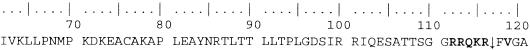Abstract
Newcastle disease virus isolates from chickens in eastern Uganda in 2001 were found to be velogenic by fusion protein cleavage site sequence analysis and biological characterization; the intracerebral pathogenicity index was 1.8. Analysis of their hemagglutinin-neuraminidase protein gene sequences revealed a novel genotype unrelated to those that caused previous outbreaks.
Newcastle disease (ND), caused by ND virus (NDV), is a serious illness of birds, particularly chickens, and has been one of the major causes of economic losses in the poultry industry (3). NDV is a single-stranded, negative-sense enveloped RNA virus of the Paramyxoviridae family in the order Mononegavirales (20), which includes avian paramyxovirus type 1 (PMV-1). The viral genome is 15,186 nucleotides long (10) and contains six genes encoding six major polypeptides: nucleocapsid protein, phosphoprotein, matrix protein, fusion (F) protein, hemagglutinin-neuraminidase (HN), and large RNA-dependent polymerase protein (17).
Pathotyping in chickens is used to classify NDV strains into highly velogenic, intermediate, or lentogenic strains (6). F protein, which is synthesized as nonfunctional precursor F0 and proteolytically cleaved to yield polypeptides F1 and F2 by host proteases (18), is an important determinant of NDV pathogenicity (24). Different pathotypes (21) are characterized by differences in the amino acid sequences surrounding the F0 cleavage site, which hosts the molecular marker for virulence. Previous studies comparing the precursor F0 amino acid sequences of NDVs varying in virulence for chickens showed that viruses that were virulent for chickens had the amino acid sequence 112R/K-R-Q-K/R-R116 at the C terminus of the F2 protein and phenylalanine at residue 117, the N terminus of the F1 protein, whereas viruses of low virulence had the sequence 112G/E-K/R-Q-G/E-R116 at the C terminus of the F2 protein and leucine at residue 117 (9). The amino acid sequence in a virulent virus renders the F protein susceptible to cleavage by an omnipotent protease, resulting in a fatal systemic infection (21). Along with biological virulence determinations, the Office International des Epizooties accepts reporting of F protein cleavage site sequences of NDV isolates as a virulence criterion (7).
Restriction enzyme site mapping of the F protein gene and sequence analysis have been used to classify 45 NDV isolates into seven genotypes. Isolates from outbreaks in western Europe between 1992 and 1996 belonged to genotypes VI and VII (15). Two novel genetic groups, VIIb and VIII, were recently identified from ND outbreaks in southern Africa (13). Phylogenetic studies of both the F protein and the HN protein genes of NDV have been used for molecular epidemiologic analysis and characterization of NDV (5, 14) and to group NDV into specific lineages (28).
In Uganda, the first ND outbreak was documented in 1955, and in 1986, a virulent NDV isolate was characterized by using monoclonal antibodies (19). However, ND remains endemic in Uganda.
The aim of the present study was to genetically characterize and phylogenetically group NDV isolates from ND outbreaks in Uganda in 2001.
Filtrates of processed tissues from the lungs, trachea, heart, liver, spleen, kidneys, and intestines of chickens with suspected ND in 2001 were used to inoculate specific-pathogen-free eggs (Lohmann Tierzucht, Cuxhaven, Germany) as previously reported (22, 23). Allantoic fluids from the eggs were used for serological analyses and RNA extraction.
Four antisera—PMV-1 polyclonal antibodies, LaSota monoclonal antibodies, PMV-3 polyclonal antibodies, and PMV-1 monoclonal antibodies—were used to characterize the isolates by the hemagglutination inhibition test (2, 16) performed according to European Community directive 92/66/EC (8).
For reverse transcription (RT)-PCR and nucleotide sequence analysis, RNA was extracted from allantoic fluids by using an RNeasy minikit (Qiagen GmbH, Hilden, Germany). Degenerate oligonucleotide RT-PCR primers (DNA Technology, Aarhus, Denmark) were designed to amplify regions of the genes for the F protein cleavage site and the HN protein, representing bases 7561 to 7938 of the complete genome (10), in a one-tube RT-PCR carried out according to the Titan One tube RT-PCR system procedure (Roche, Mannheim, Germany). Purified PCR products were sequenced by BigDye Terminator cycle sequencing according to the manufacturer's automated DNA sequencing chemistry guide (Applied Biosystems, Foster City, Calif.). The final products were analyzed by using an ABI 377 automated DNA sequencer.
Nucleotide sequence editing, analysis, and prediction of amino acid sequences for the F protein and the HN protein were conducted with BioEdit (www.mbio.ncsu.edu/BioEdit/bioedit.html), while alignment was carried out with CLUSTAL X (DNAStar, Madison, Wis.) (27). The robustness of the groupings was assessed by bootstrap resampling of 100 replicates by using the PHYLIP package (11) on the Pasteur server (bioweb.pasteur.fr/seqanal/phylogeny/intro-uk.html). The phylogenetic tree distance matrix was created by using the Jukes-Cantor model and was constructed by neighbor joining (11).
The intracerebral pathogenicity index (4) of 1.8 and the F protein sequence (112RRQKRF117) around the F2-F1 cleavage site of all 16 isolates suggest a high level of virulence (1, 9) for the Ugandan NDV isolates. The sequence 85NRT87 (Fig. 1), which is a potential glycosylation site, was conserved in all of the Ugandan NDV isolates.
FIG. 1.
F protein cleavage site. All 16 Ugandan NDV isolates had the same amino acid residues (RRQKRFVG) around the cleavage site, which is indicated by an arrow. The sequence in bold represents the F protein cleavage site motif.
Comparison of the nucleotide sequences of the HN protein genes of the 16 isolates with those of genotype VIa isolates (Y19016 and Y18725, predominantly from the Middle East) and vaccine strains used in Uganda (V4 and LaSota) showed the highest similarities, 87.5 to 89.1%, to VIa; the similarities to LaSota were 84.8 to 86.1%.
Because of the geographical proximity between eastern Africa and southern Africa, the Ugandan NDV isolates would have been expected to be more closely related to either genotype VII from southern African countries or genotype VIII from South Africa. Genotypes V, VIa, and VIII were responsible for the second ND panzootic, and subgenotypes VIb, VIc, and VId of genotype VI were mainly responsible for the third ND panzootic (30).
Our results and those of Lomniczi et al. (15) and Herczeg et al. (13) show that so far, all of the NDV isolates that have caused outbreaks in eastern Africa and southern Africa originated from the Middle East and Asia. These results support the hypotheses that NDVs had a long history of evolution in southern Asia and that this region was one of the original locations for the transmission of ND and an ND panzootic (30).
Strict sequestration of isolates based upon the length of the HN protein gene sequence (25), a factor which is not clearly demonstrated in phylogenetic analyses of F protein, matrix protein, or phosphoprotein protein gene sequences (26), was previously reported by Gould et al. (12). For this reason and because the Ugandan NDV isolates had the same apparent biological properties and cleavage site sequences, phylogenetic analyses were based on the HN protein gene nucleotide sequences and revealed that the Ugandan NDV isolates belong to the same genotype. The sequestration of these isolates into a monophyletic group, supported by a bootstrap value of 100% and separate from the currently known genotypes (Fig. 2), suggests that they belong to a novel genotype. Although on the basis of the results of pairwise alignments of the HN protein genes the isolates formed two subclades, each supported by a moderate bootstrap value of 58% (Fig. 2), there were high similarities among them, 97 to 100%, indicating that the isolates are very closely related and share a common ancestry. As previously noted (13, 15, 29), viruses sharing temporal, geographical, antigenic, or epidemiological parameters tend to fall into specific lineages, a fact that has proved valuable in assessing the spread of ND. However, phylogenetic analyses of the presently known NDV genotypes and the Ugandan isolates revealed no common ancestry for these isolates.
FIG. 2.
Phylogenetic tree of the nucleotide sequences of the Ugandan NDV isolates based on a 377-bp region of the HN protein gene. Genotypes and subtypes are indicated at the right. Numbers at nodes indicate support by bootstrap resampling of 100 replicates.
Nucleotide sequence accession numbers.
The accession numbers for the F protein and the HN protein genes submitted to GenBank are AY367559 and AY371991 to AY372006, respectively (Table 1).
TABLE 1.
GenBank accession numbers for 16 Ugandan NDV isolates
| NDV isolatea | Accession no.b |
|---|---|
| Chicken/Pallisa/0405/01 | AY367559 |
| Chicken/Pallisa/0208/01 | AY371991 |
| Chicken/Soroti/0104/01 | AY371992 |
| Chicken/Pallisa/0406/01 | AY371993 |
| Chicken/Pallisa/0321/01 | AY371994 |
| Chicken/Pallisa/0305/01 | AY371995 |
| Chicken/Soroti/0509/01 | AY371996 |
| Chicken/Tororo/0222/01 | AY371998 |
| Chicken/Pallisa/0405/01 | AY371999 |
| Chicken/Pallisa/0408/01 | AY372000 |
| Chicken/Soroti/0814/01 | AY372001 |
| Chicken/Pallisa/0609/01 | AY372002 |
| Chicken/Pallisa/0206/01 | AY372003 |
| Chicken/Pallisa/0102/01 | AY372004 |
| Chicken/Pallisa/0407/01 | AY372005 |
| Chicken/Pallisa/0515/01 | AY372006 |
| Chicken/Pallisa/0103/01 | AY371997 |
Isolate designations are given as species host/geographical location of isolate/isolate number/year of isolation. All 16 Ugandan NDV isolates had an identical intracerebral pathogenicity index of 1.8.
All accession numbers were for the HN protein gene, except AY367559, which was for the F protein gene.
Acknowledgments
This study was funded by the Danish International Development Agency under the Livestock Systems Research Program in Uganda.
We thank Lisbeth Nielsen and Søs Hinge Siem of the Danish Veterinary Institute and Mukiibi Muka of the Livestock Health Research Institute for invaluable contributions to this study.
REFERENCES
- 1.Aldous, E. W., and D. J. Alexander. 2001. Detection and differentiation of Newcastle disease virus (avian paramyxovirus type 1). Avian Pathol. 30:117-128. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, D. J., R. J. Manvell, J. P. Lowings, K. M. Frost, M. S. Collins, P. H. Russell, and J. E. Smith. 1997. Antigenic diversity and similarities detected in avian paramyxovirus type 1 (Newcastle disease virus) isolates using monoclonal antibodies. Avian Pathol. 26:399-418. [DOI] [PubMed] [Google Scholar]
- 3.Alexander, D. J. 1988. Newcastle disease diagnosis, p. 147-160. In D. J. Alexander (ed.), Newcastle disease. Kluwer Academic Publishers, Boston, Mass.
- 4.Alexander, D. J. 1989. Newcastle disease, p. 114-120. In H. G. Purchase, L. H. Arp, C. H. Domermuth, and J. E. Pearson (ed.), A laboratory manual for isolation and identification of avian pathogens, 3rd ed. American Association of Avian Pathologists, Inc., Kennett Square, Pa.
- 5.Ballagi-Pordany, A., E. Wehmann, J. Herczeg, S. Belak, and B. Lomniczi. 1996. Identification and grouping of Newcastle disease virus strains by restriction site analysis of a region from the F gene. Arch. Virol. 141:243-261. [DOI] [PubMed] [Google Scholar]
- 6.Beard, C. W., and R. P. Hanson. 1984. Newcastle disease, p. 425-470. In M. S. Hofstad, H. J. Barnes, B. W. Calnek, W. M. Reid, and H. W. Yonder (ed.), Diseases of poultry, 8th ed. Iowa State University Press, Ames.
- 7.Berinstein, A., H. S. Sellers, D. J. King, and B. S. Seal. 2001. Use of a heteroduplex mobility assay to detect differences in the fusion protein cleavage site coding sequence among Newcastle disease virus isolates. J. Clin. Microbiol. 39:3171-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.CEC. 1992. Council Directive 92/66/EC of 14 July 1992 introducing community measures for the control of Newcastle disease. Offic. J. Eur. Comm. L260:1-20. [Google Scholar]
- 9.Collins, M. S., I. Strong, and D. J. Alexander. 1994. Evaluation of the molecular basis of pathogenicity of the variant Newcastle disease viruses termed “pigeon PMV-1 viruses.” Arch. Virol. 134:403-411. [DOI] [PubMed] [Google Scholar]
- 10.de Leeuw, O., and B. Peeters. 1999. Complete nucleotide sequence of Newcastle disease virus: evidence for the existence of a new genus within the subfamily Paramyxovirinae. J. Gen. Virol. 80:131-136. [DOI] [PubMed] [Google Scholar]
- 11.Felsentein, J. 1993. PHYLIP (phylogeny inference package), version 3.57c. University of Washington, Seattle.
- 12.Gould, A. R., E. Hansson, K. Selleck, J. A. Kattenbelt, M. Mackenzie, and A. J. Della-Porta. 2003. Newcastle disease viral fusion and haemagglutinin-neuraminidase gene motifs as markers for viral lineage. Avian Pathol. 32:361-373. [DOI] [PubMed] [Google Scholar]
- 13.Herczeg, J., E. Wehmann, R. R. Bragg, P. M. Travassos Dias, G. Hadjiev, O. Werner, and B. Lomniczi. 1999. Two novel genetic groups (VIIb and VIII) responsible for recent Newcastle disease outbreaks in southern Africa, one (VIIb) of which reached southern Europe. Arch. Virol. 144:2087-2099. [DOI] [PubMed] [Google Scholar]
- 14.Ke, M. G., J. H. Liu, Y. M. Lin, H. J. Chen, S. S. Tsai, and C. P. Chang. 2001. Molecular characterization of Newcastle disease viruses isolated from recent outbreaks in Taiwan. J. Virol. Methods 97:1-11. [DOI] [PubMed] [Google Scholar]
- 15.Lomniczi, B., E. Wehmann, J. Herczeg, A. Ballagi-Pordany, E. F. Kaleta, O. Werner, G. Meulemans, P. H. Jorgensen, A. P. Manté, A. L. J. Gielkens, I. Capua, and J. Damoser. 1998. Newcastle disease outbreaks in recent years in western Europe were caused by an old (VI) and a novel genotype (VII). Arch. Virol. 143:49-64. [DOI] [PubMed] [Google Scholar]
- 16.Meulemans, G., M. Gonze, M. C. Carlier, P. Petit, A. Burny, and Le Long. 1987. Evaluation of the use of monoclonal antibodies to haemagglutination and fusion glycoproteins of Newcastle disease virus for virus identification and strain differentiation purposes. Arch. Virol. 92:55-62. [DOI] [PubMed] [Google Scholar]
- 17.Millar, N. S., P. Chambers, and P. T. Emmerson. 1988. Nucleotide sequence of the fusion and haemagglutination neuraminidase glycoprotein genes of Newcastle disease virus, strain Ulster: molecular basis of variations of pathogenicity between strains. J. Gen. Virol. 69:613-620. [DOI] [PubMed] [Google Scholar]
- 18.Morrison, T., C. McQuain, T. Sergel, L. McGinnes, and J. Reitter. 1993. The role of the amino terminus F1 of the Newcastle disease virus fusion protein in cleavage and fusion. Virology 193:997-1000. [DOI] [PubMed] [Google Scholar]
- 19.Mukiibi, M. G. 1992. Epidemiology of Newcastle disease and the need to vaccinate local chickens in Uganda, p. 155-163. In P. B Spradbrow (ed.), Newcastle disease in village chickens: control with thermostable oral vaccines. Proceeding no. 39 of the International Workshop, Australia Centre for International Agricultural Research. Australian Center for International Agricultural Research, Canberra, Australia.
- 20.Murphy, F. A., C. M. Fauquet, D. H. L. Bishop, S. A. Ghabrial, A. W. Jarvis, G. P. Martelli, M. A. Mayo, and M. D. Summers. 1995. Virus taxonomy: classification and nomenclature of viruses. Sixth report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 21.Nagai, Y., H. Ogura, and H. Klenk. 1976. Studies on the assembly of the envelope of Newcastle disease virus. Virology 69:523-538. [DOI] [PubMed] [Google Scholar]
- 22.Palmeri, S., and M. L. Perdue. 1989. An alternative method of oligonucleotide fingerprinting for resolving Newcastle disease virus-specific RNA fragments. Avian Dis. 33:345-350. [PubMed] [Google Scholar]
- 23.Palmeri, S. 1989. Genetic relationships among lentogenic strains of Newcastle disease virus. Avian Dis. 33:351-356. [PubMed] [Google Scholar]
- 24.Rott, R. 1992. Molecular aspects of Newcastle disease virus pathogenicity, p. 139-144. In Proceedings of the Workshop on Avian Paramyxoviruses. Rauischholzhausen, Marburg, Germany.
- 25.Sakaguchi, T., T. Toyoda, B. Gotoh, N. M. Inocencio, K. Kuma, T. Miyata, and Y. Nagai. 1989. Newcastle disease virus evolution. 1. Multiple lineages defined by sequence variability of the haemagglutinin-neuraminidase gene. Virology 169:260-272. [DOI] [PubMed] [Google Scholar]
- 26.Seal, B. S., D. J. King, D. P. Locke, D. A. Senne, and M. W. Jackwood. 1998. Phylogenetic relationships among highly virulent Newcastle disease virus isolates obtained from exotic birds and poultry from 1989 to 1996. J. Clin. Microbiol. 36:1141-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G Higgins. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westbury, H. 2001. Newcastle disease virus: an evolving pathogen? Avian Pathol. 30:5-11. [DOI] [PubMed] [Google Scholar]
- 29.Yang, C. Y., P. C. Chang, J. M. Hwang, and H. K. Sheih. 1997. Nucleotide sequence and phylogenetic analysis of Newcastle disease virus isolates from recent outbreaks in Taiwan. Avian Dis. 41:365-373. [PubMed] [Google Scholar]
- 30.Yu, L., Z. Wang, Y. Jiang, L. Chang, and J. Kwang. 2001. Characterization of newly emerging Newcastle disease virus isolates from the People's Republic of China and Taiwan. J. Clin. Microbiol. 39:3512-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]