Abstract
A reverse transcriptase PCR was developed to detect 50 or 5,000 RNA copies of influenza A virus per ml in throat swab specimens. The assay was more sensitive than the Directigen Flu A test. The technique was also used to detect amantadine-resistant isolates.
Influenza A virus causes annual epidemic disease resulting in approximately 20,000 to 36,000 deaths per year in the United States (10, 12). Patients at increased risk include the elderly and those with cardiovascular, pulmonary, or renal disease, diabetes, and immunosuppression, as well as residents of nursing homes (1, 5, 6). In these cases, rapid tests, such as Directigen Flu A (8), have greatly improved the timely detection and management of influenza virus, although such tests lack adequate sensitivity. We studied the use of a reverse transcriptase PCR (RT-PCR) assay as a rapid, sensitive, and inexpensive technique to diagnose influenza and detect amantadine or rimantadine resistance (9, 13).
Throat swab and nasal swab specimens were collected for influenza diagnosis during the course of routine patient care at the Edward Hines Jr. Veterans Administration Hospital in the winter of 1999 to 2000. Specimens were processed and tested with a Directigen Flu A kit (Becton Dickinson, Cockeysville, Md.) with subsequent viral isolation performed with specimens with a negative result. Specimens were subjected to RT-PCR using RNA isolated (with a QIAamp kit) from 100 μl of each specimen and of positive and negative controls. Eukaryotic RNA (300 ng) was spiked into each sample as an internal control for RNA degradation by the coamplification of β2-microglobulin. cDNA was synthesized (at 42°C for 60 min) in a standard 20-μl reaction mixture containing 11 μl of each RNA preparation, 20 U of RT (Seikagaku America, East Falmouth, Mass.), and 1 μg of random primers (Pharmacia, Piscataway, N.J.). Mock reaction mixtures were run without RT (contamination control). PCR was performed with 50-μl reaction mixtures coamplifying the 212-bp segment of the influenza virus matrix gene (forward primer, 5′-CAGAGACTTGAAGATGTCTTTGCTG; reverse primer, 5′-GCTCTGTCCATGTTATTTGGATC) (3) and a 300-bp segment of the control β2-microglobulin gene (forward primer, 5′-GCCTGCCGTGTGAACCACGTGAC; reverse primer, 5′-TACCTGTGGAGCAACCTGCTCAGA) (11). Each reaction mixture contained 10 μl of either the cDNA or mock cDNA reaction mixture for each specimen and 40 μl of a master mix containing 0.5 U of Taq polymerase (Applied Biosystems, Foster City, Calif.), 2.5 mM (each) dATP, dCTP, and dGTP, 5.0 mM dUTP (Pharmacia), 4 mM MgCl2, a 1.5 μM concentration of each PCR primer, and 0.5 U of Amperase (Applied Biosystems). A positive and negative control for the master mix was included with each assay. The cycling parameters were 50°C for 5 min; 94°C for 10 min; and 36 cycles of 94°C for 10 s, 54°C for 30 s, and 72°C for 30 s. To detect PCR products, 20 μl of each RT-PCR product was incubated at 37°C for 15 min with 20 μl of an internal matrix gene probe (5′-TCCTGTCACCTCTGACTAAGGGGATTTTG) end labeled with [γ-32P]ATP (Amersham) (3). PCR products and size markers (Promega, Madison, Wis.) were separated on 0.8% agarose gels, which were subsequently stained with ethidium bromide for the direct detection of PCR products on a transilluminator. The gels were then exposed to X-ray film for 24 to 48 h, and influenza virus PCR products were detected by autoradiography. Results were deemed valid only if they met the following control conditions: detection of the β2-microglobulin PCR product in the specimen cDNA reaction mixtures (for the RNA extraction control), no detection of PCR products in the mock cDNA or negative control reaction mixture (for the DNA contamination controls), and detection of PCR products in the low- and high-positive influenza virus control mixtures (for the sensitivity controls).
To detect amantadine- and rimantadine-resistant influenza A virus, the RT-PCR was performed as described above using specific cycling conditions (50°C for 5 min; 94°C for 10 min; and 42 cycles of 45°C for 1 min, 72°C for 1 min, and 94°C for 30 s) and primers (forward primer, 5′-GGGACTCATCCTAGCTCCAGTGCTGGTCTAAA; reverse primer, 5′-CGATCAAGAATCCACAATATCAAGTGCAAGATCCCAATAATA) (4, 9). A 164-bp segment that contained nucleotide changes resulting in amino acid substitutions at positions 26, 27, 30, and 31 of the M2 protein (2, 7) was amplified. Restriction fragment length polymorphism analysis of the amplified products was used to detect mutations at the four different positions as follows: AciI for position 26 (where wild-type DNA is digested), BspWI for position 27 (where mutant DNA is digested), ItaI for position 30 (where wild-type DNA is digested), and SspI for position 31 (where mutant DNA is digested). Digested products were analyzed by agarose gel electrophoresis as described above.
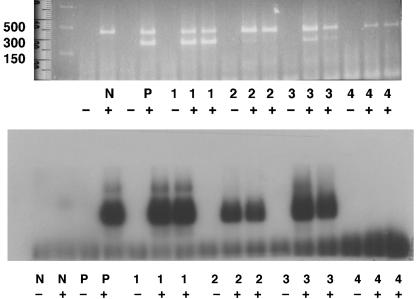
The assay was specific for influenza A virus (H1N1 and H3N2 subtypes) with a sensitivity of 5,000 copies/ml by gel staining or 50 copies/ml by autoradiography (data not shown). Of the 119 specimens collected, 29 (24%) were positive by Directigen Flu A, 0 were positive by shell vial assay, 6 were positive by tissue culture, and 56 (47%) were positive by RT-PCR. All specimens that were positive by RT-PCR were also positive by Directigen Flu A and/or viral isolation. A typical RT-PCR assay is shown in Fig. 1.
FIG. 1.
RT-PCR detection of influenza A virus by direct observation of ethidium-stained PCR products (top) and by autoradiography (bottom). Results are shown for influenza virus-negative (N) and -positive (P) controls and for patient samples 1 to 4. −, mock RT reaction mixtures; +, reaction mixtures containing RT. Molecular weight markers are shown for the top panel. The upper band is the 300-bp β2-microglobulin PCR product, and the lower band is the 212-bp influenza A virus PCR product.
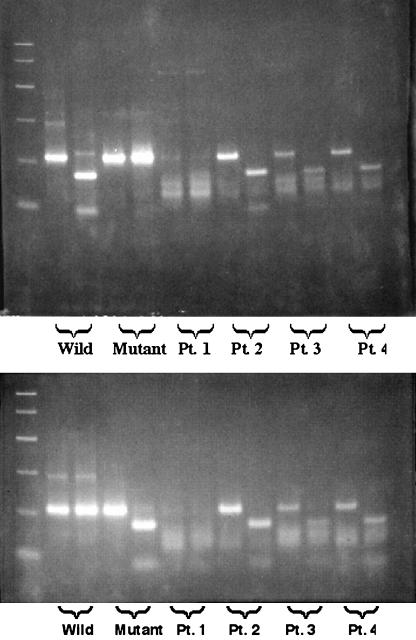
Specimens taken from patients who developed influenza virus infection while receiving rimantadine prophylaxis were tested for the presence of resistance mutations (at amino acid positions 26, 27, 30, and 31) in the transmembrane domain of the M2 protein of influenza A virus (Fig. 2) (4, 9). PCR products were left undigested or were digested with ItaI at position 30 (Fig. 2, top) or SspI at position 31 (Fig. 2, bottom). For position 30, the wild-type DNA was cut by the enzyme and shows a reduction in size, while the mutant DNA was resistant to digestion. At position 31, the reverse was found; only the mutant DNA was digested with the enzyme. For position 30, the specimens from patients 2 to 4 failed to show any mutation, while the PCR product for the patient 1 sample was resistant to digestion, indicating the presence of the amantadine resistance mutation. For position 31, mutations were found for the specimens from patients 2 to 4 but not for the patient 1 specimen. No mutations were found for amino acid positions 26 and 27 (data not shown).
FIG. 2.
Detection of amantadine resistance mutations at amino acid positions 30 (top) and 31 (bottom) by using restriction fragment length polymorphism of RT-PCR products. Results are shown for wild-type (Wild) and mutant control viruses and for patient samples (Pt.) 1 to 4. Undigested PCR products are shown in the left lane of each doublet. The right lane of each doublet shows the results of digesting PCR products with either ItaI (top) or SspI (bottom).
RT-PCR can provide increased sensitivity in the detection of influenza virus, which can be used to enhance community and hospital surveillance programs and hasten the specific diagnosis of influenza among ill patients. The use of this technology may also provide help in identifying antiviral resistance.
Acknowledgments
The Research Service at Hines VA Hospital supported this work.
We are grateful for the help of Alexander I. Klimov from the Centers for Disease Control and Prevention, who provided us with the amantadine-resistant isolates of influenza A virus.
REFERENCES
- 1.Barker, W. H., and J. P. Mullooly. 1982. Pneumonia and influenza deaths during epidemics: implications for prevention. Arch. Intern. Med. 142:85-89. [PubMed] [Google Scholar]
- 2.Black, R. A., P. A. Rota, N. Gorodkova, A. Cramer, H. D. Klenk, A. P. Kendal. 1993. Production of the M2 protein of influenza A virus in insect cells is enhanced in the presence of amantadine. J. Gen. Virol. 74:1673-1677. [DOI] [PubMed] [Google Scholar]
- 3.Donofrio, J. C., D. Coonrod, J. N. Davidson, and R. F. Betts. 1992. Detection of influenza A and B in respiratory secretions with the polymerase chain reaction. PCR Methods Appl. 1:263-268. [DOI] [PubMed] [Google Scholar]
- 4.Englund, J. A., R. E. Champlin, P. R. Wyde, H. Kantarjian, R. L. Atmar, J. Tarrand, H. Yousuf, H. Regnery, A. I. Klimov, N. J. Cox, and E. Whimbey. 1998. Common emergence of amantadine- and rimantadine-resistant influenza A viruses in symptomatic immunocompromised adults. Clin. Infect. Dis. 26:1418-1424. [DOI] [PubMed] [Google Scholar]
- 5.Glezen, W. P., M. Decker, and D. M. Perrotta. 1987. Survey of underlying conditions of persons hospitalized with acute respiratory disease during influenza epidemics in Houston, 1978-1981. Am. Rev. Respir. Dis. 136:550-555. [DOI] [PubMed] [Google Scholar]
- 6.Glezen, W. P., S. B. Greenberg, R. L. Atmar, P. A. Predia, and R. B. Couch. 2000. Impact of respiratory virus infections on persons with chronic underlying conditions. JAMA 283:499-505. [DOI] [PubMed] [Google Scholar]
- 7.Hay, A. J. 1992. The action of adamantanamines against influenza A viruses: inhibition of the M2 ion channel protein. Semin. Virol. 3:21-30. [Google Scholar]
- 8.Kaiser, L., M. S. Briones, and F. G. Hayden. 1999. Performance of virus isolation and Directigen Flu A to detect influenza A virus in experimental human infection. J. Clin. Virol. 14:191-197. [DOI] [PubMed] [Google Scholar]
- 9.Klimov, A. I., E. Rocha, F. G. Hayden, P. Shult, L. F. Roumillat, and N. J. Cox. 1995. Prolonged shedding of amantadine-resistant influenza A viruses by immunodeficient patients: detection by polymerase chain reaction-restriction analysis. J. Infect. Dis. 172:1352-1355. [DOI] [PubMed] [Google Scholar]
- 10.Liu, K.-J., and A. P. Kendal. 1987. Impact of influenza epidemics on mortality in the United States from October 1972 to May 1985. Am. J. Public Health 77:712-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu, P., J. Garcia-Sanz, M. Lichtenheld, and E. R. Podack. 1992. Perforin expression in human peripheral blood mononuclear cells. J. Immunol. 148:3354-3360. [PubMed] [Google Scholar]
- 12.Thompson, W. W., D. K. Shay, E. Weintraub, L. Brammer, N. Cox, L. J. Anderson, and K. Fukuda. 2003. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289:179-186. [DOI] [PubMed] [Google Scholar]
- 13.Valassina, M., A. M. Cuppone, M. G. Cusi, and P. E. Valensin. 1997. Rapid detection of different RNA respiratory virus species by multiplex RT-PCR: application to clinical specimens. Clin. Diagn. Virol. 8:227-232. [DOI] [PubMed] [Google Scholar]