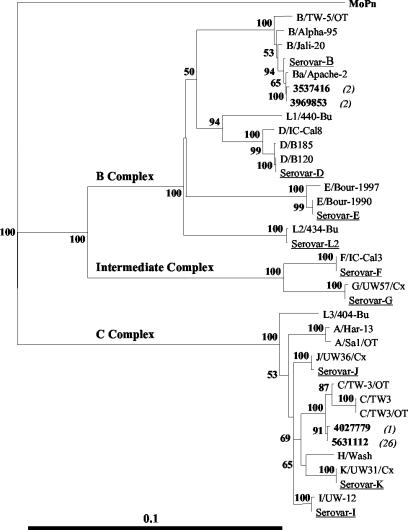
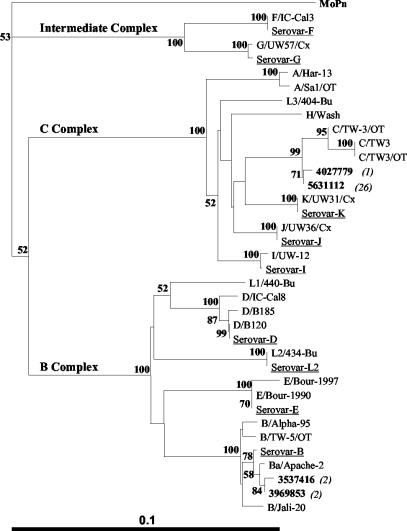
Distance neighbor-joining phylogenetic trees based on the nucleotide or deduced amino acid sequences spanning VDI to VDIV of the omp1 gene or MOMP from the 31 conjunctival samples and various reference strains are illustrated in Fig. 2 and 3, respectively. Phylogenetic analyses of the nucleotide and amino acid sequences generated trees having significant levels of confidence and illustrating a clear segregation of the C. trachomatis serovars into three distinct clusters. These clusters were consistent with the three described serocomplexes, B, C, and the intermediate complex (5, 32). The C. trachomatis sequences derived from the conjunctival samples showed a phylogenetic spread across two of the three serocomplexes, with predominance within the larger and more genetically diverse C complex.
FIG. 2.
Distance neighbor-joining phylogenetic tree based on C. trachomatis nucleotide sequences spanning VDI to VDIV for the conjunctival samples. For clarity, only one representative from each genetic variant of the samples is illustrated (in bold type). Values in parentheses indicate the number of samples of that variant. Selected reference isolates stored in our laboratory are also included (underlined). The tree was rooted by using the equivalent MoPn omp1 nucleotide sequence region. Numbers at the nodes are bootstrap values (percentages) from 1,000 replicates. The bar corresponds to a graphic distance equivalent to 0.1 nucleotide substitution per site.
FIG. 3.
Distance neighbor-joining phylogenetic tree based on C. trachomatis amino acid sequences spanning VDI to VDIV for the conjunctival samples. For clarity, only one representative from each genetic variant of the samples is illustrated (in bold type). Values in parentheses indicate the number of samples of that variant. Selected reference isolates stored in our laboratory are also included (underlined). The tree was rooted by using the equivalent MoPn MOMP amino acid sequence region. Numbers at the nodes are bootstrap values (percentages) from 1,000 replicates. The bar corresponds to a graphic distance equivalent to 0.1 amino acid substitution per site.
DISCUSSION
In this study, a combination of PCR and nucleotide sequencing was used to genotype eye swab samples from individuals exhibiting clinical signs of trachoma in remote Australian communities. Analysis of circulating C. trachomatis genotypes among individuals exhibiting clinical signs of trachoma in Australia has not been described before, despite the important association of repetitive infections with preventable blindness (16, 27, 28). Subtle sequence differences in the variable domains of the C. trachomatis omp1 genes between serovars provide a valuable and sensitive means for molecular epidemiological analysis. In addition to containing serotyping determinants, the surface-exposed portions of the MOMP variable domains represent the major target sites for neutralizing antibodies, responsible for antigenic differences between serovars (1, 24, 31). The variability in chlamydial MOMP sequences suggests an adaptive evolutionary approach to escape host immune responses. C. trachomatis serovars A to C are commonly associated with trachoma, serovars D to K are associated with urogenital infections, and serovars L1 to L3 are associated with systemic lymphogranuloma venereum (26, 31). Importantly, serovars B and C have been infrequently identified in urogenital tract infections (21). In an unpublished study of antenatal women in northern Australia in 1990, 48% of C. trachomatis cervical isolates (n = 31) were serovar B (S. H. Hutton, personal communication).
In contrast to the extensive findings associated with the typing of C. trachomatis isolates from urogenital or urine samples, there are minimal reports describing C. trachomatis serovar distributions among conjunctival isolates, despite the widespread occurrence of trachoma in developing countries and its association with preventable blindness (16, 27, 28). Minimal trachoma prevalence data have been reported for the Australian Northern Territory in recent years. However, despite an unknown prevalence of trachoma in the communities described in this study, a recent report described trachoma prevalence in children aged 4 to 15 years across seven communities in the Northern Territory as ranging from 17% (endemic) to 38% (hyperendemic) (19). Additionally, trachoma prevalence rates have been shown to significantly rise during the wet season, correlating with increased numbers of bush flies, likely transmission vectors of trachoma (9). Notably, the rise in trachoma cases on which this study was based occurred during the dry season.
Genotyping analyses of 31 C. trachomatis conjunctival samples collected from remote communities in the Australian Northern Territory revealed a predominance of serovar C (87.1%) in all 6 coastal communities; the remaining samples constituted serovar Ba (12.9%) and were all from 1 inland community. Considering the lack of epidemiological data associated with C. trachomatis genotype distributions among trachoma-associated strains, limited conclusions can be made as to their phylogenetic relationships. Nonetheless, these findings are consistent with previously published results in which all of the Northern Territory C. trachomatis conjunctival samples tested were classified as belonging to serovars A to C, which are causative of trachoma (5, 32). It seems unlikely from these findings that the Northern Territory trachoma infections are related to urogenital C. trachomatis. In a study in progress, we are examining the circulating genotypes of C. trachomatis urogenital samples from one of these communities for comparison with the ocular samples.
We found low levels of sequence variations between clinical samples of a given chlamydial genotype. In fact, 26 of the 27 serovar C samples (from different communities) contained identical sequences. The one variant exhibited a nucleotide change at position 697, located in the CD between VDII and VDIII. Similar findings of sequence variations outside the variable domains have been reported and have been described as distinguishing features between trachoma and urogenital isolates of the same C. trachomatis serovar (11). The possibility of sequencing errors was eliminated through the generation of identical sequences by using two independent DNA extractions in conjunction with multiple PCR primer sets. These low levels of sequence divergence within the omp1 genes were not surprising, considering the relatively small number of samples, which were collected from clusters of clinical cases in only six remote communities in one geographical territory. In addition, these samples were collected over a relatively short period of time (6 months).
The sequences from the four serovar Ba samples could be divided into two groups based on a single nucleotide difference (nucleotide position 662), also located in the CD between VDII and VDIII. Interestingly, the phylogenetic trees based on the nucleotide and amino acid sequences (Fig. 2 and 3, respectively) show close relationships between these clinical samples and reference strains isolated from conjunctival sources. Despite these close relationships, the omp1 sequences generated from all samples were distinct from their most closely related reference sequences. In particular, the 27 serovar C sequences exhibited at least three or four amino acid changes, as demonstrated by the distinct phylogenetic branch. Various assumptions can be made through analysis of the C. trachomatis phylogenetic trees. First, it appears likely that the four serovar Ba conjunctival strains evolved from strain Ba/Apache-2, be it only recently. Second, the serovar C conjunctival strains appear to have evolved from a more distinct lineage, illustrative of their broader geographical spread across the various coastal and inland communities.
The geographical isolation of Australia must be an important factor contributing to the uniqueness of the C. trachomatis strains tested here relative to other reference strains. An interesting epidemiological finding in this study was that all of the serovar Ba clinical samples were from the one inland community. The majority of serovar C samples (96.3%) were from coastal communities; only one sample was from the inland community. Importantly, genotyping was performed blind, in that sample details were unknown until the conclusion of the sequence analysis. Despite the small number of samples successfully genotyped (n = 31), an epidemiological pattern was apparent.
The chlamydial COBAS Amplicor assay uses a multicopy cryptic plasmid target and thus offers higher levels of sensitivity than conventional PCR, which uses a single-copy gene target (17). Therefore, it was considered highly likely that the 6 of 38 conjunctival samples that repeatedly failed to generate omp1 PCR products, despite positive C. trachomatis identification by the COBAS Amplicor, contained levels of bacteria that were below the detection limit of the omp1 PCR. Similar findings of a positive plasmid-directed PCR result and a negative omp1 PCR result have been reported for various genital samples (20, 30).
This application of PCR-based genotyping of C. trachomatis conjunctival swab samples provides a convenient and sensitive method for future epidemiological studies to address the pathogenicity and transmission of chlamydial eye infections. Such studies would prove of great value given the current prevalence of chlamydial infections and preventable blindness in both rural Australia and developing countries worldwide. The results presented in this study provide an understanding of the circulating C. trachomatis genotypes in the Australian Northern Territory and their evolutionary relationships to known reference strains. These findings provide the foundation for a more widespread study which could consist of both larger numbers of samples and sample collection over a larger area, including Western Australia and central Australia.
Acknowledgments
We thank Peter O'Loughlin (Queensland Medical Laboratory, Brisbane, Queensland, Australia), Gary Lum (Royal Darwin Hospital), and the members of the Serology Department, Western Diagnostic Pathology, Myaree, Western Australia, Australia, for assistance with sample processing and storage. We also thank Bart Currie, Susan Hutton (Menzies School of Health Research, Darwin, Northern Territory, Australia), and Scott Cameron (National Centre for Epidemiology and Population Health) for input into the Northern Territory part of the study. Additionally, we thank Nichole Lister for the generous supply of primers NL-F and NL-R used for PCR amplification of the C. trachomatis omp1 gene.
We thank the Centre for Disease Control and The Royal Women's Hospital for supplying funding for this study.
REFERENCES
- 1.Baehr, W., Y. X. Zhang, T. Joseph, H. Su, F. E. Nano, K. D. Everett, and H. D. Caldwell. 1988. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc. Natl. Acad. Sci. USA 85:4000-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandea, C. I., K. Kubota, T. M. Brown, P. H. Kilmarx, V. Bhullar, S. Yanpaisarn, P. Chaisilwattana, W. Siriwasin, and C. M. Black. 2001. Typing of Chlamydia trachomatis strains from urine samples by amplification and sequencing the major outer membrane protein gene (omp1). Sex. Transm. Infect. 77:419-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowden, F. J., B. A. Paterson, J. Mein, J. Savage, C. K. Fairley, S. M. Garland, and S. N. Tabrizi. 1999. Estimating the prevalence of Trichomonas vaginalis, Chlamydia trachomatis, Neisseria gonorrhoeae, and human papillomavirus infection in indigenous women in northern Australia. Sex. Transm. Infect. 75:431-434. [PMC free article] [PubMed] [Google Scholar]
- 4.Cabral, T., A. M. Jolly, and J. L. Wylie. 2003. Chlamydia trachomatis omp1 genotypic diversity and concordance with sexual network data. J. Infect. Dis. 187:279-286. [DOI] [PubMed] [Google Scholar]
- 5.Caldwell, H. D., and J. Schachter. 1982. Antigenic analysis of the major outer membrane protein of Chlamydia spp. Infect. Immun. 35:1024-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cates, W., Jr., and J. N. Wasserheit. 1991. Genital chlamydial infections: epidemiology and reproductive sequelae. Am. J. Obstet. Gynecol. 164:1771-1781. [DOI] [PubMed] [Google Scholar]
- 7.Cevenini, R., M. Donati, and V. Sambri. 2002. Chlamydia trachomatis—the agent. Best Pract. Res. Clin. Obstet. Gynaecol. 16:761-773. [DOI] [PubMed] [Google Scholar]
- 8.Cohen, C. R., and R. C. Brunham. 1999. Pathogenesis of Chlamydia induced pelvic inflammatory disease. Sex. Transm. Infect. 75:21-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.da Cruz, L., I. R. Dadour, I. L. McAllister, A. Jackson, and T. Isaacs. 2002. Seasonal variation in trachoma and bush flies in north-western Australian Aboriginal communities. Clin. Exp. Ophthalmol. 30:80-83. [DOI] [PubMed] [Google Scholar]
- 10.Ewald, D. P., G. V. Hall, and C. C. Franks. 2003. An evaluation of a SAFE-style trachoma control program in central Australia. Med. J. Aust. 178:65-68. [DOI] [PubMed] [Google Scholar]
- 11.Frost, E. H., S. Deslandes, D. Gendron, D. Bourgaux-Ramoisy, and P. Bourgaux. 1995. Variation outside variable segments of the major outer membrane protein distinguishes trachoma from urogenital isolates of the same serovar of Chlamydia trachomatis. Genitourin. Med. 71:18-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garland, S. M., A. Malatt, S. Tabrizi, D. Grando, M. I. Lees, J. H. Andrew, and H. R. Taylor. 1995. Chlamydia trachomatis conjunctivitis. Prevalence and association with genital tract infection. Med. J. Aust. 162:363-366. [PubMed] [Google Scholar]
- 13.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 14.Jonsdottir, K., M. Kristjansson, J. H. Olafsson, and O. Steingrimsson. 2003. The molecular epidemiology of genital Chlamydia trachomatis in the greater Reykjavik area, Iceland. Sex. Transm. Dis. 30:249-256. [DOI] [PubMed] [Google Scholar]
- 15.Jurstrand, M., L. Falk, H. Fredlund, M. Lindberg, P. Olcen, S. Andersson, K. Persson, J. Albert, and A. Backman. 2001. Characterization of Chlamydia trachomatis omp1 genotypes among sexually transmitted disease patients in Sweden. J. Clin. Microbiol. 39:3915-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalayoglu, M. V. 2002. Ocular chlamydial infections: pathogenesis and emerging treatment strategies. Curr. Drug Targets Infect. Disord. 2:85-91. [DOI] [PubMed] [Google Scholar]
- 16a.Lister, N. A., S. N. Tabrizi, C. K. Fairley, A. Smith, P. H. Janssen, and S. Garland. 2004. Variability of the Chlamydia trachomatis omp1 gene detected in samples from men tested in male-only saunas in Melbourne, Australia. J. Clin. Microbiol. 42:2596-2601. [DOI] [PMC free article] [PubMed]
- 17.Mahony, J. B., K. E. Luinstra, J. W. Sellors, and M. A. Chernesky. 1993. Comparison of plasmid- and chromosome-based polymerase chain reaction assays for detecting Chlamydia trachomatis nucleic acids. J. Clin. Microbiol. 31:1753-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ossewaarde, J. M., M. Rieffe, A. de Vries, R. P. Derksen-Nawrocki, H. J. Hooft, G. J. van Doornum, and A. M. van Loon. 1994. Comparison of two panels of monoclonal antibodies for determination of Chlamydia trachomatis serovars. J. Clin. Microbiol. 32:2968-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paterson, B. 2002. Trachoma: new problem or old dilemma? Northern Territory Dis. Control Bull. 9:1-5. [Online.] http://www.nt.gov.au/health/cdc/bulletin. [Google Scholar]
- 20.Pedersen, L. N., H. O. Kjaer, J. K. Moller, T. F. Orntoft, and L. Ostergaard. 2000. High-resolution genotyping of Chlamydia trachomatis from recurrent urogenital infections. J. Clin. Microbiol. 38:3068-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poole, E., and I. Lamont. 1992. Chlamydia trachomatis serovar differentiation by direct sequence analysis of the variable segment 4 region of the major outer membrane protein gene. Infect. Immun. 60:1089-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 23.Stamm, W. E. 1999. Chlamydia trachomatis infections: progress and problems. J. Infect. Dis. 179(Suppl. 2):380-383. [DOI] [PubMed] [Google Scholar]
- 24.Stephens, R. S., R. Sanchez-Pescador, E. A. Wagar, C. Inouye, and M. S. Urdea. 1987. Diversity of Chlamydia trachomatis major outer membrane protein genes. J. Bacteriol. 169:3879-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephens, R. S., E. A. Wagar, and G. K. Schoolnik. 1988. High-resolution mapping of serovar-specific and common antigenic determinants of the major outer membrane protein of Chlamydia trachomatis. J. Exp. Med. 167:817-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stothard, D. R., G. Boguslawski, and R. B. Jones. 1998. Phylogenetic analysis of the Chlamydia trachomatis major outer membrane protein and examination of potential pathogenic determinants. Infect. Immun. 66:3618-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tabbara, K. F. 2001. Trachoma: a review. J. Chemother. 13(Suppl. 1):18-22. [DOI] [PubMed] [Google Scholar]
- 28.Taylor, H. R. 2001. Trachoma in Australia. Med. J. Aust. 175:371-372. [DOI] [PubMed] [Google Scholar]
- 29.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vincelette, J., J. Schirm, M. Bogard, A. Bourgault, D. S. Luijt, A. Bianchi, P. C. van Voorst Vader, A. Butcher, and M. Rosenstraus. 1999. Multicenter evaluation of the fully automated COBAS AMPLICOR PCR test for detection of Chlamydia trachomatis in urogenital specimens. J. Clin. Microbiol. 37:74-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, S. P., C. C. Kuo, R. C. Barnes, R. S. Stephens, and J. T. Grayston. 1985. Immunotyping of Chlamydia trachomatis with monoclonal antibodies. J. Infect. Dis. 152:791-800. [DOI] [PubMed] [Google Scholar]
- 32.Yuan, Y., Y. X. Zhang, N. G. Watkins, and H. D. Caldwell. 1989. Nucleotide and deduced amino acid sequences for the four variable domains of the major outer membrane proteins of the 15 Chlamydia trachomatis serovars. Infect. Immun. 57:1040-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]