Abstract
Implantation of biomaterials elicits a foreign body response characterized by fusion of macrophages to form foreign body giant cells and fibrotic encapsulation. Studies of macrophage polarization in this response have suggested that alternative (M2) activation is associated with more favorable outcomes. Here we investigated this process in vivo by implanting mixed cellulose ester filters or PDMS disks in the peritoneal cavity of WT and MCP-1 KO mice. We analyzed classical (M1) and alternative (M2) gene expression via Q-PCR, immunohistochemistry, and ELISA in both non-adherent cells isolated by lavage and implant-adherent cells. Our results show that macrophages undergo unique activation that displays features of both M1 and M2 polarization including induction of TNF, which induces the expression and nuclear translocation of p50 and RelA determined by immunofluorescence and western blot. Both processes were compromised in fusion-deficient MCP-1 KO macrophages in vitro and in vivo. Furthermore, inclusion of BAY 11-7028, an inhibitor of NFκB activation, reduced nuclear translocation of RelA and fusion in WT macrophages. Our studies suggest that peritoneal implants elicit a unique macrophage polarization phenotype leading to induction of TNF and activation of the NFκB pathway.
Keywords: Macrophage, Inflammation, Foreign body response, Foreign body giant cell
1. Introduction
Macrophage polarization in inflammatory settings has been linked to various outcomes including fibrosis [1–3]. Specifically, it has been suggested that macrophages can engage one of two major activation pathways, the classical (M1) or alternative (M2), which are associated with infections and tissue repair, respectively [4]. Typically, M1 macrophages are induced in response to interferon-γ (IFN) and express pro-inflammatory IL-6. In contrast, M2 macrophages are induced in response to IL-4 and express anti-inflammatory IL-10. However, it is now appreciated that macrophages can display variable activation states in a continuum between M1 and M2 [5–8]. Consistent with this suggestion, several studies have shown that macrophages can acquire intermediate activation states in tissue injury models [4, 7, 9, 10]. Therefore, our understanding of macrophage activation and its contribution to outcomes and resolution is incomplete.
In the foreign body response (FBR), which is elicited following the implantation of biomaterials and devices in vascularized tissues, macrophages encounter a complex wound-like environment that includes the surface of the implant [11–14]. It is appreciated that adsorbed proteins mediate the interactions of cells with such surfaces and are capable of inducing macrophage activation, perhaps skewed towards the M2 phenotype, following the acute phase [1]. In fact, a recent study has demonstrated attenuation of the foreign body response and a predominant M2 macrophage phenotype following implantation of low protein adsorbing zwitterion-based hydrogels [15]. Moreover, macrophages on the surface of implants can undergo fusion to form foreign body giant cells (FBGC), which are a hallmark of the FBR [11, 16]. Because FBGC formation can be induced in vitro by exposure of macrophages to IL-4, it is tempting to consider significant overlap between FBGC and M2 macrophages. In fact, several studies of macrophage phenotype have suggested that a higher percentage of M2 macrophages is inversely proportional to scar tissue formation and implant encapsulation [15, 17–21]. Moreover, predominantly M2 macrophages were observed in the response to porous materials that elicit tissue integration and increased neovascularization or to cross-linked collagen disks [22, 23].
We have previously shown that mice deficient in MCP-1 fail to mount a fibrotic reaction in a foreign body response model [24]. Specifically, we showed minimal fibrotic encapsulation of foreign bodies implanted in the peritoneal cavity. Ultrastructural evaluation of MCP-1 KO macrophages revealed reduced activation despite extensive contact with implants. In addition, these cells failed to induce MMP-9, which is required for fusion and is considered a marker for M2 macrophages. Parallel in vitro studies allowed us to describe a defect in the induction of TNF, which was required for the expression of MMP9. In addition, we showed that addition of TNF to MCP-1 KO macrophages could induced MMP9 and rescue the adhesion defect. Therefore, we postulated that MCP-1 KO macrophages failed to undergo proper activation and assumed a state that was incapable of inducing pro-fibrotic signals. In the present study, we examined the recruitment and activation of MCP-1 KO and WT macrophages in the context of the FBR. We discovered that WT macrophages undergo complex activation including induction of TNF and activation of the canonical NFκB pathway. In contrast, MCP-1 KO macrophages failed to induce TNF or activate NFκB. Pharmacological inhibition of NFκB in WT macrophages limited induction and nuclear translocation of RelA and overall fusion indicating the importance of this pathway. Moreover, the levels of implant-associated TGF-β in MCP-1 KO mice were reduced, providing a possible explanation for the diminished fibrotic response.
2. Materials and Methods
2.1 Materials
Anti-mouse F4/80 Ab (Cl:A3-1) and anti-TGF-β (AHP1734) was purchased from Serotec (Raleigh, NC). Anti-p50 (ab7971), anti-p65/RelA (ab7970), anti-iNOS (ab3523), and anti- TNF-α (ab 6671) were purchased from Abcam (Cambridge, Massachusetts). F4/80-FITC-conjugate (BM8), CD36-PE-conjugate (No.72-1) were purchased from eBioscience (San Diego, California). Anti- IL-1β Ab (AF-401-NA), recombinant mouse (rm) GM-CSF (rm GM-CSF), rm IL-4, and Duoset TNF-α ELISA kit were purchased from R&D Systems (Minneapolis, MN). Anti-phospho p65/RelA (3033) was purchased from Cell Signaling (Danvers, MA). Anti-Arg1 (sc-18354) was purchased from Santa Cruz Biotechnology Inc. (Dallas, TX). Vectastain ABC kit for immunohistochemistry and Vectashield mounting medium was purchased from Vector Laboratories (Burlingame, CA). May-Grunwald and Wright-Giemsa stains were obtained from Sigma (St. Louis, Missouri). Filters (0.45-μm pore size, mixed cellulose ester) were acquired from Millipore (Billerica, MA) and PDMS from Invotec International (Jacksonville, FL). PDMS disks (4 mm in diameter and 1 mm thick) were cut with the aid of a biopsy punch (Acuderm; Fort Lauderdale, FL).
2.2 Implantation of Biomaterials
All procedures were performed in accordance with the regulations adopted by the National Institutes of Health and approved by the Animal Care and Use Committee of Yale University. IP implantations were performed as described previously [24]. A total of 60 MCP-1 KO and 60 WT mice were used for in vivo experiments. Mice were age-matched (3–4 mo of age) in all experiments. For implantation of filters or PDMS disks, each mouse received one 25 mm2 Millipore filter or a 4 mm diameter disk in the peritoneum through a 0.7 cm incision in the skin and peritoneum. The peritoneum was closed with sterile sutures (silk, size 4.0) and the skin with sterile clips. Implants were excised en bloc at 2, 4 and 7 d after implantation. For each experiment, five mice per time point per genotype per implant type were used (n = 5). Filters were used predominantly for histological analysis due to their ability to remain in situ during sectioning and staining and thus preserving the tissue-biomaterial interface. PDMS disks were used for histology and confocal microscopy.
2.3 Histology, Immunohistochemistry, and confocal microscopy
Explanted biomaterials were processed and stained as described previously [25]. PDMS disks and Millipore filter implants were analyzed by confocal microscopy. Millipore filter implants were processed for histological analysis and sectioned as described previously [24]. As stated above, we have shown previously that the use of filters is preferable to preserve the tissue-implant interface. Confocal microscopy was performed on d 4 samples to detect Mac3, Arg1, and iNOS. Immunohistochemistry was performed to detect IL-1β, TNF-α, TGF-β, p50 and p65/RelA. All primary antibodies were diluted in 1% BSA in PBS. Secondary FITC- or TRITC-conjugated antibodies were used at a 1:500 dilution (Life Technologies). Controls included sections stained with either isotype control non-specific Ab, only primary, or secondary Ab and revealed minimal background (not shown). Nuclei and cytoskeleton were visualized with 4,6 diamidino-2-phenylindole (DAPI) (1:1000 dilution; Sigma) and rhodamine-phalloidin (1:1000 dilution; Molecular Probes), respectively.
2.4 Flow Cytometry
Flow cytometry analysis was performed on samples retrieved by peritoneal lavage at 2, 4, and 7 d following implantation as described previously [26]. Briefly, 5 × 105 cells were resuspended in FACS buffer (1% BSA in PBS, 0.01% NaN3) and placed in CD16/CD32 (Fc block; Becton Dickinson) for 20 min at 4°C. Cells were fixed in 1% formaldehyde and then stained with anti-mouse F4/80 FITC Ab or an isotype control Ab to evaluate macrophage accumulation into the peritoneal cavity. For in vivo experiments, five mice per time point per genotype were used and the experiment was performed in triplicate. Dead/apoptotic cells were detected by 7AAD viability staining solution (eBioscience) according to supplier’s instructions. Peritoneal macrophages isolated on d2 post-implantation (5×105cells per sample), were also stained for CD-36 (CD36-PE (eBioscience)) following exposure to 10 ng/ml IL-4. Flow cytometry samples were acquired on an LSRII (BD Biosciences) and analysis was performed with FlowJo software (Tree Star, Ashland, OR, USA).
2.5 QPCR analysis
Mice bearing implants were lavaged with 2 ml PBS at 2, 4 and 7 d post implantation and the implants and peritoneal lavage fluid were collected. Cell isolates from lavage samples were prepared as described previously [26]. RNA was extracted from implants or lavage cell isolates with the RNeasy Mini Kit from Qiagen (Valecia, CA). 1μg of total RNA per sample was translated into single-stranded cDNA using the Superscript II cDNA synthesis kit and Oligo-dT both from Invitrogen (Carlsbad, CA), according to supplier’s instructions. Gene expression levels were determined using the IQ5 multi color real-time PCR and SYBR green supermix from Bio-Rad Laboratories, Inc. (Hercules, CA). The amplification of each sample was performed in duplicate. Data was normalized to the levels of the housekeeping gene GAPDH and displayed normal distribution. Mouse-specific primers used to amplify mRNA sequences were:
TNF-α forward 5′-CCCTCACACTCAGATCATCTTCT- 3′ and reverse 5′-GCTACGACGTGGGCTACAG -3′;
IL-1β forward: 5′-CTCCATGAGCTTTGTACAAGG-3′; and reverse: 5′-TGCTGATGTACCAGTTGGGG-3′;
IL-10 forward 5′-CCAAGCCTTATCGGAAATGA -3′ and reverse 5′TTTTCACAGGGGAGAAATCG -3′;
IL-6 forward 5′ AGTTGCCTTCTTGGGACTGA -3′ and reverse 5′ – TCCACGATTTCCCAGAGAAC -3′;
Arg1 forward 5′ TTGGGTGGATGCTCACACTG -3′and reverse 5′ – TTGCCCATGCAGATTCCC -3′;
iNOS forward 5′ CAGCTGGGCTGTACAAACCTT -3′ and reverse 5′ – CATTGGAAGTGAAGCGTTTCG -3′;
CD36 forward 5′ TCCTCTGACATTTGCAGGTCTATC -3′ and reverse 5′ – AAAGGCATTGGCTGGAAGAA -3′;
Ym1 forward 5′ GGCTCAGTGGCTCAAGGACAAC -3′ and reverse 5′ – AAAGTAGATGTCAGAGGGAAAT -3′;
GAPDH forward 5′-TGGCATTGTGGAAGGGCTCATGAC-3′ and reverse 5′ ATGCCAGTGAGCTTCCCGTTCAGC-3′.
Fold-change was determined in comparison to WT samples at 24 h. A total of five mice per time point per genotype were analyzed and the experiment was performed twice.
2.6 In Vitro Fusion Assay, immunofluorescence, and western blot
Mouse macrophages were expanded from the bone marrow of MCP-1 KO and WT mice as described previously [24]. Briefly, marrow was flushed from the femurs of mice and collected in Iscove’s modified Dulbecco’s medium (Life Technologies, Inc., Grand Island, NY) supplemented with 10% fetal bovine serum and penicillin / streptomycin / fungizone. Subsequently, the mononuclear cell fraction was collected by centrifugation over Lympholyte-M (Accurate Chemical, Westbury, NY) according to supplier’s instructions. Mononuclear cells were expanded in non-tissue culture treated polystyrene plates (BD, Franklin Lakes, NJ) in the presence of 1.5 ng/ml huM-CSF (R&D Systems, Minneapolis, MN) and 100 ng/ml flt-3 ligand (R&D Systems). Cells were fed on d5 and collected by scraping on d 10. 5 × 105 of expanded cells/well were plated in 24-well non-tissue culture treated polystyrene plates (BD) in media containing 10 ng/ml rm IL-4. Numerous studies have demonstrated robust macrophage fusion utilizing this protocol [24, 27, 28]. In select experiments, macrophages were plated on tissue cultured-treated polystyrene, which is suboptimal for IL-4-induced macrophage fusion [16]. At d 1, 2, or 3 cells were fixed and permeabilized, blocked, and incubated overnight at 4°C with anti-mouse p50 or anti-mouse RelA (1:500, Abcam). FITC-conjucated secondary antibodies (1:500; Life Sciences) were used for detection. Nuclei and cytoskeleton were stained with DAPI and rhodamine-phalloidin, respectively, as described above. Cells were imaged with the aid of an Axiovert 200M Zeiss microscope equipped with fluorescent optics. Confocal images were taken using a Leica TCS SP5 Spectral confocal microscope and Volocity software (Perkin Elmer). In addition, cell lysates were collected on d3 and analyzed by western blot for RelA. Blots were also analyzed for β-actin as a loading control.
2.7 Pharmacological inhibition of NFκB
Expanded macrophages were induced to fuse with 10ng/ml rmIL-4 (R&D) as described above. Selected wells were treated with either 5 μM BAY 11-7082 (BAY 11) (Santa Cruz) dissolved in DMSO or DMSO alone (vehicle control). A range of BAY 11 doses (1–20 μM) was tested and 5 μM was found to be the lowest that could inhibit fusion (not shown). Fusion was evaluated at d 5 in wells stained with Wright-Giemsa and May-Grunwald (Sigma) as described previously [24]. In addition, induction and subcellular localization of RelA was detected in d 3 samples by confocal microscopy as described above. For quantification of fusion, 10 images per well and three wells per condition were analyzed to determine the number of FBGC, the number of nuclei per FBGC, and % Fusion.
2.8 Determination of cytokine concentration by ELISA
Lavage fluid was collected on d 2, 4, and 7, centrifuged to remove cells and analyzed by ELISA for the levels of TNF, IL-6, and IL-10 according to supplier’s instructions (eBioscience). Protein extracts were generated by lysis of tissues surrounding d7 implants in RIPA buffer and analyzed by ELISA for TGF-β according to supplier’s instructions (R&D Systems).
3. Statistical Analysis
Data are expressed as the mean ± the standard error of the mean. The student t-test test and ANOVA were used to assess statistical significance. p-values ≤ 0.05 were considered significant.
4. Results
4.1 Macrophage polarization following interaction with biomaterials
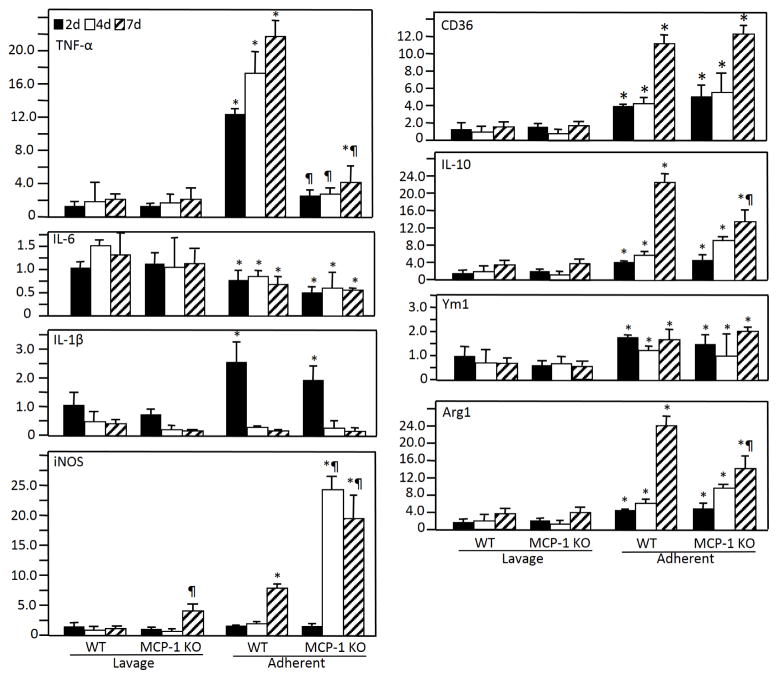
Monocyte recruitment to the peritoneal cavity was monitored by FACS of lavage fluid collected on d 2, 4, and 7 post implantation with F4/80 antibody. Accumulation of F4/80-positive cells increased over time and was not impaired in MCP-1 KO mice (Supplemental Figure 1A). In addition, analysis of CD36 surface expression following exposure of peritoneal macrophages to IL-4 indicated similar activation phenotypes in WT and MCP-1 KO recruited cells (Supplemental Figure 1B). Based on our previous observation of normal initial macrophage accumulation on implants in MCP-1 KO mice, we hypothesized that post-adhesion macrophage defects could influence the FBR phenotype [24]. Therefore, we compared the ability of WT and MCP-1 KO cells to undergo activation following adherence to implants. We focused on molecular markers associated with M1 (iNOS, IL-1β, IL-6, and TNF) and M2 (arginase 1, CD36, IL-10, and Ym1) activation and determined their expression by Q-PCR (Figure 1). This approach allowed the detection of differences between non-adherent and adherent cells, as well as between WT and MCP-1 KO cells. Comparison of non-adherent and adherent WT cells indicated a distinct activation in the latter in a manner that included features of both M1 and M2. Specifically, expression of CD36, Arg1, and IL-10 were elevated in adherent WT cells whereas IL-6, which is M1-associated, was not. However, WT adherent cells also induced expression of IL-1β, iNOS and TNF, which are typically associated with M1. MCP-1 KO cells exhibited an activation profile similar to WT cells with two notable exceptions involving a significant increase in iNOS and lack of TNF induction. Consistent with an increase in iNOS expression, MCP-1 KO cells induced Arg1 to a lesser extent than WT cells. A reduction in IL-10 levels was also observed at d 7 in MCP-1 KO cells but the overall trend in expression from d 2 to d 7 was similar to that in WT cells. Collectively, these observations indicate the inability of MCP-1 KO macrophages to induce TNF expression following adhesion to biomaterial surfaces.
Figure 1.
mRNA expression of M1 and M2 polarization markers in non-adherent (lavage) and implant-adherent cells at 2, 4, and 7 days after implantation. At each time point, five mice per genotype were used. Analysis was performed in triplicate. Fold-change was determined in comparison to WT samples at 24 h. Results are given as mean (SEM) fold change and are representative of two independent experiments that were normalized relative to the housekeeping gene GAPDH (n = 5). * indicates p ≤ 0.05 when comparing lavage and adherent cells and ¶ indicates p ≤ 0.05 when comparing WT and MCP-1 KO samples, respectively.
Previously, we demonstrated by immunohistochemical analysis that the cellular accumulation on d 4 and d 7 implants in this model is dominated by macrophages [24]. Consistent with our previous findings, confocal microscopy analysis showed the bulk of the cells on d 4 implants were Mac3-positive (Supplemental Figure 2 A–F). Therefore, we conclude that the bulk of the gene expression, especially in d 4 samples, is derived from macrophages. In addition, double immunofluorescence detection of iNOS and Arg1 revealed no overlap suggesting that these molecules are expressed on different cells (Supplemental Figure 2 G–H).
4.2 Cytokine expression during the FBR
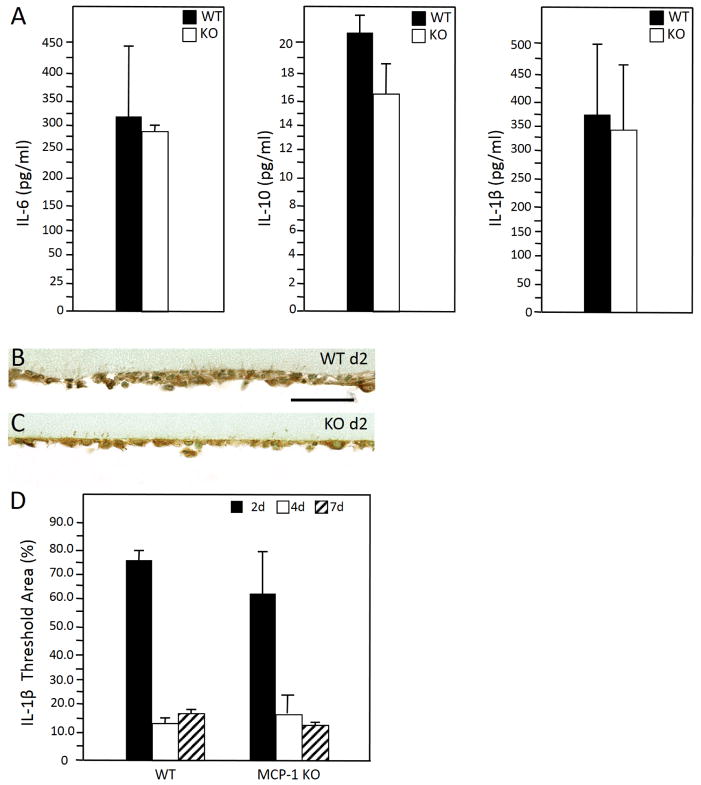
To confirm the Q-PCR data for transcripts associated with inflammation, we analyzed the levels of IL-6, IL-10, and IL-1β at 48 hr in lavage fluids via ELISA. Similar to Q-PCR analysis of non-adherent and adherent cells, we did not detect significant differences between WT and MCP-1 KO cells (Figure 2A).
Figure 2.
(A) Enzyme-linked immunosorbent assay analysis of d 2 lavage fluid revealed similar levels of IL-6, IL-10 and IL-1β in WT and MCP1-KO mice. (B – C) Representative images of d 2 peritoneal implants from WT (B) and MCP-1 KO (C) mice stained with anti-IL-1β Ab. Immunoreactivity was detected using the peroxidase reaction (brown), and nuclei were counterstained with methyl green. (D) Image analysis of samples from 2,4, and 7 d revealed similar IL-1β expression in WT and MCP-1 KO mice. (n = 5). Scale bar = 50 μm.
Previously, we observed reduced encapsulation of implants in the IP model in MCP-1 KO mice. Here we observed reduced accumulation of mature collagen fibers around collagen implants in these mice (Supplemental Figure 3). Based on our findings above, we speculate that the reduced fibrosis was primarily to a defect in macrophage activation and not recruitment. Because cell- biomaterial interactions have been shown to induce IL-1β via inflammasome activation [29], we also analyzed its expression in adherent cells by immunohistochemistry at d 2, 4, and 7 and observed similar levels in WT and MCP-1 KO mice (Figure 2 B–D).
4.3 TNF and the NFκB pathway
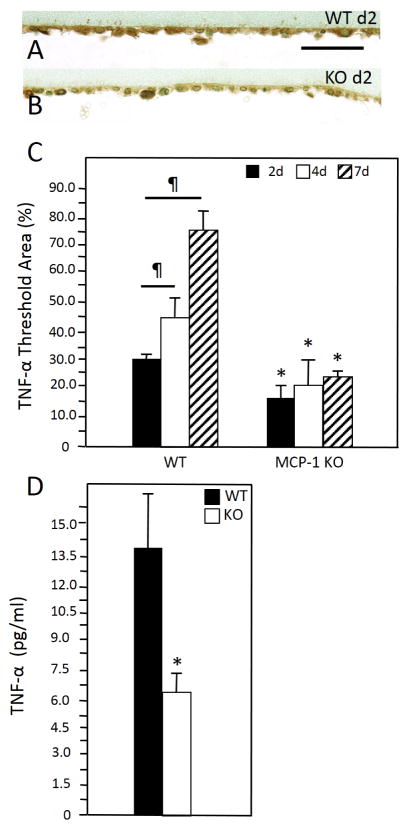
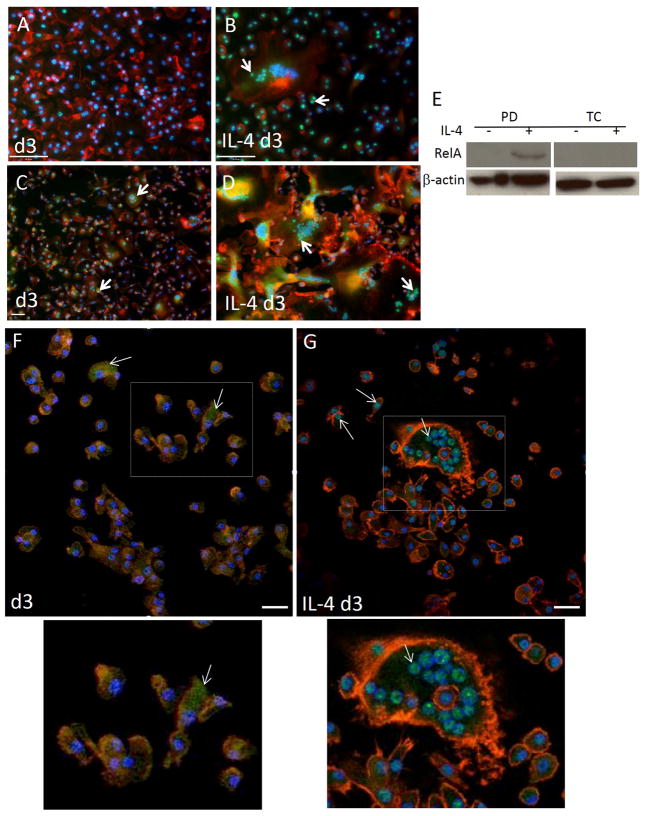
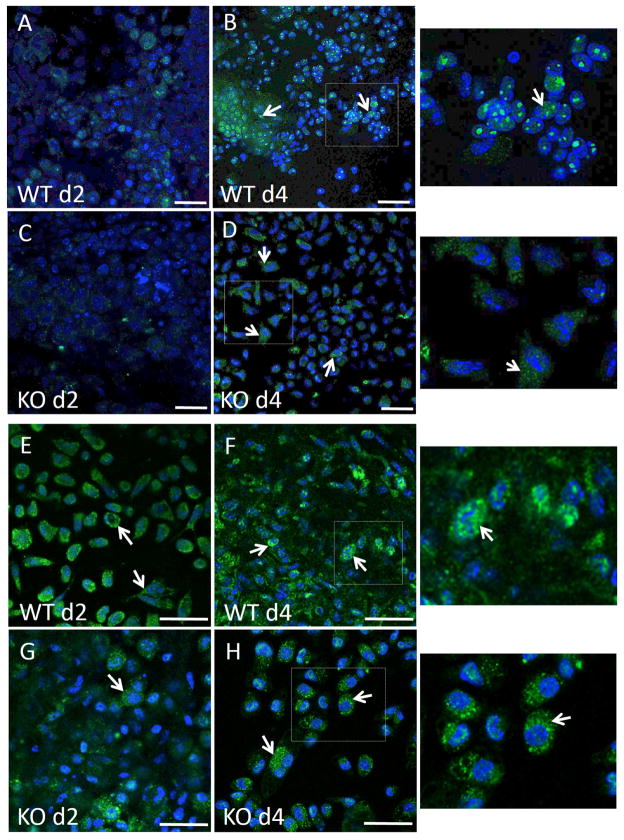
We have previously shown that MCP-1 KO macrophages were compromised in their ability to express TNF following exposure to IL-4 in vitro, which is consistent with Q-PCR data shown here [24]. In addition, we detected TNF by immunohistochemical analysis of implant-adherent cells and observed a pattern of expression that increased from d 2 to d 7 post implantation in WT mice (Figure 3 A–C). In contrast, MCP-1 KO cells displayed reduced immunoreactivity without induction of TNF. Finally, ELISA analysis of d 2 lavage fluid revealed reduced TNF levels in MCP-1 KO mice (Figure 3 D). Because TNF can activate the NFκB pathway, we investigated the engagement of this pathway during fusion by detecting p50 and RelA subcellular localization via immunofluorescence in vitro and in vivo. We were not able to detect robust p50 expression in either control or IL4-treated WT cells at 24 and 48 hr (Supplemental Figure 4). In contrast, IL4-treated WT cells displayed significant p50 expression and extensive nuclear localization at 72 hr (Figure 4B), which was not observed in untreated cells (Figure 4 A). Analysis of RelA induction and subcellular localization was similar to p50 and occurred predominantly in IL-4 treated cells (Figure 4 C, D). Confocal microscopy to detect RelA in 72 hr samples showed that it was predominantly nuclear and cytoplasmic in IL-4-treated and non-treated cells, respectively (Figure 4 F–G). Moreover, western blot analysis of d 3 samples revealed the presence of phosphorylated RelA in cells treated with IL-4 on bacteriological plastic but not on petri dish (Figure 4 E). Since induction and nuclear localization of p50 were not observed in MCP-1 KO cells (Supplemental Figure 5 G–I), or WT cells plated on a non-fusogenic tissue culture-treated plates (Supplemental Figure 5 A–F), we conclude that this event is associated with fusion.
Figure 3.
Representative images of d 2 peritoneal implants from WT (A) and MCP-1 KO (B) mice stained with anti-TNF-α Ab. Immunoreactivity was detected using the peroxidase reaction (brown), and nuclei were counterstained with methyl green. (C) Image analysis of samples from 2, 4, and 7 d implants revealed increased expression of TNF in a temporal fashion in WT mice that was reduced in MCP-1 KO mice at all time points. (D) Enzyme-linked immunosorbent assay analysis of confirmed reduced levels of TNF in d 2 lavage fluid from MCP-1 KO. ¶ and * indicate P ≤ 0.05 when comparing levels within a group and between WT and MCP-1 KO samples, respectively. (n = 5) Scale bar = 50 μm.
Figure 4.
IL-4-induced activation of NFkB. Representative images of untreated bone marrow derived macrophages (A, C, F) or treated with IL-4 (B, D, G) for 3 d and stained with anti p50 (A–B) or anti-RelA (C–D,F–G) are shown. Images A–D are from conventional fluorescence and F–G from confocal microscopy. Immunoreactivity was detected with FITC-conjugated secondary Ab. Cytoskeleton and nuclei were stained with rhodamine-phalloidin and DAPI, respectively. Arrows in B, C, D, G indicate nuclear translocation and cytoplasmic localization in F. Bottom images show enlarged areas defined by rectangles in F and G. (E) Western blot analysis of macrophages at d 3 following IL-4 treatment. Cells plated on either petri dish (PD) or tissue culture plastic (TC) were analyzed for the expression of phosphor-RelA. Scale bar = 50 μm (A–D, F, G).
4.4 NFκB pathway in vivo
To confirm the engagement of this pathway in vivo, we analyzed the distribution of p50 and RelA on implant-adherent cells by confocal microscopy. Specifically, immunohistochemical detection of p50 (Figure 5 A–D) and RelA (Figure 5 E–H) was observed by confocal microscopy in cells adherent to PDMS disks that were implanted in the peritoneal cavity of mice for 2 or 4 d. Similar to our in vitro observations, we observed significant p50 and RelA expression at d 4 in WT mice, which was primarily co-localized with nuclei. In contrast, p50 and RelA nuclear localization was reduced in MCP-1 KO mice. Induction of p52 was not detected in either WT or MCP-1 KO cells suggesting that the non-canonical NFκB pathway is not engaged in this model (not shown). Consistent with this suggestion, it was previously shown that p52 and RelB were not necessary for IL-4-induced macrophage fusion [30].
Figure 5.
Induction and nuclear localization of NFκB in vivo. Representative images of PDMS disks implanted in the peritoneum of WT (A, B, E, F) and MCP-1 KO (C, D, G, H) mice for 2 or 4 d and stained with anti-p50 Ab (A–D) or anti-RelA Ab (E–H). Images on the right show enlarged areas defined by rectangles in B, D, F, and H. Immunoreactivity was detected with FITC-conjugated secondary Ab. Nuclei were stained with DAPI. (A–D) Arrows indicate p50 induction (in B,D) and nuclear translocation (in B). (E–H) Arrows indicate RelA induction (in E–H) and nuclear translocation (in F). Scale bar = 50 μm.
4.5 Inhibition of NFκB reduces fusion
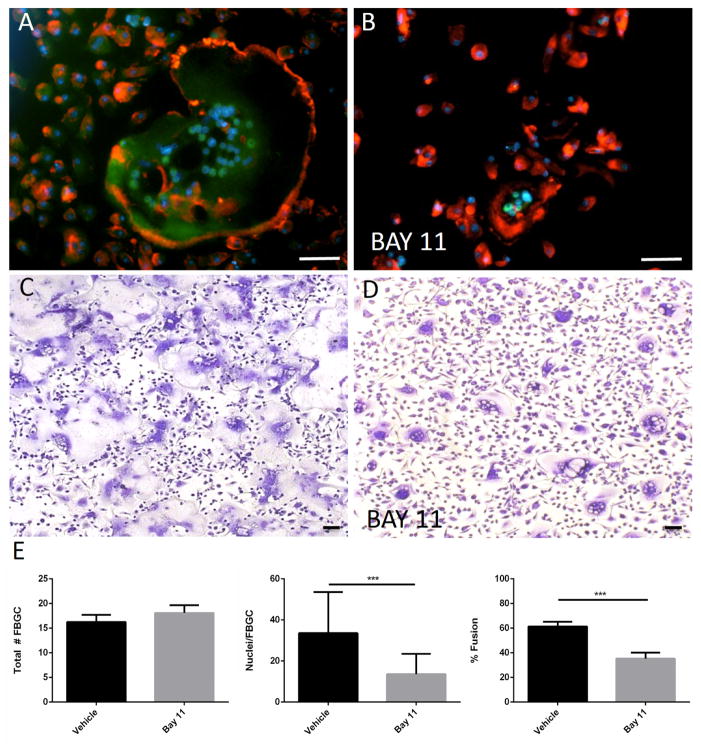
To test the role of NFκB activation in fusion, macrophages were induced to fuse with IL-4 in the presence of the NFκB inhibitor BAY 11. This inhibitor is known to inhibit the TNF-induced phosphorylation of activity of IκBα and prevent the nuclear translocation of NFκB components. Inclusion of BAY 11 in the fusion assay reduced the nuclear translocation of RelA and inhibited the formation of large FBGC (Figure 6 B, D). Quantification of fusion parameters revealed that BAY 11 did not cause a reduction in the number of FBGC but reduced the number of nuclei per FBGC and overall fusion (Figure 6 E). These observations suggest that NFκB activation is required for the normal progression of FBGC formation.
Figure 6.
Inhibition of NFκB reduces IL-4-induced fusion. Representative images of WT macrophages treated with IL-4 for 3 d (A, B) or 5 d (C, D) in the presence of BAY 11 (B, D) and stained with anti-RelA (A, B) or Giemsa and May Grunwald (C, D) are shown. RelA immunoreactivity was detected with FITC-conjugated secondary Ab. Cytoskeleton and nuclei were stained with rhodamine-phalloidin and DAPI, respectively. Scale bar = 50 μm. (E) Quantification of fusion was performed from randomly selected (10 per well) in triplicate wells from three independent experiments. Values represent mean + SEM, n = 3. ** * indicates p ≤ 0.0001.
4.6 TGF-β levels in the FBR
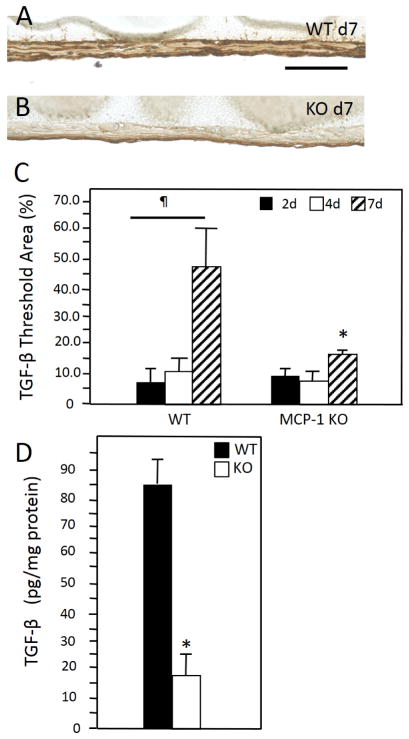
Activation of the NFκB has been linked to numerous cellular processes including fibrosis. We previously showed that MCP-1 KO mice display attenuated foreign body response in the peritoneal implant model characterized by reduced capsule thickness [24]. Consistent with this observation, here we observed reduced accumulation of mature collagen fibers around collagen implants in these mice (Supplemental Figure 3A–B). Because MCP-1 KO mice fail to induce a fibrotic response despite the recruitment and initial adhesion of macrophages to implants, we asked whether the failure to fully engage this pathway was associated with alterations in the levels of proteins involved in matrix production. Immunohistochemical analysis of implant-adherent cells revealed that the deposition of TGF-β increased by d7 in WT mice (Figure 7). Unlike their WT counterparts, levels of TGF-β in MCP-1 KO cells did not increase. Moreover, ELISA analysis of protein extracts from d 7 implants showed a significant reduction in TGF-β in MCP-1 KO mice.
Figure 7.
Representative images of d 2 peritoneal implants from WT (A) and MCP-1 KO (B) mice stained with anti-TGF-β Ab. Immunoreactivity was detected using the peroxidase reaction (brown), and nuclei were counterstained with methyl green. (C) Image analysis of samples from 2, 4, and 7 d implants revealed increased expression of TGF-β at d 7 in WT mice, which was not evident in MCP-1 KO mice. (D) Enzyme-linked immunosorbent assay analysis of confirmed reduced levels of TGF-β in d d7 protein extracts in MCP-1 KO. ¶ and * indicate P ≤ 0.05 when comparing levels within a group and between WT and MCP-1 KO samples, respectively. (n = 5) Scale bar = 50 μm.
5. Discussion
Collagenous encapsulation of biomaterials and devices represents a major obstacle in their successful employment in numerous applications indicating that strategies that limit this process are highly desirable. Based on our previous observations of attenuation of this response in MCP-1 KO mice, we hypothesized that abnormalities in macrophage activation might contribute to reduced fibrosis. Therefore, in this study we focused on the characterization and comparison of the activation of macrophages in WT and MCP-1 KO mice.
Despite extensive characterization of macrophage activation in vitro in response to IFN-γ/LPS or IL-4, it is now appreciated that macrophages undergo complex activation in vivo that reflects spatiotemporal cues and differs between species [7]. For example, detailed analysis of human and mouse macrophages from various sources via genomic and proteomic approaches revealed minimal overlap and identified only one protein, tissue transglutaminase (TTG), which could reliably detect alternatively activated human and mouse macrophages [31]. In addition, cross-talk between macrophages and other cell types has been shown to enhance cell signaling in the context of the FBR in vitro [32]. Based on our present findings, we speculate that macrophage classification in vivo in the context of the FBR is complex and reflects a distinct activation state. Others have determined the gene expression profile of lavaged and implant-associated macrophages from mice that received boiled egg whites as implants in the peritoneal cavity and demonstrated induction of both M1 and M2 markers [10]. Similarly, our experimental approach allowed us to detect the induction of genes, such as TNF, CD36, IL-1β, IL-10, iNOS, Ym1, and Arg1 in the FBR. In contrast, IL-6 expression was reduced in comparison to lavage cells. Therefore, we conclude that the interaction of cells with implants induces both M1- and M2-associated genes.
It should be noted that the in vivo environment is very complex and, unlike in vitro experiments where pure cell populations can be analyzed, our analysis most likely includes multiple macrophage subtypes and perhaps other cells. Specifically, lavage preparations could contain macrophages with variable activation states. Moreover, extraction of RNA from implants could include other cell types especially in the early time point (24 hr) when the response is not largely dominated by macrophages. Previous studies have demonstrated cell variability in the foreign body response but the production of inflammatory cytokines was attributed mainly to macrophages [33]. We have previously shown that at d 4 the response on peritoneal implants is dominated by macrophages and our studies here are consistent with our previous findings. Despite the inherent complexity of the in vivo environment, we believe our analysis is informative in identifying critical differences between WT and MCP-1 KO cells. Moreover, immunohistochemistry and ELISA were used to confirm the induction of critical cytokines. Our observations were somewhat unexpected because macrophages on the surface of implants undergo IL-4-dependent fusion to form FBGC and it was tempting to assume that their phenotype will resemble predominantly M2 polarization. It has also been shown that IL-4 can inhibit the classical activation of macrophages via p50 [34, 35]. However, macrophages adherent to implants did not acquire an exclusive M2 phenotype and our findings are consistent with a recent study showing variable activation of macrophages [10].
Even though they exhibit compromised activation and fusion, MCP-1 KO macrophages were able to produce some M2-associated factors. For example, lavage cells induced surface expression of CD36 in response to IL-4 and analysis of implants revealed the induction of Arg1, Ym1, and IL-10. Immunohistochemical analysis also revealed that the expression of iNOS and arginase did not overlap, suggesting that cells with distinct activation profiles are present within the capsule. More importantly, MCP-1 KO mice did not induce TNF, suggesting that expression of this single factor is more critical than the acquirement of a M1 or M2 phenotype.
We previously described the existence of at least two distinct IL-4-induced pathways in macrophage fusion [24]. Specifically, one involving induction of E-cadherin and β-catenin, which like CD36 expression shown here, was intact in MCP-1 KO cells. A second pathway involved the induction of MCP-1, Rac-1-dependent cytoskeletal remodeling, TNF, and MMP-9. As expected, all events downstream of MCP-1 were compromised in IL-4 treated MCP-1 KO cells in vitro. Moreover, addition of exogenous TNF to these cells could rescue the fusion defect highlighting the importance of this cytokine [24]. TNF mediates its effects, in part, via NFκB that can be activated by either the canonical (p50/RelA) or non-canonical pathway (p52/RelB) [36]. Previously, engagement of the non-canonical pathway was shown to be critical for CD44-dependent RANKL-induced macrophage fusion [37].
In the present study, we observed induction and nuclear localization of p50 and RelA in WT cells in response to IL-4 in vitro. Consistent with a previous demonstration of IL-4-induced activation of NFκB in macrophages in vitro, we observed nuclear localization of p50 after 48 hr [30]. More importantly, we observed the same phenomenon in implants in vivo in a temporal pattern consistent with TNF induction. In MCP-1 KO mice, we detected suboptimal nuclear translocation of both p50 and RelA, which was consistent with low TNF levels in these mice. The importance of NFκB activation in fusion was demonstrated by the ability of BAY 11 to reduce the extent of multinucleation and overall fusion of WT macrophages.
One of the many outcomes of NFκB nuclear translocation is the induction of genes involved in matrix remodeling, including MMP-9 and TTG [31, 38–41]. We previously demonstrated the significance of MMP-9 in fusion and the nature of fibrotic encapsulation and showed that MCP-1 KO macrophages fail to secrete MMP-9 in response to IL-4 [24, 25]. More importantly, we showed in in vitro studies that TNF was required for induction of MMP9 and addition of TNF to MCP-1 KO macrophages induced MMP9 and rescued their fusion defect. Here we expand on these observations to show in WT mice significant TGF-β production in vivo at 7 d following implantation in a temporal pattern downstream of NFκB. In contrast, levels of TGF-β did not increase in MCP-1 KO mice. It should be noted that our observations are limited to the peritoneal implantation model.
We have previously shown normal encapsulation of implants in a subcutaneous model in these mice [42]. More importantly, we show that a reduction in TNF and activation of the NFκB pathway are associated with reduced TGF-β levels suggesting a putative link between macrophage activation and fibrosis. A similar connection has been described in a mouse model of experimental autoimmune thyroiditis where a reduction in TNF activity was associated with decreased fibrosis [43]. In separate studies, systemic inhibition of NFκB with BAY 11 in mouse models of silica-induced lung injury or liver injury induced by carbon tetrachloride resulted in reduced fibrosis [44, 45]. In addition, a link between TNF and TGF-β expression has been demonstrated in TIMP3 KO mice. Specifically, these mice display increased myocardial fibrosis due to high levels of TGF-β and this defect is rescued in TIMP3/TNF-α double knockout mice [46]. However, a mechanism showing direct modulation of TGF-β by TNF has not been described. Similarly, the association between MCP-1 and TNF expression is complex with numerous studies demonstrating induction of both proteins in macrophages following activation. Moreover, the promoter region of MCP-1 contains NFκB binding sites and TNF has been shown to induce MCP-1 [47]. On the other hand, MCP-1 signaling involves multiple kinases (ERK1/2, JAK2, JNK1, p38, PI3K) but has not been shown to directly induce TNF in macrophages. In ongoing studies we are identifying key intermediate events downstream of MCP-1.
6. Conclusion
Our studies show that during the FBR macrophages undergo complex activation with features of both M1 and M2 including induction of TNF and activation of the canonical NFκB pathway. The latter is limited to cells undergoing fusion and does not occur in cells plated on non-fusogenic surfaces or in fusion-defective MCP-1 KO macrophages. In addition, a pharmacological inhibitor of NFκB reduced fusion in WT macrophages. In vivo, induction of TNF and activation of NFκB were compromised in MCP-1 KO mice and these mice displayed reduced levels of implant-associated TGF-β and diminished fibrotic response.
Supplementary Material
Acknowledgments
We thank Sarah Titman for technical assistance. This work was funded by National Institutes of Health Grant (GM-072194), NSF-GRFP (L.B.M.), and National Institutes of Health Vascular Research Training Grant (5T32HL007950-12) (A.J.S).
Abbreviations
- BAY 11
BAY 11-7028
- FBR
foreign body response
- FBGC
foreign body giant cell
- MCP
monocyte chemoattractant protein
- TNF
tumor necrosis factor α
- TGF
transforming growth factor
- NFκB
nuclear factor kappa-light-chain-enhancer of activated B cells
- MMP
matrix metalloproteinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brown BN, Ratner BD, Goodman SB, Amar S, Badylak SF. Macrophage polarization: an opportunity for improved outcomes in biomaterials and regenerative medicine. Biomaterials. 2012;33:3792–802. doi: 10.1016/j.biomaterials.2012.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–37. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mantovani A, Biswas SK, Galdiero MR, Sica A, Locati M. Macrophage plasticity and polarization in tissue repair and remodelling. J Pathol. 2013;229:176–85. doi: 10.1002/path.4133. [DOI] [PubMed] [Google Scholar]
- 5.Mosser DM, Zhang X. Activation of murine macrophages. Curr Protoc Immunol. 2008;Chapter 14(Unit 14):2. doi: 10.1002/0471142735.im1402s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stout RD, Jiang C, Matta B, Tietzel I, Watkins SK, Suttles J. Macrophages sequentially change their functional phenotype in response to changes in microenvironmental influences. J Immunol. 2005;175:342–9. doi: 10.4049/jimmunol.175.1.342. [DOI] [PubMed] [Google Scholar]
- 7.Novak ML, Koh TJ. Macrophage phenotypes during tissue repair. J Leukoc Biol. 2013 doi: 10.1189/jlb.1012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–69. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deonarine K, Panelli MC, Stashower ME, Jin P, Smith K, Slade HB, et al. Gene expression profiling of cutaneous wound healing. J Transl Med. 2007;5:11. doi: 10.1186/1479-5876-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mooney JE, Summers KM, Gongora M, Grimmond SM, Campbell JH, Hume DA, et al. Transcriptional switching in macrophages associated with the peritoneal foreign body response. Immunol Cell Biol. 2014 doi: 10.1038/icb.2014.19. [DOI] [PubMed] [Google Scholar]
- 11.Anderson JM, Rodriguez A, Chang DT. Foreign body reaction to biomaterials. Semin Immunol. 2008;20:86–100. doi: 10.1016/j.smim.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franz S, Rammelt S, Scharnweber D, Simon JC. Immune responses to implants - a review of the implications for the design of immunomodulatory biomaterials. Biomaterials. 2011;32:6692–709. doi: 10.1016/j.biomaterials.2011.05.078. [DOI] [PubMed] [Google Scholar]
- 13.Williams DF. On the mechanisms of biocompatibility. Biomaterials. 2008;29:2941–53. doi: 10.1016/j.biomaterials.2008.04.023. [DOI] [PubMed] [Google Scholar]
- 14.Williams DF. On the nature of biomaterials. Biomaterials. 2009;30:5897–909. doi: 10.1016/j.biomaterials.2009.07.027. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Cao Z, Bai T, Carr L, Ella-Menye J-R, Irvin C, et al. Zwitterionic hydrogels implanted in mice resist the foreign-body reaction. Nat Biotech. 2013 doi: 10.1038/nbt.2580. advance online publication. [DOI] [PubMed] [Google Scholar]
- 16.Helming L, Gordon S. Molecular mediators of macrophage fusion. Trends Cell Biol. 2009;19:514–22. doi: 10.1016/j.tcb.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Badylak SF, Valentin JE, Ravindra AK, McCabe GP, Stewart-Akers AM. Macrophage phenotype as a determinant of biologic scaffold remodeling. Tissue Eng Part A. 2008;14:1835–42. doi: 10.1089/ten.tea.2007.0264. [DOI] [PubMed] [Google Scholar]
- 18.Brown BN, Londono R, Tottey S, Zhang L, Kukla KA, Wolf MT, et al. Macrophage phenotype as a predictor of constructive remodeling following the implantation of biologically derived surgical mesh materials. Acta Biomater. 2012;8:978–87. doi: 10.1016/j.actbio.2011.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valentin JE, Stewart-Akers AM, Gilbert TW, Badylak SF. Macrophage participation in the degradation and remodeling of extracellular matrix scaffolds. Tissue Eng Part A. 2009;15:1687–94. doi: 10.1089/ten.tea.2008.0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valentin JE, Turner NJ, Gilbert TW, Badylak SF. Functional skeletal muscle formation with a biologic scaffold. Biomaterials. 2010;31:7475–84. doi: 10.1016/j.biomaterials.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mueller CK, Schultze-Mosgau S. Histomorphometric analysis of the phenotypical differentiation of recruited macrophages following subcutaneous implantation of an allogenous acellular dermal matrix. Int J Oral Maxillofac Surg. 2011;40:401–7. doi: 10.1016/j.ijom.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 22.Madden LR, Mortisen DJ, Sussman EM, Dupras SK, Fugate JA, Cuy JL, et al. Proangiogenic scaffolds as functional templates for cardiac tissue engineering. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15211–6. doi: 10.1073/pnas.1006442107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Putten SM, Ploeger DT, Popa ER, Bank RA. Macrophage phenotypes in the collagen-induced foreign body reaction in rats. Acta Biomater. 2013;9:6502–10. doi: 10.1016/j.actbio.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 24.Skokos EA, Charokopos A, Khan K, Wanjala J, Kyriakides TR. Lack of TNF-alpha-induced MMP-9 production and abnormal E-cadherin redistribution associated with compromised fusion in MCP-1-null macrophages. The American journal of pathology. 2011;178:2311–21. doi: 10.1016/j.ajpath.2011.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacLauchlan S, Skokos EA, Meznarich N, Zhu DH, Raoof S, Shipley JM, et al. Macrophage fusion, giant cell formation, and the foreign body response require matrix metalloproteinase 9. J Leukoc Biol. 2009;85:617–26. doi: 10.1189/jlb.1008588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jay SM, Skokos EA, Zeng J, Knox K, Kyriakides TR. Macrophage fusion leading to foreign body giant cell formation persists under phagocytic stimulation by microspheres in vitro and in vivo in mouse models. Journal of biomedical materials research Part A. 2010;93:189–99. doi: 10.1002/jbm.a.32513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNally AK, Anderson JM. Beta1 and beta2 integrins mediate adhesion during macrophage fusion and multinucleated foreign body giant cell formation. Am J Pathol. 2002;160:621–30. doi: 10.1016/s0002-9440(10)64882-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jay SM, Skokos E, Laiwalla F, Krady MM, Kyriakides TR. Foreign body giant cell formation is preceded by lamellipodia formation and can be attenuated by inhibition of Rac1 activation. The American journal of pathology. 2007;171:632–40. doi: 10.2353/ajpath.2007.061213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malik AF, Hoque R, Ouyang X, Ghani A, Hong E, Khan K, et al. Inflammasome components Asc and caspase-1 mediate biomaterial-induced inflammation and foreign body response. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20095–100. doi: 10.1073/pnas.1105152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu M, Qi X, Moreno JL, Farber DL, Keegan AD. NF-kappaB signaling participates in both RANKL- and IL-4-induced macrophage fusion: receptor cross-talk leads to alterations in NF-kappaB pathways. J Immunol. 2011;187:1797–806. doi: 10.4049/jimmunol.1002628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez FO, Helming L, Milde R, Varin A, Melgert BN, Draijer C, et al. Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences. Blood. 2013;121:e57–69. doi: 10.1182/blood-2012-06-436212. [DOI] [PubMed] [Google Scholar]
- 32.Holt DJ, Chamberlain LM, Grainger DW. Cell-cell signaling in co-cultures of macrophages and fibroblasts. Biomaterials. 2010;31:9382–94. doi: 10.1016/j.biomaterials.2010.07.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higgins DM, Basaraba RJ, Hohnbaum AC, Lee EJ, Grainger DW, Gonzalez-Juarrero M. Localized immunosuppressive environment in the foreign body response to implanted biomaterials. The American journal of pathology. 2009;175:161–70. doi: 10.2353/ajpath.2009.080962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porta C, Rimoldi M, Raes G, Brys L, Ghezzi P, Di Liberto D, et al. Tolerance and M2 (alternative) macrophage polarization are related processes orchestrated by p50 nuclear factor kappaB. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14978–83. doi: 10.1073/pnas.0809784106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saccani A, Schioppa T, Porta C, Biswas SK, Nebuloni M, Vago L, et al. p50 nuclear factor-kappaB overexpression in tumor-associated macrophages inhibits M1 inflammatory responses and antitumor resistance. Cancer Res. 2006;66:11432–40. doi: 10.1158/0008-5472.CAN-06-1867. [DOI] [PubMed] [Google Scholar]
- 36.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–62. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 37.Cui W, Ke JZ, Zhang Q, Ke HZ, Chalouni C, Vignery A. The intracellular domain of CD44 promotes the fusion of macrophages. Blood. 2006;107:796–805. doi: 10.1182/blood-2005-05-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bond M, Fabunmi RP, Baker AH, Newby AC. Synergistic upregulation of metalloproteinase-9 by growth factors and inflammatory cytokines: an absolute requirement for transcription factor NF-kappa B. FEBS Lett. 1998;435:29–34. doi: 10.1016/s0014-5793(98)01034-5. [DOI] [PubMed] [Google Scholar]
- 39.Farina AR, Tacconelli A, Vacca A, Maroder M, Gulino A, Mackay AR. Transcriptional up-regulation of matrix metalloproteinase-9 expression during spontaneous epithelial to neuroblast phenotype conversion by SK-N-SH neuroblastoma cells, involved in enhanced invasivity, depends upon GT-box and nuclear factor kappaB elements. Cell Growth Differ. 1999;10:353–67. [PubMed] [Google Scholar]
- 40.Yan C, Wang H, Aggarwal B, Boyd DD. A novel homologous recombination system to study 92 kDa type IV collagenase transcription demonstrates that the NF-kappaB motif drives the transition from a repressed to an activated state of gene expression. Faseb J. 2004;18:540–1. doi: 10.1096/fj.03-0960fje. [DOI] [PubMed] [Google Scholar]
- 41.Chen CS, Wu CH, Lai YC, Lee WS, Chen HM, Chen RJ, et al. NF-kappaB-activated tissue transglutaminase is involved in ethanol-induced hepatic injury and the possible role of propolis in preventing fibrogenesis. Toxicology. 2008;246:148–57. doi: 10.1016/j.tox.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 42.Kyriakides TR, Foster MJ, Keeney GE, Tsai A, Giachelli CM, Clark-Lewis I, et al. The CC chemokine ligand, CCL2/MCP1, participates in macrophage fusion and foreign body giant cell formation. Am J Pathol. 2004;165:2157–66. doi: 10.1016/S0002-9440(10)63265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen K, Wei Y, Sharp GC, Braley-Mullen H. Decreasing TNF-alpha results in less fibrosis and earlier resolution of granulomatous experimental autoimmune thyroiditis. J Leukoc Biol. 2007;81:306–14. doi: 10.1189/jlb.0606402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Giuseppe M, Gambelli F, Hoyle GW, Lungarella G, Studer SM, Richards T, et al. Systemic inhibition of NF-kappaB activation protects from silicosis. PLoS One. 2009;4:e5689. doi: 10.1371/journal.pone.0005689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang Y, Wei Y, Demarco V, Chen K, Sharp GC, Braley-Mullen H. Murine FLIP transgene expressed on thyroid epithelial cells promotes resolution of granulomatous experimental autoimmune thyroiditis in DBA/1 mice. The American journal of pathology. 2007;170:875–87. doi: 10.2353/ajpath.2007.060816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kassiri Z, Defamie V, Hariri M, Oudit GY, Anthwal S, Dawood F, et al. Simultaneous transforming growth factor beta-tumor necrosis factor activation and crosstalk cause aberrant remodeling response and myocardial fibrosis in Timp3-deficient heart. The Journal of biological chemistry. 2009;284:29893–904. doi: 10.1074/jbc.M109.028449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res. 2009;29:313–26. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.