Abstract
Recently, we developed an immunoglobulin G (IgG)-capture BED-enzyme immunoassay (BED-CEIA) to identify recent human immunodeficiency virus (HIV) type 1 (HIV-1) seroconversion for use in incidence estimates. We have established an algorithm for its use; developed quality control reagents to monitor the assay; and evaluated its performance for interrun, intrarun, and operator variability. Analysis of 144 individual plates, which involved multiple plate lots and several operators over more than a year, indicated that the coefficients of variation (CVs) were between 10 and 15% for raw optical density (OD) values in the dynamic range between 0.5 and 2.0 OD units; the CVs decreased to 5 to 10% when the OD was normalized (OD-n; OD-n = specimen OD/calibrator OD). The intrarun CVs were generally in the range of 5 to 10% for specimens with ODs <0.5 and less than 5% for specimens with ODs >0.5. The level of concordance between multiple plate lots (n = 6) and multiple operators (n = 7) was quite high (R2 > 0.9). Comparison of the results of the initial and the confirmatory tests with specimens with OD-n values ≤1.5 demonstrated a high degree of correlation (R2 = 0.92); 566 (92%) of 615 of specimens tested in the two modes retained the same classification (recent or long-term infection). The values for those specimens with changed classifications (n = 49) were close to the cutoff (OD-n = 1.0), as expected. The twofold difference in the HIV IgG contents between the controls and the calibrator reagents was exploited to monitor individual plate runs by using a control plot, which was incorporated into the spreadsheet for data entry and run monitoring. This information provides baseline data for the successful transfer of BED-CEIA to other laboratories and the use of BED-CEIA for the detection of recent HIV seroconversion and the calculation of incidence estimates worldwide.
Recent emphasis on laboratory methods that can be used to detect and distinguish recent human immunodeficiency virus (HIV) type 1 (HIV-1) infections from long-term infections has resulted in exploration of a variety of approaches for estimation of the incidence of HIV-1 infection (1, 2, 4, 6, 10-13; J. Jenner, M. Grazioplene, A. Kazianis, K. Phinney, and B. Werner, Abstr. 10th Conf. Retrovir. Opportunistic Infect., abstr. no. 659, 2003). Although some efforts have been directed at detecting p24 antigen or HIV-1 RNA in the absence of HIV antibodies (1, 4, 10, 12, 13), other approaches are based on qualitative and quantitative differences in the evolution of HIV antibodies following seroconversion (2, 9, 11; Jenner et al., 10th Conf. Retrovir. Opportunistic Infect.).
The development and application of a less sensitive (LS) 3A11 assay provided a first practical approach and permitted detection of recent HIV-1 infection to estimate incidence (2). This assay was based on the increasing HIV antibody titers following seroconversion and distinguished recent from long-term infections on the basis of the antibody levels measured at a 1/20,000 dilution in conjunction with a predefined cutoff (determined with a calibrator specimen). This simple algorithm involving sensitive or LS assays provided a tool that could be used to test a single specimen to detect recent HIV seroconversion. However, the three-step, labor-intensive process of diluting the specimen 1/20,000, the need for dedicated equipment, the subtype-dependent assay performance (7), and the lack of availability of the assay resulted in the evaluation of other approaches, including a less sensitive modification of a 96-well HIV-1 enzyme immunoassay (EIA; Vironostika HIV-1 EIA; Organon Teknika Corp., Durham, N.C.). Although the LS Vironostika EIA works reasonably well with samples from HIV-1 subtype B-infected individuals (3), it does not address the issues related to 1/20,000 dilution and subtype-dependent performance (15). None of the commercial assays are likely to have similar performances with different subtypes because they use an antigen(s) derived from a single subtype.
We evaluated a number of alternative approaches for distinguishing recent from long-term infections (9) and have recently developed a new assay, an immunoglobulin G (IgG)-capture BED-EIA (BED-CEIA), that captures and detects an increasing proportion of HIV IgG in the serum (8). The BED-CEIA was designed by using a branched gp41 peptide (BED) with sequences derived from multiple subtypes to achieve similar performances with the different subtypes. An exhaustive evaluation of more than 600 longitudinal specimens from 139 incident infections allowed us to define a threshold cutoff that detects recent infection in which seroconversion occurred within the previous 160 days. This assay was developed in-house and is not yet commercially available. However, there is considerable interest in the use and application of this assay in various settings.
To enable training and to permit the transfer of the methodology to other laboratories, over the past year we have further refined the protocol and have included additional control reagents to monitor the performance of the assay. We describe here the performance characteristics of the BED-CEIA by assessment of the interrun, intrarun, batch, and operator variabilities and other parameters. These data will assist in the use and interpretation of the results of the assay and should provide a basis for the implementation of BED-CEIA in other laboratories.
MATERIALS AND METHODS
BED-CEIA.
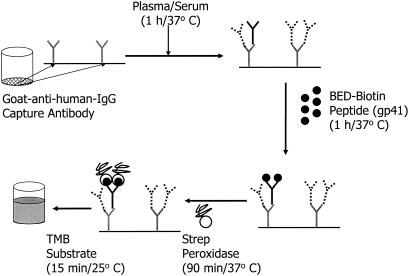
The assay procedure and the rationale for this approach have been described earlier (8). A simplified schematic of the BED-CEIA with the essential steps is shown in Fig. 1. The assay uses a 1/100 dilution of serum or plasma and can be completed in about 4 h with a standard 96-well microplate washer and reader. Due to the quantitative nature of this assay, as opposed to the qualitative nature of diagnostic assays, the procedure, incubation times, and temperatures established for the assay were strictly followed.
FIG. 1.
Schematic showing the various steps of BED-CEIA. Among the antibodies captured, HIV IgG is shown by solid lines, while non-HIV IgG is shown by broken lines. TMB, tetramethylbenzidine; Strep, streptavidin.
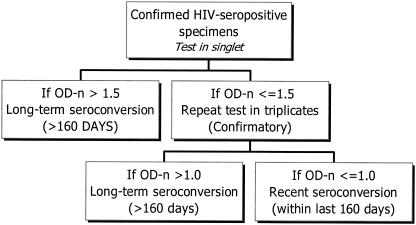
To test cross-sectional specimens, we followed the algorithm shown in Fig. 2. It is similar to the serologic testing algorithm for recent HIV seroconversion by an LS EIA (3). Test sera were initially tested singly. The calibrator (CAL) and control (see below) specimens were tested in triplicate on every plate, and median values were used to calculate the normalized OD (OD-n; OD-n = specimen OD/calibrator OD). Use of the median values ensured that if one of the three replicates was an outlier, it did not affect the results. If the OD-n was >1.5, the specimen was classified as being from a long-term seroconverter. Those specimens with OD-n values ≤1.5 were retested in triplicate to confirm their status (confirmatory mode) by using the median values for the triplicate values to calculate OD-n during the confirmatory mode. During the retesting, if the OD-n was >1.0, the seroconversion was assumed to have occurred more than 160 days before serum collection. Individuals with specimens with OD-n values ≤1.0 were considered to be recently infected, with seroconversion occurring within the last 160 days (8).
FIG. 2.
Algorithm for testing cross-sectional specimens to detect recent HIV-1 infection for incidence estimate purposes. See Materials and Methods for a description of OD-n.
Controls and CAL specimens.
Two plasma specimens, one from an HIV-1-seropositive individual and one from an HIV-seronegative individual, were used to generate controls and a CAL. These specimens were selected because of their adequate availability in our inventory (∼500 sealed vials of each type of lyophilized specimen that had been stored frozen). The contents of the vials were reconstituted to the original volume by adding 200 μl of deionized water. The vials were vortexed and warmed to 37°C to solubilize the contents. High-positive control (HPC), CAL, and low-positive control (LPC) specimens were prepared by using 1/10, 1/20, and 1/40 dilutions of the HIV-1-positive specimen in HIV seronegative plasma, respectively. The relative proportions of HIV IgG in the HPC, CAL, and LPC specimens were 1×, 0.5×, and 0.25× (where the multiplication factor is related to the actual proportion of HIV IgG), respectively, because of the twofold dilution differences among these controls. The negative control (NC) was the HIV-seronegative plasma used to prepare the other controls.
Serum specimens.
During this study, about 3,000 blinded HIV-positive serum specimens from various cross-sectional populations were tested by the BED-CEIA. The specimens were from the United States, Thailand, Cote d'Ivoire, and Uganda and also included commercial sera (Boston Biomedica Inc., Boston, Mass.), which were used for the training. No specimen-specific data are presented here. Data for assessment of the reproducibility and performance characteristics of the BED-CEIA are presented here.
RESULTS
The mean OD-n values for the controls and the CAL for 144 individual plates are shown in Table 1. The CAL, by definition, has a mean OD-n of 1.0, while the HPC and LPC exhibited mean OD-n values of 1.460 and 0.597, respectively. which bracketed the value for the CAL. The coefficients of variation (CVs) for the HPC, CAL, and LPC were 10 to 15% for raw OD values (data not shown) but were reduced to between 5 and 10% when the OD-n was used. The CV was >25% for the NC, which had a mean OD-n close to 0.1. Upper and lower 99% limits were calculated (standard deviation, ±2.58) as shown. The mean OD-n values for the control specimens on multiple plate lots (n = 6) were very similar (data not shown separately), suggesting a stable performance of the assay over multiple plate lots coated at various times and run by seven different operators.
TABLE 1.
Mean OD-n, standard deviation, CV, and 99% confidence intervals for controls and CAL specimensa
| Reagent | Mean (SD) OD-n | % CV | OD-n limit
|
|
|---|---|---|---|---|
| 99% upper | 99% lower | |||
| NC | 0.072 (0.020) | 27.2 | 0.124 | 0.020 |
| LPC | 0.597 (0.048) | 8.0 | 0.721 | 0.473 |
| CAL | 1.000 (0.000) | |||
| HPC | 1.460 (0.079) | 5.41 | 1.664 | 1.256 |
A total of 144 plates were tested.
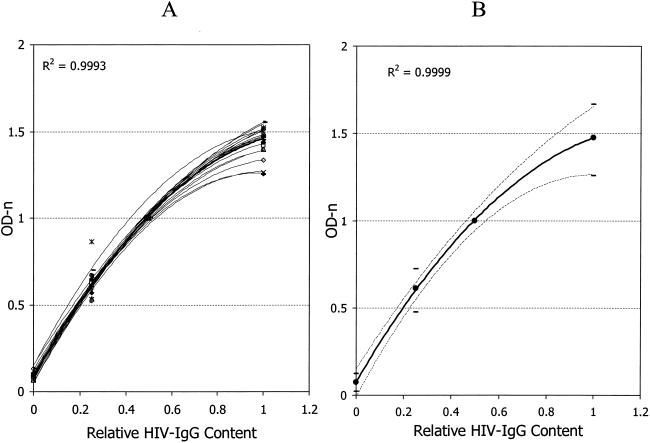
Twofold differences in HIV IgG titers among HPC, CAL, and LPC (1.0-, 0.5-, and 0.25-fold, respectively) were exploited to make control plots (Fig. 3). Data from 24 plates with the best-fit lines are shown in Fig. 3A. Twenty-three of the 24 lines shown are typical of most runs. Figure 3 also shows data for the only plate among the 144 plates for which the test failed. Mean data for 144 plates were used to generate the mean best-fit plot for the controls and CAL and the 99% confidence intervals, as shown in Fig. 3B. The control or CAL values that are out of range would result in a plot outside the 99% limits, and a run with such results should be repeated.
FIG. 3.
Control plots showing the best-fit lines for 24 plates (A) and mean control plot for 144 plates with 99% limits (B). The x axis represents the relative levels of HIV IgG in controls and CAL. R2 values are shown for the best-fit lines for a mean of 24 plates (A) or a mean of 144 plates (B).
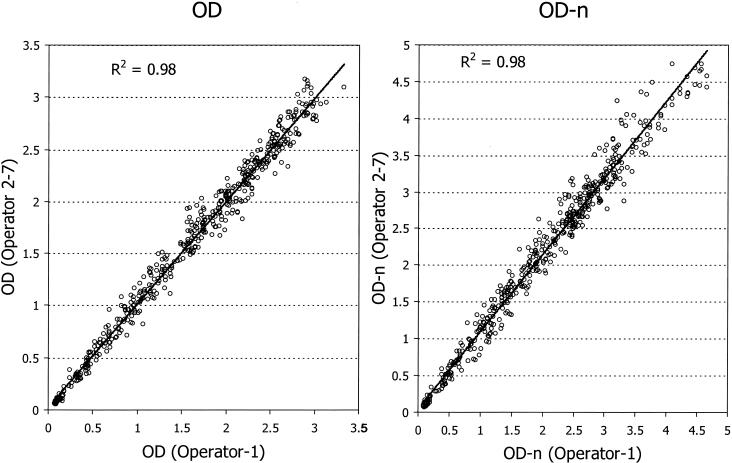
Interoperator variability was examined by comparing raw OD and OD-n values for 504 specimens tested by an experienced technician and a subset tested by one or more trainees (Fig. 4). The reproducibility between two or more operators was outstanding, with R2 equal to 0.98 for both raw OD and OD-n values. Incidentally, these plates were the first set run by new trainees during their training period, suggesting that the BED-CEIA is a simple and highly reproducible assay.
FIG. 4.
Interoperator reproducibility of BED-CEIA runs. Six operators tested a total of 504 specimens during their training periods, and raw OD (left) or OD-n (right) values were compared with the values generated by an experienced operator (operator 1) at the Centers for Disease Control and Prevention.
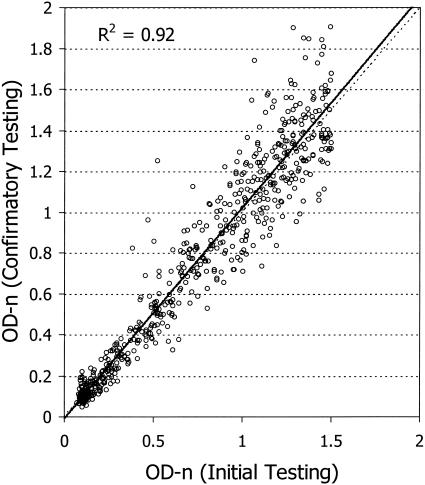
According to our algorithm (Fig. 2), 615 (20.5%) of 3,000 specimens had OD-n values ≤1.5 and therefore were further tested in triplicate for confirmation of the results. The level of concordance between the initial test results and the confirmatory test results was excellent (R2 = 0.92; Fig. 5), with 566 (92%) of 615 specimens retaining the same classification (recent or long-term seroconversion). To put it another way, 385 (12.8%) of 3,000 specimens were classified as being from individuals with recent seroconversion (OD-n, ≤1.0) on the basis of initial testing alone and 388 (12.9%) specimens were classified as being from individuals with recent seroconversion after the confirmatory testing, indicating a high degree of reproducibility. The classification changed for only 49 (8%) specimens upon confirmatory testing, and the values for all specimens were close to the cutoff, with mean values for the initial tests and the confirmatory tests of 1.007 ± 0.28 and 1.004 ± 0.27, respectively. As expected, the values for about equal number of specimens moved from below 1.0 to above 1.0 (n = 23), and vice versa (n = 26).
FIG. 5.
Concordance between the results of initial and confirmatory tests for 625 specimens (R2 = 0.92). The solid line represents the best-fit line, while the broken line represents 100% concordance (R2 = 1.0).
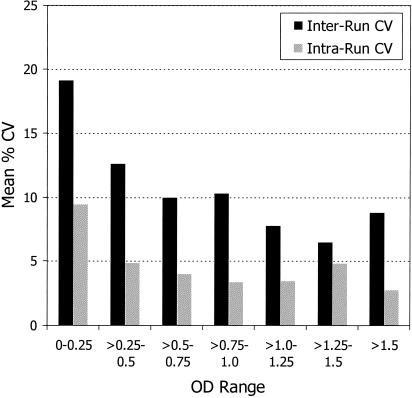
Interrun CVs between the initial test results and the confirmatory test results and the intrarun CVs for triplicates in the confirmatory mode were examined for various OD ranges (Fig. 6). As expected, the mean interrun CVs were generally higher than the mean intrarun CVs for all ranges. Interrun CVs were 10 to 20% for OD values <0.5 but were about 10% or less when specimen OD values were >0.5. Mean intrarun CVs were about 10% for specimens with OD values <0.25 but were less than 5% for specimens with OD values >0.25. A set of eight specimens with various OD values was also run in replicates of 12. Again, the intrarun CVs were similar to those shown in Fig. 6 (data not shown).
FIG. 6.
Mean interrun CVs between initial and confirmatory tests and mean intrarun CVs for specimens tested in triplicate at various ranges of OD values.
DISCUSSION
The application of laboratory methods to detect incident infections gained increased attention following a report by Janssen et al. (2) demonstrating that simple modifications in the assay protocol of commercial EIAs can be cleverly used to detect recent HIV infection. Many laboratories in the United States and elsewhere have been using versions of LS EIAs (3, 5, 11, 14, 15) to detect recently infected people and to estimate the incidence of HIV infections. However, there are limitations to this approach. These include (i) significant variability between runs and among laboratories performing the assays (3); (ii) the need to perform a three-step, 1/20,000 dilution, which is labor-intensive and which contributes to high CVs; and (iii) subtype-dependent performance (7, 15), which limits its use in international settings where the populations are infected with divergent HIV subtypes.
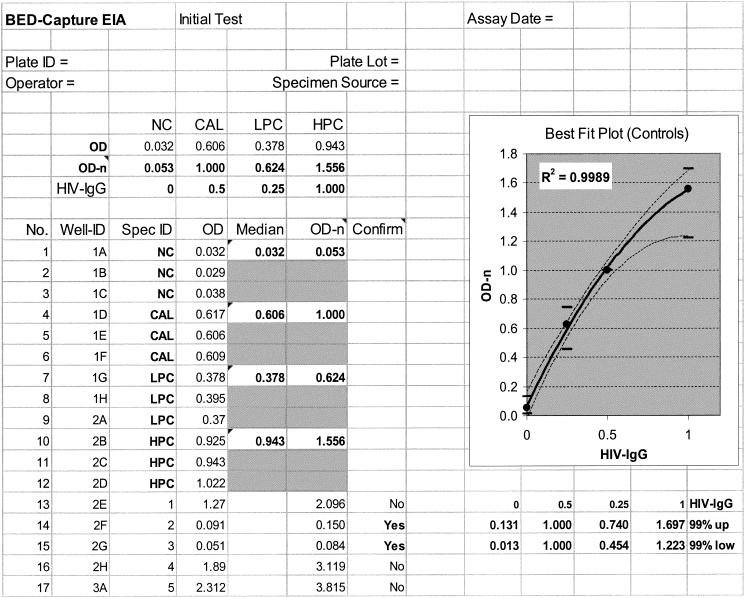
Development of the BED-CEIA to detect recent HIV infection by indirectly measuring the increasing proportion of HIV IgG following seroconversion addresses many of these issues (8). With data collected over 18 months, we demonstrate here that BED-CEIA is a very stable assay with minimal variation and outstanding interrun, intrarun, and interoperator CVs (Table 1; Fig. 4 to 6). Not only does the assay format permit the use of a 1/100 dilution of serum, but also more than twofold differences in dilutions are well tolerated, as long as the proportions of HIV IgG and non-HIV IgG do not change (data not shown). A multisubtype antigen (branched gp41 peptide; BED) was used in the assay to permit the equivalent detection of increasing antibody levels in individuals infected with each of the different subtypes. Our recent work with individuals infected with divergent HIV-1 subtypes (subtypes A through E) suggests that the assay performs similarly for individuals infected with different subtypes (unpublished data). It has recently been reported that both the LS 3A11 EIA (7) and the LS Vironostika EIA (15) have subtype-dependent performances; hence, their usefulness in countries where the different subtypes are present remains questionable. We have developed a CAL and other appropriate control reagents (HPC, LPC, and NC) that were run in triplicate on every plate. The ODs for the HPC and LPC reagents were designed to bracket the OD for the CAL, which is used to calibrate the assay and define the threshold cutoff value. These reagents are also used to validate the runs, to calculate the interrun CVs and 99% confidence limits, and to define the criteria for the acceptance or rejection of runs. Only 1 of 144 plates run over the 18 months of the study was rejected (the ODs were beyond the 99% limits). In addition, we have developed a spreadsheet that incorporates a control plot for quality assurance and analysis of the raw data (Fig. 7). This spreadsheet allows instant validation of the results for each plate and a systematic approach to data management. We recommend use of the control and CAL OD-n values derived from the last 20 plates instead of the values derived from the first 20 plates to establish the mean and the 99% confidence limits. This process accounts for lot-to-lot changes in one or more reagents or controls. Any major changes in raw OD values for the controls should be carefully examined to assess the performance of the assay.
FIG. 7.
Representation of a spreadsheet designed for data processing and quality assurance. Raw OD values are transferred electronically, and the best-fit plot indicating the acceptance or rejection of the data for the plate is generated.
Our analysis of longitudinal specimens containing subtypes B and E has shown that when an OD-n of 1.0 is used as the cutoff, HIV seroconversion is likely to have occurred in the last 160 days (8). As reported earlier (8), use of a more stringent cutoff or seroconversion period may be used for specific applications to increase the predictive value of the detection of recent infection. However, changes in the cutoff or seroconversion duration do not affect the overall performance or CV of the assay.
It is noteworthy that the commercial assays being used or considered for use for detection of recent infection are designed as qualitative assays. Therefore, the kit reagents are prepared for a qualitative answer (yes or no) and have a wider range of values for acceptability. This can result in increased variability when such assays are modified (as in the LS EIAs) to gain a quantitative answer. The BED-CEIA, in contrast, is designed to be a quantitative EIA and therefore should be more reproducible. Since most cross-sectional specimen sets include specimens from individuals exhibiting antibodies at various levels, the precision of the methodology becomes more important. In addition, our procedure for normalizing the OD value by a simple ratio (specimen OD/CAL OD) resulted in better reproducibility. Subtraction of the OD value for the NC from the OD values for the specimen and CAL (as is done by the LS EIA protocol) resulted in more variability (data not shown).
Although the BED-CEIA has outstanding performance characteristics, it is still an in-house assay with limited availability. Some of the crucial reagents (e.g., the BED-biotin peptide and the CAL and control reagents) are available only from our laboratory; however, we plan to develop an assay kit for wider availability. This will allow us to conduct studies of the stabilities of the reagents in a kit format. We anticipate that the actual variability of the test among laboratories will be higher than that shown here as more laboratories begin to perform the assay. However, these data provide guidance for monitoring the performance of the test to minimize assay variations. Personnel from several laboratories have successfully been trained over the last year, and our data indicate that the reproducibility during the training was excellent (Fig. 4). Subsequently, the assay has successfully been transferred to six laboratories. This description of the performance characteristics of BED-CEIA should further help with training and the transfer of this technology to other laboratories and expansion of the use of this approach for the detection of recent HIV seroconversion.
Acknowledgments
We thank Jon Kaplan for comments and Kim Distel for editorial assistance.
REFERENCES
- 1.Courouce, A. M., F. Barin, M. Maniez, C. Janot, L. Noel, M. H. Elghouzzi, et al. 1992. Effectiveness of assays for antibodies to HIV and p24 antigen to detect very recent HIV infections in blood donors. AIDS 6:1548-1550. [DOI] [PubMed] [Google Scholar]
- 2.Janssen, R. S., G. A. Satten, S. L. Stramer, B. D. Rawal, T. R. O'Brien, B. J. Weiblen, F. M. Hecht, N. Jack, F. R. Cleghorn, J. O. Kahn, M. A. Chesney, and M. P. Busch. 1998. New testing strategy to detect early HIV-1 infection for use in incidence estimates and for clinical and prevention purposes. JAMA 280:42-48. [DOI] [PubMed] [Google Scholar]
- 3.Kothe, D., R. H. Byers, S. P. Caudill, G. A. Satten, R. J. Janssen, H. Hannon, and J. V. Mei. 2003. Performance characterstics of a new less-sensitive HIV-1 EIA for use in estimating HIV seroincidence. J. Acquir. Immune Defic. Syndr. 33:625-634. [DOI] [PubMed] [Google Scholar]
- 4.More, D., K. O'Brien, and E. Walter. 2000. Utility of an HIV-1 RNA assay in the diagnosis of acute retroviral syndrome. South. Med. J. 93:1004-1006. [PubMed] [Google Scholar]
- 5.Murphy, G., J. V. Parry, S. B. Gupta, C. Graham, L. F. Jordan, A. N. Nicoll, and O. N. Gill. 2001. Test of HIV incidence shows continuing HIV transmission in homosexual/bisexual men in England and Wales. Communicable Dis. Public Health 4:33-37. [PubMed] [Google Scholar]
- 6.Parekh, B. S., and J. S. McDougal. 2001. New approaches for detecting recent HIV-1 infection. AIDS Rev. 3:183-193. [Google Scholar]
- 7.Parekh, B. S., D. J. Hu, S. Vanichseni, G. A. Satten, D. Candal, N. L. Young, D. Kitayaporn, L. O. Srisuwanvilai, S. Rakhtam, R. Janssen, K. Choopanya, and T. D. Mastro. 2001. Evaluation of a sensitive/less-sensitive testing algorithm using the 3A11-LS assay for detecting recent HIV seroconversion among individuals with HIV-1 subtype B or E infection in Thailand. AIDS Res. Hum. Retrovir. 17:453-458. [DOI] [PubMed] [Google Scholar]
- 8.Parekh, B. S., M. S. Kennedy, T. Dobbs, C. P. Pau, R. Byers, T. Green, D. J. Hu, S. Vanichseni, N. L. Young, K. Choopanya, T. D. Mastro, and J. S. McDougal. 2002. Quantitative detection of increasing HIV type 1 antibodies after seroconversion: a simple assay for detecting recent HIV infection and estimating incidence. AIDS Res. Hum. Retrovir. 18:295-307. [DOI] [PubMed] [Google Scholar]
- 9.Parekh, B. S., C. P. Pau, M. S. Kennedy, T. L. Dobbs, and J. S. McDougal. 2001. Assessment of antibody assays for identifying and distinguishing recent from long-term HIV type 1 infection. AIDS Res. Hum. Retrovir. 17:137-146. [DOI] [PubMed] [Google Scholar]
- 10.Quinn, T. C., R. Brookmeyer, R. Kline, M. Shepherd, R. Paranjape, S. Mehendale, D. A. Gadkari, and R. Bollinger. 2000. Feasibility of pooling sera for HIV-1 viral RNA to diagnose acute primary HIV-1 infection and estimate HIV incidence. AIDS 14:2751-2757. [DOI] [PubMed] [Google Scholar]
- 11.Rawal, B. D., A. Degula, L. Lebedeva, R. S. Janssen, F. M. Hecht, H. W. Sheppard, and M. P. Busch. 2003. Development of a new less-sensitive enzyme immunoassay for detection of early HIV-1 infection. J. Acquir. Immune Defic. Syndr. 33:349-355. [DOI] [PubMed] [Google Scholar]
- 12.Thies, K., C. Anders, M. Baldus, T. Schleiffer, B. Weber, H. Rabenau, and P. Hellstern. 1994. Detection of primary HIV infection by a second-generation HIV (p24) antigen test. Infusionsther. Transfusionsmed. 21:333-336. [DOI] [PubMed] [Google Scholar]
- 13.Thomas, H. I., S. Wilson, C. M. O'Toole, C. M. Lister, A. M. Saeed, R. P. Watkins, and P. Morgan-Capner. 1996. Differential maturation of avidity of IgG antibodies to gp41, p24 and p17 following infection with HIV-1. Clin. Exp. Immunol. 103:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinstock, H., M. Dale, M. Gwinn, G. A. Satten, D. Kothe, J. Mei, J. Royalty, L. Linley, C. Fridlund, B. Parekh, B. D. Rawal, M. P. Busch, and R. S. Janssen. 2002. HIV seroincidence among patients at clinics for sexually transmitted diseases in nine cities in the United States. J. Acquir. Immune Defic. Syndr. 29:478-483. [DOI] [PubMed] [Google Scholar]
- 15.Young, C. L., D. J. Hu, R. Byers, S. Vanichseni, N. L. Young, R. Nelson, P. A. Mock, K. Choopanya, R. Janssen, T. D. Mastro, and J. V. Mei. 2003. Evaluation of a sensitive/less-sensitive testing algorithm using the bioMerieux Vironostika-LS assay for detecting recent HIV-1 subtype B′ or E infection in Thailand. AIDS Res. Hum. Retrovir. 19:481-486. [DOI] [PubMed] [Google Scholar]