Abstract
We report a case of human immunodeficiency virus (HIV) type 1 infection not detected by a highly sensitive combined antigen-antibody assay. The virus was a subtype B strain harboring a unique sequence within the immunodominant epitope of the transmembrane glycoprotein. Immunochemical analysis indicated that this sequence was probably responsible for the failure to detect HIV antibodies.
CASE REPORT
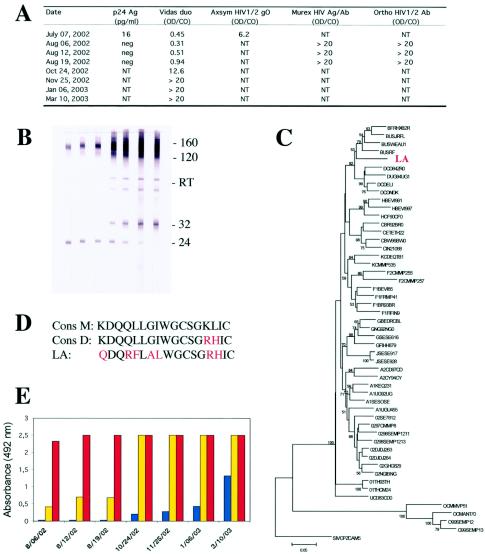
A 31-year-old man (patient LA) consulted for febrile pharyngitis at the beginning of July 2002. He had had unprotected homosexual relationships during the preceding months. The routine test for the detection of anti-human immunodeficiency virus (anti-HIV) antibodies, the Axsym HIV1/2 gO test (Abbott Laboratories, North Chicago, Ill.), gave a positive result, whereas the Vidas Duo test (BioMérieux, Marcy l'Étoile, France) was negative (Fig. 1A). p24 antigen (Vidas HIV p24 II; BioMérieux) was detected (16 pg/ml), but the Western blot (new LAV blot; Bio-Rad, Marnes-la-Coquette, France) was negative. This serological profile, together with clinical and epidemiological data, unambiguously suggested primary HIV infection. The patient was referred to J.-M. Descamps 3 weeks later. At that time, the Vidas Duo test still gave negative results, whereas another combined assay (HIV Ag/Ab; Murex, Dartford, United Kingdom) and a third-generation assay for HIV antibody (HIV1/2 Ab; Ortho Diagnostics, Inc., Raritan, N.J.) both gave strong positive results (Fig. 1A). The viral load was 64,000 copies/ml (Roche Monitor, Branchburg, N.J.). Western blot analysis (Genelabs, Singapore, Singapore) revealed the presence of antibodies against gp160, gp120, and p24. Changes in the serological profile for subsequent blood samples clearly confirmed that primary HIV infection had occurred a few weeks previously (Fig. 1A and B). The Vidas Duo test began to give positive results at the lower limit of the cutoff value (0.33 for a cutoff value equal to 0.35) a few days later, more than a month after the initial detection. Results with this test were clearly positive 2 months later, even though the signal was not strong (Fig. 1A), which is surprising given the recognized high performance of this reagent (4, 6).
FIG. 1.
Serological and molecular characteristics of the reported case of HIV-1 infection not detected by a highly sensitive combined antigen-antibody assay. (A) Results observed with the various screening assays. The ratio of the absorbance of the sample to the cutoff value (OD/CO) is shown for four assays. Values above 1 are positive. The serum sample collection dates are given. Abbreviations: neg, negative; NT, not tested. (B) Western blot analysis of sequential serum samples from patient LA from 6 August 2002 to 10 March 2003. From left to right, the strips are shown in chronological order, with the leftmost strip collected 6 August 2002 (sample collection dates given in panel A). The positions of gp160, gp120, gp32, and p24 (160, 120, 32, and 24, respectively) and reverse transcriptase (RT) are shown to the right of the blot. (C) Phylogenetic analysis of the LA strain. The LA strain is a variant of subtype B. The value at each node indicates the percentage of bootstraps in which the cluster to the right was found. Only values above 50 are shown. (D) Sequences of the immunodominant region of gp41. The consensus sequence of HIV-1 group M (Cons M), the consensus sequence of HIV-1 subtype D (Cons D), and the sequence of the LA strain are shown. Substitutions are shown in red. (E) Binding properties of antibodies present in sequential serum samples from patient LA to peptides of the Cons M (blue), Cons D (yellow), and LA strain (red). The serum sample collection dates are given and start at 6 August 2002.
As all immunoassays designed to detect anti-HIV antibodies contain at least the immunodominant epitope of the transmembrane glycoprotein (gp41) in its native form or a recombinant form or as a synthetic peptide (as in the Vidas Duo test) on the solid phase, we investigated the amino acid sequence of this major antigenic region in the strain carried by this patient. We amplified the gp41 region of the env gene of the LA strain by a nested PCR protocol designed to detect phylogenetically diverse HIV variants (8). The amplified segment was sequenced (469 nucleotides), and the subtype of the strain was determined by the neighbor-joining method. The sequence was compared with 50 reference sequences corresponding to the nine subtypes and major circulating recombinant forms (CRF01-AE and CRF02-AG) of HIV type 1 (HIV-1) groups M and O, available from the HIV sequence database (http://hiv-web.lanl.gov). Distances were calculated with the Kimura two-parameter method, as implemented in the MEGA program. Bootstrap analysis with 100 simulations was used to test the reliability of branching. The LA strain clearly belonged to subtype B (Fig. 1C). Additional phylogenetic analysis of the pol gene (1,269 nucleotides encompassing the protease plus reverse transcriptase regions) confirmed that this strain belonged to subtype B (data not shown). However, although the LA strain was clearly a subtype B variant, it possessed a unique sequence within the immunodominant region, which differed from the group M consensus sequence by seven amino acid substitutions (Fig. 1D). Two of these substitutions, located in the cysteine loop (K601R and L602H), are rare in subtype B but are frequently found in subtype D. Another two of these substitutions, located upstream, were very rare (L592F) or unique (G594A). The G594A mutation has never before been reported in the National Center for Biotechnology Information (NCBI) Protein Database. The other three mutations are frequently encountered (3).
We investigated whether this unique, highly divergent sequence was responsible for the delayed detection of HIV antibodies by the Vidas Duo test by preparing three synthetic peptides overlapping the immunodominant epitope: one corresponded to the group M consensus sequence, one corresponded to the subtype D consensus sequence (containing the K601R and L602H mutations), and one corresponded to the LA strain sequence. Sequential serum samples from patient LA were tested in parallel for these three peptides (1 μg/ml) by an indirect enzyme-linked immunosorbent assay based on a procedure described previously (2). The patient's serum samples reacted strongly with the peptide from the strain with which the patient was infected, right from the first available sample, whereas they did not react with the group M consensus peptide until at least 2 months later and never reached the same intensity of reaction, even several months later (Fig. 1E). Intermediate binding results were obtained with the subtype D peptide that had two important substitutions within the cysteine loop in common with the LA strain.
Combined antigen-antibody assays that simultaneously detect the p24 antigen and HIV-specific antibodies were recently developed as a means of shortening the delay between infection with HIV and laboratory diagnosis. These assays, often referred to as fourth-generation assays, are now widely used to screen for HIV infection because they are clearly the most sensitive assays for the detection of HIV-1 infection (4, 5, 6, 7). It is clear that although considerable progress has been made in the serological detection of HIV infection over the past 18 years, the continuous and rapid genetic evolution of HIV will provide a permanent, ongoing challenge to the efficacy of tools developed by the highly creative researchers working in diagnostics. Previous reports have shown that infections with divergent non-subtype B variants might delay antibody detection (1). This report shows that even HIV infections due to subtype B variants may be detected with some delay, even with the most sensitive combined assays, if genetic drift involves the highly conserved immunodominant region of gp41. Our findings indicate the following. (i) Physicians and clinical biologists must interpret the results of HIV screening assays critically, always taking into account the clinical and epidemiological background for each individual case. (ii) National and international agencies should promote the continuous evaluation and reevaluation of screening assays for HIV infection and monitoring of the genetic evolution of the virus. (iii) Manufacturers of diagnostic tools might be able to improve their assays by including additional consensus sequences representing divergent HIV-1 group M variants. The last point is illustrated here by the ability of the antibody present in early serum samples from our patient to bind the divergent serotype D consensus peptide.
Acknowledgments
This work was supported by funds from the French Ministry of Health and the Agence Nationale de Recherche sur le Sida (ANRS, France).
REFERENCES
- 1.Apetrei, C., I. Loussert-Ajaka, D. Descamps, F. Damond, S. Saragosti, F. Brun-Vézinet, and F. Simon. 1996. Lack of screening test sensitivity during HIV-1 non-subtype B seroconversions. AIDS 10:F57-F60. [DOI] [PubMed] [Google Scholar]
- 2.Barin, F., Y. Lahbabi, L. Buzelay, B. Lejeune, A. Baillou-Beaufils, F. Denis, C. Mathiot, S. M'Boup, V. Vithayasai, U. Dietrich, and A. Goudeau. 1996. Diversity of antibody binding to V3 peptides representing consensus sequences of HIV type 1 genotypes A to E: an approach for HIV type 1 serological subtyping. AIDS Res. Hum. Retrovir. 12:1279-1289. [DOI] [PubMed] [Google Scholar]
- 3.Dorn, J., S. Masciotra, C. Yang, R. Downing, B. Biryahwaho, T. D. Mastro, J. Nkengasong, D. Pieniazek, M. A. Rayfield, D. J. Hu, and R. B. Lal. 2000. Analysis of genetic variability within the immunodominant epitopes of envelope gp41 from human immunodeficiency virus type 1 (HIV-1) group M and its impact on HIV-1 antibody detection. J. Clin. Microbiol. 38:773-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ly, T. D., L. Martin, D. Daghfal, A. Sandridge, D. West, R. Bristow, L. Chalouas, X. Qiu, S. C. Lou, J. C. Hunt, G. Schochetman, and S. G. Devare. 2001. Seven human immunodeficiency virus (HIV) antigen-antibody combination assays: evaluation of HIV seroconversion sensitivity and subtype detection. J. Clin. Microbiol. 39:3122-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saville, R., N. T. Constantine, F. R. Cleghorn, N. Jack, C. Bartholomew, J. Edwards, P. Gomez, and W. A. Blattner. 2001. Fourth-generation enzyme-linked immunosorbent assay for the simultaneous detection of human immunodeficiency virus antigen and antibody. J. Clin. Microbiol. 39:2518-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber, B., E. H. M. Fall, A. Berger, and H. W. Doerr. 1998. Reduction of diagnostic window by new fourth-generation human immunodeficiency virus screening assays. J. Clin. Microbiol. 36:2235-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber, B., L. Gürtler, R. Thorstensson, U. Michl, A. Mühlbacher, P. Bürgisser, R. Villaescusa, A. Eiras, C. Gabriel, H. Stekel, S. Tanprasert, S. Oota, M. J. Silvestre, C. Marques, M. Ladeira, H. Rabeneau, A. Berger, U. Schmitt, and W. Melchior. 2002. Multicenter evaluation of a new automated fourth-generation human immunodeficiency virus screening assay with a sensitive antigen detection module and high specificity. J. Clin. Microbiol. 40:1938-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang, C., D. Piezanek, S. M. Owen, C. Fridlund, J. Nkengasong, T. D. Mastro, M. A. Rayfield, R. Downing, B. Biryawaho, A. Tanuri, L. Zekeng, G. van der Groen, F. Gao, and R. Lal. 1999. Detection of phylogenetically diverse human immunodeficiency virus type 1 groups M and O from plasma by using highly sensitive and specific generic primers. J. Clin. Microbiol. 37:2581-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]