Abstract
Pythium insidiosum, the only species in the genus that infects mammals, is the etiological agent of pythiosis, a granulomatous disease characterized by cutaneous and subcutaneous lesions and vascular diseases. Accurate diagnosis of pythiosis and identification of its causal agent are often inconsistent with current immunological diagnostic methods. A species-specific DNA probe was constructed by using a 530-bp HinfI fragment from the ribosomal DNA intergenic spacer of P. insidiosum. When the probe was incubated with dot blots of genomic DNA from 104 Pythium species, it hybridized only to the DNA of P. insidiosum and P. destruens—two species that have been considered conspecific. The probe also hybridized to DNA from 22 P. insidiosum isolates in this study, regardless of their geographic origin or animal host. When tested against genomic DNA from other pathogenic organisms (Aspergillus fumigatus, Basidiobolus ranarum, Conidiobolus coronatus, Lagenidium giganteum, Paracoccidioides brasiliensis, and Prototheca wickerhamii), no cross-hybridization of the probe was detected. The specificity of the probe to hybridize to genomic DNA from all isolates of P. insidiosum and not cross-react with DNA from other Pythium species or pathogens that cause symptoms similar to pythiosis in their hosts makes it a powerful tool for the accurate diagnosis of pythiosis. In addition, the probe has the potential for pathological and environmental diagnostic applications.
Pythium insidiosum (11) is the etiological agent of pythiosis, a granulomatous disease that affects animals including equines, canines, felines, cattle, and humans. The symptoms of pythiosis include cutaneous and subcutaneous infections, bone lesions, esophagitis, gastrointestinal diseases, and pulmonary infections. Pythium is a genus in the class Oomycetes in the Kingdom Straminipila (39) that is comprised of over 120 described species (53). However, P. insidiosum is the only species in the genus capable of infecting mammalian hosts, whereas the other species are saprophytic or pathogenic to plants. Results from restriction fragment length polymorphism analysis (46) and sequence analysis (47) revealed three clades of P. insidiosum isolates representing geographical variants or cryptic speciation.
Like most Pythium species, P. insidiosum is difficult to identify morphologically. Gomori's methenamine-silver nitrate stains P. insidiosum hyphae in tissue nonspecifically. In the infected tissues, short, septate hyphae with perpendicular branching are rarely visible, but members of the Zygomycetes have a similar appearance in histological sections (12). Perhaps the most important cultural identification feature is the production of motile biflagellate zoospores (33), but this is relatively nonspecific and may take weeks to observe, and for some strains it is difficult to induce zoospore production in culture. Another special feature of pythiosis in equines is the presence of numerous “kunkers” (yellow-white, coral-like necrotic masses of tissue that contain hyphae) in lesions that exhibit an intense eosinophilic inflammatory reaction (10, 50). However, similar structures are encountered in habronemiasis and subcutaneous zygomycosis caused by the nematode Habronema and the Zygomycetes (Entomophthorales) Basidiobolus and Conidiobolus, respectively (10, 43), which can lead to a misdiagnosis of the disease. In dogs, the presence of tumor-like masses is often confused with neoplasia (7, 52).
Numerous serological methods have been developed to diagnose and monitor pythiosis, including a complement fixation test (36), immunoperoxidase test (6), and a fluorescein isothiocyanate-conjugated rabbit antiglobulin for staining hyphal elements in tissue (32). Immunodiffusion (ID) tests have been the most widely used serological diagnostic methods for pythiosis (31). Although the ID test is entirely specific for pythiosis, negative ID test results have been reported in cases of proven pythiosis (38, 51). An enzyme-linked immunosorbent assay (ELISA) method with hyphal antigens has been shown to be a more sensitive and accurate method for detecting pythiosis in the early and chronic stages of the disease and is especially useful when the ID test fails (19, 25, 35).
Due to the often contradictory results from serological tests, it was our goal to find an additional approach for diagnosing pythiosis and detecting P. insidiosum by molecular biology techniques. DNA probes are particularly useful for the identification of pathogenic agents because probes can often detect DNA in mixed cultures, so pure cultures are unnecessary. Ribosomal DNA (rDNA) is an especially useful target for DNA probes since it is a highly abundant target in cells, representing a preamplification step that increases test sensitivity, and produces a strong hybridization signal. Some investigators have successfully used regions of rDNA to diagnose some cases of pythiosis in humans and animals (3, 18, 41).
In the present study, a species-specific DNA probe from the rDNA intergenic spacer (IGS) of the P. insidiosum ex-type culture was designed to distinguish P. insidiosum from all other Pythium species was from other pathogens that cause diseases that produce symptoms that resemble pythiosis. The potential applications for the DNA probe are discussed.
MATERIALS AND METHODS
Culture conditions and DNA isolation.
A collection of 128 Pythium isolates representing 104 species was used in the present study (Tables 1 and 2). Pythium isolates were maintained on cornmeal agar (CMA) prepared according to Gams et al. (16). P. insidiosum strains and all other fungal and algal strains were grown on Sabouraud dextrose agar (2.0% dextrose, 1.0% peptone).
TABLE 1.
List of Pythium species used in Fig. 2 to determine the specificity of the P. insidiosum probe
| Pyrhium species | Designationa | Accession no.b | Pyrhium species | Designationa | Accession no.b | |
|---|---|---|---|---|---|---|
| P. acanthicum | acmc | CBS 284.31 | P. mamillatum | mamd | CBS 251.28 | |
| P. acanthophoron | acn | CBS 337.29 | P. marinum | mar | CBS 312.93 | |
| P. acrogynum | acrc | CBS 549.88 | P. marsipium | msp | CBS 773.81 | |
| P. adhaerens | adh | CBS 520.74 | P. mastophorum | masd | CBS 375.72 | |
| P. amasculinum | ama | CBS 552.88 | P. middletonii | midd | CBS 528.74 | |
| P. anandrum | anac | CBS 258.31 | P. minus | minc | CBS 226.88 | |
| P. angustatum | angd | CBS 522.74 | P. monospermum | mone | CBS 158.73 | |
| P. aphanidermatum | aphc | CBS 118.80 | P. multisporum | mulc | CBS 470.50 | |
| P. apleroticum | aple | CBS 772.81 | P. myriotylum | myre | CBS 254.70 | |
| P. aquatile | aque | CBS 215.80 | P. nagaii | nag | IMI 308183 | |
| P. aristosporum | aric | CBS 263.38 | P. nodosum | nod | CBS 102274 | |
| P. arrhenomanes | arrc | CBS 324.62 | P. nunn | nune | CBS 808.96 | |
| P. australe | aus | IMI 332970 | P. oedochilum | oedf | CBS 292.37 | |
| P. boreale | borc | CBS 551.88 | P. okanaganense | okac | CBS 315.81 | |
| P. buismaniae | buic | CBS 288.31 | P. oligandrum | olid | CBS 382.34 | |
| P. capillosum | capf | CBS 222.94 | P. orthogonon | orte | CBS 376.72 | |
| P. catenulatum | catd (a) | CBS 843.68 | P. ostracodes | ostd | CBS 768.73 | |
| cat (b) | CBS 842.68 | P. pachycaule | pacc | CBS 227.88 | ||
| P. chamaehyphon | chac | CBS 259.30 | P. paddicum | pad | CBS 698.83 | |
| P. chondricola | choc | CBS 203.85 | P. paroecandrum | pard | CBS 157.64 | |
| P. coloratum | cole | CBS 154.64 | P. parvum | pvmc | CBS 225.88 | |
| P. conidiophorum | con | CBS 224.88 | P. periilum | perd | CBS 169.68 | |
| P. contiguanum | ctg | CBS 221.94 | P. periplocum | pcmc | CBS 289.31 | |
| P. cucurbitacearum | cuc | CBS 748.96 | P. perplexum | plx | CBS 674.85 | |
| P. cylindrosporum | cylc | CBS 218.94 | P. pleroticum | ple | CBS 776.81 | |
| P. debaryanum | deb | CBS 752.96 | P. plurisporium | plue | CBS 100530 | |
| P. deliense | dele | CBS 314.33 | P. polymastum | pold | CBS 811.70 | |
| P. destruens | dese | ATCC 64221 | P. polymorphon | pmn | CBS 751.96 | |
| P. diclinum | dice | CBS 664.79 | P. porphyrae | pord | CBS 369.79 | |
| P. dimorphum | dimc | CBS 406.72 | P. prolatum | proc | CBS 845.68 | |
| P. dissimile | disc | CBS 155.64 | P. pyrilobum | pyrc | CBS 158.64 | |
| P. dissotocum | dcm | CBS 166.68 | P. radiosum | radc | CBS 217.94 | |
| P. echinulatum | echd | CBS 281.64 | P. rostratum | rose | CBS 533.74 | |
| P. erinaceus | eric | CBS 505.80 | P. salpingophorum | sald | CBS 471.50 | |
| P. flevoense | flec | CBS 234.72 | P. scleroteichum | sclf | CBS 294.37 | |
| P. folliculosum | folc | CBS 220.94 | Pythium sp. | sp-1 | CBS 739.94 | |
| P. graminicola | grae | CBS 327.62 | Pythium sp. | sp-2 | CBS 101876 | |
| P. grandisporangium | gspc | CBS 286.79 | Pythium sp. | sp-3 | CBS 223.94 | |
| P. helicandrum | helf | CBS 393.54 | P. spinosum | spid | CBS 275.67 | |
| P. helicoides | hcsd | CBS 286.31 | P. splendens | spld | CBS 462.48 | |
| P. heterothallicum | hetc (m) | CBS 450.67 | P. sulcatum | sulc | CBS 603.73 | |
| hetc (f) | CBS 451.67 | P. sylvaticum | sylc (m) | CBS 452.67 | ||
| P. hydnosporum | hydd | CBS 253.60 | sylc (f) | CBS 453.67 | ||
| P. hypogynum | hyp | CBS 692.79 | P. torulosum | tord | CBS 316.33 | |
| P. indigoferae | indd | CBS 261.30 | P. tracheiphilum | trac | CBS 323.65 | |
| P. inflatum | infe | CBS 168.68 | P. ultimum var. ultimum | ulue | CBS 398.51 | |
| P. insidiosum | inse | CBS 574.85 | P. ultimum var. sporangiiferum | ulsc | CBS 219.65 | |
| P. intermedium | int | CBS 266.38 | P. uncinulatum | uncc | CBS 518.77 | |
| P. irregulare | ine | CBS 250.28 | P. undulatum | unde | CBS 157.69 | |
| P. iwayamai | iwad | CBS 156.64 | P. vanterpoolii | vanc | CBS 295.37 | |
| P. kunmingense | kunc | CBS 550.88 | P. vexans | vexd | CBS 119.80 | |
| P. lutarium | lutc | CBS 222.88 | P. violae | viod | CBS 159.64 | |
| P. macrosporum | macc (+) | CBS 574.80 | P. volutum | vol | CBS 699.83 | |
| macc (−) | CBS 575.80 | P. zingiberis | zin | CBS 216.82 |
Designations by which isolates are referred to in the tables and figures of this study. For heterothallic species, mating types are designated by a, b, m (male), f (female), or + and − (opposite mating types) in parentheses.
Accession numbers of isolates in the following culture collections: CBS (Centraalbureau voor Schimmelcultures, Utrecht, Netherlands), IMI (International Mycological Institute, Egham, United Kingdom), ATCC (American Type Culture Collection, Manassas, Va.).
Ex-type strain.
Isolate used by Van der Plaats-Niterink (53) for species description.
Isolate designated as the neotype strain because all ex-type material is missing.
Authentic strain, identified by the author of the species.
TABLE 2.
Isolates of P. insidiosum and other pathogenic organisms used to test the specificity of the DNA probe (Fig. 3 and 4)
| Species | Designationa | Accession no.b | Host | Origind |
|---|---|---|---|---|
| Pythium insidiosum | 65c | CBS 574.85 | Equine | Costa Rica |
| 291 | CBS 673.85 | Human | Thailand | |
| 296 | CBS 777.84 | Mosquito larva | India | |
| 297 | CBS 702.83 | Equine | Japan | |
| 339 | CBS 575.85 | Equine | Costa Rica | |
| 344 | CBS 580.85 | Equine | Costa Rica | |
| 393 | CBS 101039 | Human | India | |
| 394 | CBS 101555 | Equine | Brazil | |
| M4 | Undepositedc | Feline | Florida | |
| M6 | ATCC 200269 | Human | Tennessee | |
| M7 | Undepositedc | Human | Thailand | |
| M9 | Undepositedc | Canine | Louisiana | |
| M15 | ATCC 28251 | Equine | Papua New Guinea | |
| M16 | ATCC 76049 | Human | Haiti | |
| M18 | ATCC 90478 | Tremarctos ornatus | South Carolina | |
| M19 | ATCC 90586 | Human | Texas | |
| M20 | ATCC 200268 | Canine | North Carolina | |
| M21 | Undepositedc | Human | Pennsylvania | |
| M22 | Undepositedc | Canine | Wisconsin | |
| M25 | Undepositedc | Human | Thailand | |
| Pythium destruens | M23e | ATCC 64221 | Equine | Papua New Guinea |
| M24 | ATCC 64218 | Equine | Papua New Guinea | |
| Aspergillus flavus | Af | Undepositedc | Human | United States |
| Basidiobolus ranarum | Br | Undepositedc | Human | United States |
| Conidiobolus coronatus | Cc | Undepositedc | Human | United States |
| Lagenidium giganteum | Lgf | ATCC 36492 | Mosquito larva | North Carolina |
| Paracoccidioides brasiliensis | Pb | Undepositedc | Human | Argentina |
| Prototheca wickerhamii | Pwg | Undeposited | Human | ? |
Designations by which isolates are referred to in the tables and figures of this study.
Accession numbers of isolates in the following culture collections: CBS (Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands) and ATCC (American Type Culture Collection, Manassas, Va.).
Personal collection of L. Mendoza.
?, unknown geographic origin.
Ex-type strain.
DNA provided by M. Hudspeth, Northern Illinois University, DeKalb, Ill.
Culture provided by R. Scott Pore, West Virginia University, Morgantown, W.Va.
For DNA isolation, small blocks of actively growing cultures were used to inoculate petri dishes containing pea broth (filtered and autoclaved decoction of 200 g of frozen peas, boiled for 20 min in 1 liter of deionized water, to which 5 g of glucose was added). Petri dishes were incubated at the optimum temperature for the species concerned (53). After 5 to 10 days, depending on the growth rate of the species, mycelia were harvested by filtration, washed twice with demineralized water, freeze-dried, and stored at −20°C until ready for DNA isolation. For each strain, 30 mg of freeze-dried mycelium were ground with sand and a micropestle and DNA was isolated according to the method of Möller et al. (37) with modifications as described previously (24).
PCR.
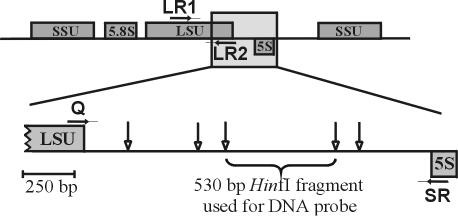
The rDNA IGS between the large subunit (LSU) and 5S rRNA genes (IGS-1) of P. insidiosum (isolate 65) was amplified with primers Q (ACGCCTCTAAGTCAGAATC) (5) and SR (GAAGCCCGGGTGCTGTCTAG) (24) (Fig. 1). In addition, primers LR1 (GCATATCAATAAGCGGAGGA) and LR2 (GACTTAGAGGCGTTCAG) (21) were used to amplify a 2,500-bp region of the LSU rRNA gene for P. insidiosum (isolate 65), Aspergillus flavus, Basidiobolus ranarum, and Prototheca wickerhamii (Fig. 1). Reactions were carried out in 50-μl volumes containing 5 μl of Taq Extender 10× reaction buffer (Stratagene, La Jolla, Calif.); 200 μM (each) dATP, dCTP, dGTP, and dTTP; 1.5 mM MgCl2; 20 pmol of each primer, 10 ng of DNA template, 1.25 U of Taq DNA polymerase, and 1.25 U of Taq Extender (Stratagene). Amplifications were done by using a Techne Unit Genius Thermocycler (Techne, Inc., Princeton, N.J.) with the following temperature cycling parameters: denaturation at 93°C for 3 min for the first cycle and 1 min for subsequent cycles, annealing for 1 min at 50°C, and elongation at 72°C for 2 min for primers Q and SR, and 5 min for primers LR1 and LR2. The parameters were repeated for 30 cycles with Q and SR and 25 cycles with LR1 and LR2, followed by a final elongation at 72°C for 8 to 10 min.
FIG. 1.
HinfI restriction site map of IGS-1 for P. insidiosum (isolate 65). Vertical arrows indicate locations of HinfI restriction sites. Rectangles represent coding regions for the LSU, SSU (small subunit), 5.8S, and 5S rRNA genes. Locations and orientations of the primers LR1, LR2, Q, and SR are indicated by horizontal arrows.
Restriction site mapping of IGS-1.
A partial digestion of the Q-SR amplicon was carried out in a total volume of 15 μl containing 8 to 10 μl of PCR product, 1.5 μl of REact 2 buffer, and 5 U HinfI (Invitrogen, Carlsbad, Calif.). Eight individual reactions were incubated at 37°C and reactions were stopped at 10 s, at 30 s, and at 1, 3, 5, 10, 30, and 60 min by the addition of 4.0 μl of gel loading buffer (40% [wt/vol] sucrose, 0.25% bromophenol blue, 20 mM EDTA). Digested DNA fragments were separated in a 1.0% agarose gel in 1× Tris-borate-EDTA buffer for subsequent Southern blotting and hybridization with the Q-primer oligonucleotide probe for the construction of the HinfI restriction site map of IGS-1.
Labeling DNA probes and blot preparation.
Fragments of DNA to be used for hybridization studies were excised and purified from agarose gels by using the GeneClean kit (Qbiogene, Inc., Carlsbad, Calif.). Restriction fragment and PCR product DNA probes were random-primed labeled with digoxigenin (DIG)-11-dUTP by using the DIG DNA LABELING Kit (Roche Diagnostics GmbH, Mannheim, Germany). The Q-primer oligonucleotide probe was 3′-end labeled with DIG-11-ddUTP by using the DIG oligonucleotide 3′-end labeling kit (Roche Diagnostics GmbH). Both labeling procedures were carried out according to the manufacturer's specifications for use in DNA-DNA hybridizations with Southern blots and genomic DNA spot blots.
Spot blots of the appropriate dilutions of genomic DNA were prepared by spotting 1.0 μl of denatured DNA (heated at 100°C for 10 min and immediately chilled on ice) on a Hybond-N membrane (Amersham Corp., Piscataway, N.J.). Southern blots of partially digested DNA were prepared according to the method of Southern (49). The DNA was fixed to the membranes from both dot blots and Southern blots by cross-linking under UV light on a transilluminator for 4 min.
DNA-DNA hybridization.
For hybridizations with DNA probes, membranes were incubated for 1 h with constant agitation in 15 ml of prehybridization solution (1% sodium dodecyl sulfate [SDS], 1 M NaCl) at 42°C for the Q-primer probe and at 60°C for restriction fragment or PCR product probes. Subsequently, the DNA probe was denatured at 100°C for 10 min and then added directly to the prehybridization solution after a 1-h incubation at a final concentration of 5 to 25 ng/ml. The hybridization reaction was conducted overnight at the corresponding prehybridization temperature with constant agitation. Membranes were then washed twice, 5 min per wash, in 2× wash solution (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate; pH 7.0]-0.1% SDS) at room temperature. Two additional washes were then done for 15 min, each in 0.1× wash solution (0.1× SSC-0.1% SDS) at the temperature used for the initial hybridization reaction. After the posthybridization washes, chemiluminescence detection was performed according to the manufacturer's protocol (Roche), and membranes were exposed to Kodak X-Omat X-ray film (Eastman Kodak Co., Rochester, N.Y.) for 1 to 30 min to detect the chemiluminescent reaction. Subsequent to further chemiluminescent detections, membranes were stripped to remove DNA probes by rinsing the membrane in water for 2 min, followed by two 15 min incubations at 37°C in stripping solution (0.2 M NaOH, 0.1% SDS) and washed two to three times in 2× SSC.
Cloning and sequencing of the DNA probe.
The 530-bp HinfI fragment that was to be used as a species-specific probe was cloned into the pPCR-Script Amp SK(+) vector (Stratagene) by using the PCR-Script AMP cloning kit. Plasmid DNA was purified from the cultures by using the QIAprep Spin Miniprep kit (Qiagen, Inc., Mississauga, Ontario, Canada) and screened for the presence of the insert by restriction endonuclease digestion. Plasmids containing the insert were sequenced in both directions with the primers T3 (AATTAACCCTCACTAAAGGG) and T7, (GTAATACGACTCACTATAGGGC) by using a 377XL ABI DNA Sequencer (University Core DNA Services, University of Calgary, Calgary, Alberta, Canada).
Nucleotide sequence accession number.
The nucleotide sequence of the 530-bp HinfI fragment from the rDNA IGS-1 of P. insidiosum (CBS 574.85) is available in the GenBank database (accession number AY484596).
RESULTS
Characterization of the P. insidiosum species-specific probe
The region between the LSU and 5S rRNA genes (IGS-1) of the P. insidiosum ex-type culture (CBS 574.85) was PCR amplified with primers Q and SR (Fig. 1). A HinfI partial digestion of the 1,900-bp Q-SR amplicon was Southern blotted and hybridized with a DIG-11-ddUTP-labeled Q-primer oligonucleotide probe to construct the HinfI restriction site map of IGS-1 (Fig. 1).
The central 530-bp HinfI fragment was selected as a potential species-specific DNA probe for P. insidiosum because it did not overlap any of the gene regions and because it gave strong hybridization signals in preliminary trials. The fragment was cloned into the pPCR-Script Amp SK(+) vector and sequenced. The resulting sequence had a G+C ratio of 44.34% and a Tm of 82.14°C. A basic local alignment search tool (BLAST) search was done on GenBank, and no significant hits were obtained. The fragment was then random-primed labeled with DIG-11-dUTP to be used in hybridization tests with genomic DNA of several Pythium species and animal pathogens.
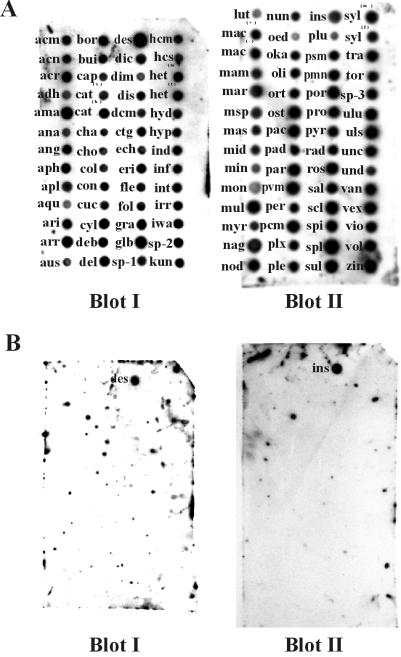
Probe specificity among Pythium species.
The specificity of the probe for P. insidiosum was tested by performing DNA-DNA hybridizations with spot blots of genomic DNA from 101 Pythium species. To ensure that approximately equal amounts of target DNA were present for each species, spot blots containing various genomic DNA concentrations for each species were hybridized with a 2,500-bp LSU rRNA gene probe (amplified with primers LR1 and LR2; Fig. 1) of the P. insidiosum ex-type culture (data not shown). Concentrations of genomic DNA that produced hybridization signals of equal intensities were selected for each species and spotted onto two membranes (blots I and II; Fig. 2), which were then hybridized with the LSU rRNA gene probe of P. insidiosum (isolate 65) (Fig. 2A). The membranes were then stripped to remove bound probe and rehybridized with the 530-bp P. insidiosum IGS-1 fragment probe (Fig. 2B). The probe hybridized to genomic DNA of P. insidiosum isolate 65 and P. destruens isolate M23 but did not cross-react with genomic DNA from any of the other 102 Pythium species.
FIG. 2.
Dot blot hybridizations of genomic DNA representing 104 Pythium species with an LSU rRNA gene probe (LR1-LR2 PCR amplicon) from P. insidiosum (isolate 65) (A) and a 530-bp IGS-1 fragment probe from IGS-1 of P. insidiosum (isolate 65) (B). Subsequent to hybridization with the LR1-LR2 probe, the membranes in panel A were stripped and rehybridized with the IGS-1 fragment probe in panel B. Hybridization conditions were identical for both probes. The 3-letter abbreviations correspond to the isolate designations in Table 1.
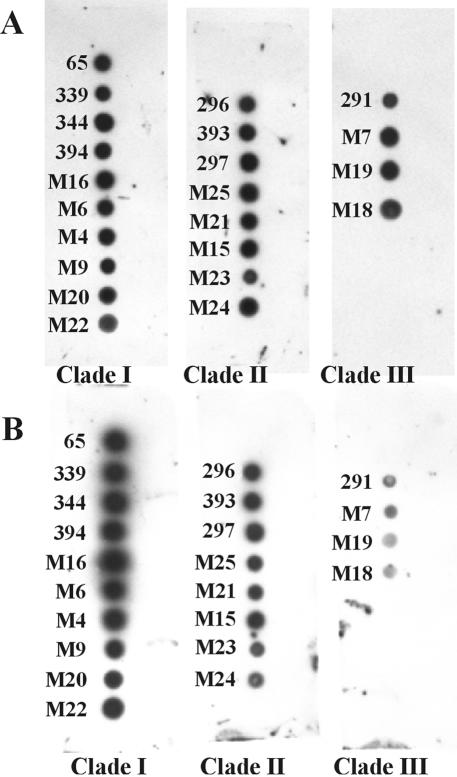
Probe sensitivity for P. insidiosum isolates.
Previously, sequence analysis of the rDNA internal transcribed spacer revealed the existence of three distinct clades of P. insidiosum isolates from various animals hosts and geographic regions (47). Clade I consisted of isolates from North, Central, and South America. Clade II contained isolates from Asia, Australia, and the United States, and clade III consisted of isolates from Thailand and the United States. To test whether the P. insidiosum probe would hybridize to genomic DNA from P. insidiosum isolates representing each of these clades, spot blots were prepared by using genomic DNA from 22 isolates (Table 2). Three individual blots were prepared, each corresponding to one of the three clades (Fig. 3). The target genomic DNA concentrations were first normalized by hybridization with the 2,500-bp LSU rRNA gene probe of P. insidiosum (Fig. 3A). The membranes were then stripped and rehybridized with the 530-bp P. insidiosum IGS-1 probe (Fig. 3B). The probe hybridized with DNA from all 22 isolates of P. insidiosum, but the hybridization signal was most intense with isolates representing clade I. The intensity of the hybridization signal was decreased among clade II isolates and was weakest with the four clade III isolates (Fig. 3B).
FIG. 3.
Dot blot hybridizations of P. insidiosum genomic DNA from isolates representing the genetic clades (clades I, II, and III) discussed by Schurko et al. (47). Blots were hybridized with the LSU rRNA gene probe (LR1-LR2 amplicon) from P. insidiosum 65 (A) and the 530-bp IGS-1 fragment probe from P. insidiosum 65 (B). Subsequent to hybridization with the LR1-LR2 probe, membranes in panel A were stripped and rehybridized with the IGS-1 fragment probe in panel B. Isolate numbers correspond to those listed in Table 2.
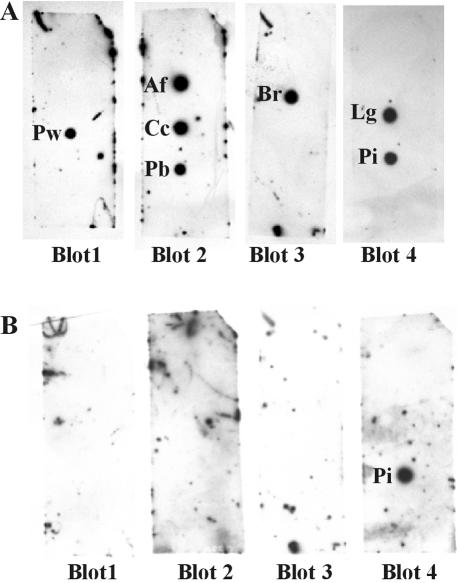
Probe specificity when tested with other pathogens.
The specificity of the P. insidiosum IGS-1 fragment probe was tested against genomic DNA from several pathogenic organisms that could be present in lesions similar to those caused by pythiosis (Table 2). Spot blots were prepared (Fig. 4) by using genomic DNA of Aspergillus flavus (Ascomycota), Basidiobolus ranarum (Zygomycota), Conidiobolus coronatus (Zygomycota), Paracoccidioides brasiliensis (Ascomycota), and Prototheca wickerhamii (Chlorophyta). An isolate of Lagenidium giganteum (Oomycota) was also included, along with the ex-type culture of P. insidiosum. Each blot was first hybridized with a LSU rRNA gene probe to ensure that hybridization signals were approximately equal (Fig. 4A). The membranes were stripped, and all four blots were simultaneously rehybridized in the same hybridization bottle with the 530-bp P. insidiosum IGS-1 fragment probe which only hybridized to genomic DNA of P. insidiosum isolate 65 and did not cross-react with DNA from any of the other pathogens (Fig. 4B).
FIG. 4.
Spot blot hybridizations of genomic DNA from organisms that produce symptoms similar to pythiosis. (A) Membranes were hybridized with the LSU rRNA gene probes (LR1-LR2 amplicons) amplified from the following organisms: Prototheca wickerhamii (blot 1), Aspergillus flavus and Basidiobolus ranarum (1:1 mixture of both probes; blots 2 and 3), and P. insidiosum (blot 4). (B) Membranes hybridized with 530-bp IGS-1 fragment probe from P. insidiosum. Subsequent to hybridization with the LR1-LR2 probes, membranes in panel A were stripped and rehybridized with the IGS-1 fragment probe in panel B. Species abbreviations are as follows: Af, Aspergillus flavus; Br, Basidiobolus ranarum; Cc, Conidiobolus coronatus; Lg, Lagenidium giganteum; Pi, Pythium insidiosum; Pb, Paracoccidioides brasiliensis; Pw, Prototheca wickerhamii.
DISCUSSION
A species-specific DNA probe for P. insidiosum was constructed by using a 530-bp HinfI fragment from IGS-1 between the LSU and 5S rRNA genes of P. insidiosum. When tested against genomic DNA of 104 Pythium species, the probe specifically hybridized to DNA of the ex-type cultures of P. insidiosum and P. destruens. This not only demonstrated the ability of the probe to distinguish P. insidiosum from other Pythium species but also supported antigenic (34) and molecular evidence (46, 47) that P. insidiosum and P. destruens should be considered conspecific. The finding that the probe hybridized to all 22 isolates in the three geographic clades described by Schurko et al. (47) supports the idea that P. insidiosum is indeed a single species or a group of very closely related species. However, the differences in intensity of hybridization between clades, and particularly the weak reactions for clade III, the most distant cluster in terms of restriction fragment length polymorphisms and internal transcribed spacer sequences (46, 47), point to significant genetic divergence within the species.
The probe was also able to distinguish P. insidiosum from other pathogenic organisms that cause diseases of humans and animals (Fig. 4). Basidiobolus ranarum and Conidiobolus coronatus are zygomycetes which cause conidiobolomycosis and basidiobolomycosis, respectively. The ascomycetes Aspergillus flavus and Paracoccidioides brasiliensis are the etiological agents of aspergillosis and paracoccidioidomycosis, respectively. Prototheca wickerhamii is a chlorophyte alga which causes protothecosis. These clinical entities can cause cutaneous and subcutaneous lesions and infections of internal organs, arteries, body cavities, and bones. Among these organisms, species-specific probes have been constructed for Paracoccidioides brasiliensis (29, 45) and A. flavus (44), but the cross-hybridization of these probes with P. insidiosum has not yet been examined. Misdiagnosis of some of these diseases may be possible due to their common symptoms and histopathological characteristics. This can be especially problematic in the case of pythiosis because this disease is often confused with those caused by members of the zygomycetes, and this would likely result in the prescription of antifungal agents. Although Shenep et al. (48) demonstrated successful synergistic treatment with a combination of itraconazole and terbinafine, therapeutic measures are not expected to have a curative effect since P. insidiosum is not a member of the true fungi and does not respond to common antifungal agents.
The isolate of Lagenidium giganteum included in the analysis was from a mosquito larva in North Carolina. Similarly, P. insidiosum 296 was from a mosquito larva in India. The hybridization of the probe to DNA from P. insidiosum 296 and not to DNA from L. giganteum supports the distinction of these two isolates as separate species, despite their similar hosts, and also confirms the ability of P. insidiosum to infect insect hosts, as well as mammalian hosts. Lagenidium is closely related to Pythium based on coxII gene sequencing (22) and LSU rRNA gene sequencing (42). This result also indicated that the probe does not cross-react with a closely related genus, although only one Lagenidium species was tested.
Lagenidiosis, a recently described disease in canines similar to pythiosis and zygomycosis, is thought to be caused by a Lagenidium species, although not necessarily L. giganteum (20). Znajda et al. (55) described a nested PCR assay to detect oomycete DNA in tissues from cases of pythiosis and lagenidiosis, but the assay was tested against a limited number of P. insidiosum isolates. Since the P. insidiosum probe described here detects genetically diverse strains of P. insidiosum, results from hybridization studies might help determine whether the etiological agent of lagenidiosis is indeed Lagenidium sp. or in fact a cryptic strain of P. insidiosum, if the probe also hybridizes with that strain.
DNA probes from regions of rDNA have been developed to identify many infectious fungal agents. Species-specific probes have been described for several Candida species (8, 9, 15), Trichophyton rubrum (14), and a variety of other fungal pathogens (13, 29, 44, 45). Species-specific probes have also been constructed to identify several plant pathogenic oomycetes, including Pythium spp. (4, 24, 27, 28, 30), Phytophthora spp. (17, 23, 26), and Personosclerospora sorghi (54).
The P. insidiosum IGS-1 fragment probe has several practical applications in clinical and diagnostic settings. The probe can be used to classify an organism as P. insidiosum from suspected cases of pythiosis and to distinguish it from pathogenically related zygomycetes, ascomycetes, and algae, provided a pure culture of the infectious agent is available. However, the ability of the probe to detect P. insidiosum in infected tissue sections or blood samples would avoid the need to obtain pure cultures. The probe could also confirm the diagnosis of pythiosis based on results from histopathological examinations and ID or ELISA tests, which have been shown to produce conflicting results (40, 51). It could also be useful for monitoring a patient's response to therapy by detecting the presence or absence of the organism in blood or tissue samples.
A common route of infection by P. insidiosum is via zoospores through small cuts or body cavities of individuals who spend time in or drink from stagnant swampy waters or fields. Many fungal species, including other Pythium species, thrive in such environments. The probe could serve as a diagnostic tool to detect P. insidiosum in such environments and to differentiate it from other Pythium species that may be present and harmless to animal hosts. Commercial kits are available that use ELISA to detect Pythium and Phytophthora zoospores in irrigation water (2). In addition, polyclonal antibodies (1) and competitive PCR (38) have been used to detect the zoospores of P. porphyrae in seawater from Porphyra cultivation farms. Similarly, the DNA probe for P. insidiosum has the potential to detect P. insidiosum zoospores in contaminated water sources that are frequented by humans and animals.
The species-specific probe for P. insidiosum presented here provides a molecular biological diagnostic method that can be used, in conjunction with present clinical tools, to diagnose cases of pythiosis with greater confidence and accuracy. Rapid identification is essential so that an appropriate clinical decision can be made concerning the identity of the isolate, the therapeutic approach to be used, and the dosage and duration of the appropriate therapy. The probe also has the potential to be used as a tool to screen tissue and blood samples and environmental sources that may harbor the infectious organism.
Acknowledgments
This study was supported by a research grant to G.R.K. from the Natural Sciences and Engineering Research Council of Canada and in part by grant 91-0545 to L.M. from the Center for Animal Production and Enhancement, Michigan State University.
We thank Ans de Cock, Roger Herr, Ayush Kumar, and Susan Cuvelier for research assistance.
REFERENCES
- 1.Addepalli, M. K., and Y. Fujita. 2001. Serological detection of red rot disease initiation stages of microbial pathogen, Pythium porphyrae (Oomycota) on Porphyra yezoensis. J. Appl. Phycol. 13:221-227. [Google Scholar]
- 2.Ali-Shtayeh, M. S., J. D. MacDonald, and J. Kabashima. 1991. A method for using commercial ELISA tests to detect zoospores of Phytophthora and Pythium species in irrigation water. Plant Dis. 75:305-311. [Google Scholar]
- 3.Badenoch, P. R., D. J. Coster, B. L. Wetherall, H. T. Brettig, M. A. Rozenbilds, A. Drenth, and G. Wagels. 2001. Pythium insidiosum keratitis confirmed by DNA sequence analysis. Br. J. Ophthalmol. 85:502-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey, A. M., D. J. Mitchell, K. L. Manjunath, G. Nolasco, and C. L. Niblett. 2002. Identification to the species level of plant pathogens Phytophthora and Pythium by using sequences of the ITS1 region of ribosomal DNA as capture probes for PCR ELISA. FEMS Microbiol. Lett. 207:153-158. [DOI] [PubMed] [Google Scholar]
- 5.Belkhiri, A., J. Buchko, and G. R. Klassen. 1992. The 5S ribosomal RNA gene in Pythium species: two different genomic locations. Mol. Biol. Evol. 9:1089-1102. [DOI] [PubMed] [Google Scholar]
- 6.Brown, C. C., J. J. McClure, P. Triche, and C. Crowder. 1988. Use of immunohistochemical methods for diagnosis of equine pythiosis. Am. J. Vet. Res. 49:1866-1868. [PubMed] [Google Scholar]
- 7.Buergelt, C. D. 2000. If it's not neoplasia, it may be pythiosis. Vet. Med. 95:198-200. [Google Scholar]
- 8.Carlotti, A., A. Couble, J. Domingo, K. Miroy, and J. Villard. 1996. Species-specific identification of Candida krusei by hybridization with the CkF1,2 DNA probe. J. Clin. Microbiol. 34:1726-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlotti, A., T. Srikantha, K. Schroppel, C. Kvaal, J. Villard, and D. R. Soll. 1997. A novel repeat sequence (CKRS-1) containing a tandemly repeated sub-element (kre) accounts for differences between Candida krusei strains fingerprinted with the probe CkF1,2. Curr. Genet. 31:255-263. [DOI] [PubMed] [Google Scholar]
- 10.Chaffin, M. K., J. Schumacher, and W. C. McMullan. 1995. Cutaneous pythiosis in the horse. Vet. Clin. North. Am. Equine Pract. 11:91-103. [DOI] [PubMed] [Google Scholar]
- 11.de Cock, A. W. A. M., L. Mendoza, A. A. Padhye, L. Ajello, and L. Kaufman. 1987. Pythium insidiosum sp. nov., the etiologic agent of pythiosis. J. Clin. Microbiol. 25:344-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dykstra, M. J., N. J. Sharp, T. Olivry, A. Hillier, K. M. Murphy, L. Kaufman, G. A. Kunkle, and C. Pucheu-Haston. 1999. A description of cutaneous-subcutaneous pythiosis in fifteen dogs. Med. Mycol. 37:427-433. [DOI] [PubMed] [Google Scholar]
- 13.Einsele, H., H. Hebart, G. Roller, J. Loffler, I. Rothenhofer, C. A. Muller, R. A. Bowden, J. van Burik, D. Engelhard, L. Kanz, and U. Schumacher. 1997. Detection and identification of fungal pathogens in blood by using molecular probes. J. Clin. Microbiol. 35:1353-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El Fari, M., H. J. Tietz, W. Presber, W. Sterry, and Y. Graser. 1999. Development of an oligonucleotide probe specific for Trichophyton rubrum. Br. J. Dermatol. 141:240-245. [DOI] [PubMed] [Google Scholar]
- 15.Elie, C. M., T. J. Lott, E. Reiss, and C. J. Morrison. 1998. Rapid identification of Candida species with species-specific DNA probes. J. Clin. Microbiol. 36:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gams, W., E. S. Hoekstra, and A. Atroot. 1998. CBS course of mycology, 4th ed. Centraalbureau voor Schimmelcultures, Baarn, The Netherlands.
- 17.Goodwin, P. H., B. C. Kirkpatrick, and J. M. Duniway. 1989. Cloned DNA probes for identification of Phytophthora parasitica. Phytopathology 79:716-721. [Google Scholar]
- 18.Grooters, A. M., and M. K. Gee. 2002. Development of a nested polymerase chain reaction assay for the detection and identification of Pythium insidiosum. J. Vet. Intern. Med. 16:147-152. [DOI] [PubMed] [Google Scholar]
- 19.Grooters, A. M., B. S. Leise, M. K. Lopez, M. K. Gee, and K. L. O'Reilly. 2002. Development and evaluation of an enzyme-linked immunosorbent assay for the serodiagnosis of pythiosis in dogs. J. Vet. Intern. Med. 16:142-146. [DOI] [PubMed] [Google Scholar]
- 20.Grooters, A. M., E. C. Hodgin, R. W. Bauer, C. J. Detrisac, N. R. Znajda, and R. C. Thomas. 2003. Clinicopathologic findings associated with Lagenidium sp. infection in six dogs: initial description of an emerging oomycosis. J. Vet. Intern. Med. 17:637-646. [DOI] [PubMed] [Google Scholar]
- 21.Henrion, B., F. Le Tacon, and F. Martin. 1992. Rapid identification of genetic variation of ectomycorrhizal fungi by amplification of rRNA genes. New. Phytol. 122:289-298. [DOI] [PubMed] [Google Scholar]
- 22.Hudspeth, D. S. S., S. A. Nadler, and M. E. S. Hudspeth. 2000. A COX2 molecular phylogeny of the Peronosporomycetes. Mycologia 92:674-684. [Google Scholar]
- 23.Judelson, H. S., and B. Messenger-Routh. 1996. Quantitation of Phytophthora cinnamomi in avocado roots using a species-specific DNA probe. Phytopathology 86:763-768. [Google Scholar]
- 24.Klassen, G. R., M. Balcerzak, and A. W. A. M. de Cock. 1996. 5S rRNA gene spacers as species-specific probes for eight species of Pythium. Phytopathology 86:581-587. [Google Scholar]
- 25.Krajaejun, T., M. Kunakorn, S. Niemhom, P. Chongtrakool, and R. Pracharktam. 2002. Development and evaluation of an in-house enzyme-linked immunosorbent assay for early diagnosis and monitoring of human pythiosis. Clin. Diagn. Lab. Immunol. 9:378-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, S. B., T. J. White, and J. W. Taylor. 1993. Detection of Phytophthora species by oligonucleotide hybridization to amplified ribosomal DNA spacers. Phytopathology 83:177-181. [Google Scholar]
- 27.Lévesque, C. A., T. C. Vrain, and S. H. De Boer. 1994. Development of a species-specific probe for Pythium ultimum using amplified ribosomal DNA. Phytopathology 84:474-478. [Google Scholar]
- 28.Lévesque, C. A., C. E. Harlton, and A. W. A. M. de Cock. 1998. Identification of some Oomycetes by reverse dot blot hybridization. Phytopathology 88:213-222. [DOI] [PubMed] [Google Scholar]
- 29.Lindsley, M. D., S. F. Hurst, N. J. Iqbal, and C. J. Morrison. 2001. Rapid identification of dimorphic and yeast-like fungal pathogens using specific DNA probes. J. Clin. Microbiol. 39:3505-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin, F. N. 1991. Selection of DNA probes useful for isolate identification of two Pythium spp. Phytopathology 81:742-746. [Google Scholar]
- 31.Mendoza, L., L. Kaufman, and P. G. Standard. 1986. Immunodiffusion test for diagnosing and monitoring pythiosis in horses. J. Clin. Microbiol. 23:813-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendoza, L., L. Kaufman, and P. Standard. 1987. Antigenic relationship between the animal and human pathogen Pythium insidiosum and nonpathogenic Pythium species. J. Clin. Microbiol. 25:2159-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mendoza, L., and J. Prendas. 1988. A method to obtain rapid zoosporogenesis of Pythium insidiosum. Mycopathologia 104:59-62. [DOI] [PubMed] [Google Scholar]
- 34.Mendoza, L., and G. Marin. 1989. Antigenic relationship between Pythium insidiosum de Cock et al. 1987 and its synonym Pythium destruens Shipton 1987. Mycoses 32:73-77. [DOI] [PubMed] [Google Scholar]
- 35.Mendoza, L., L. Kaufman, W. Mandy, and R. Glass. 1997. Serodiagnosis of human and animal pythiosis using an enzyme-linked immunosorbent assay. Clin. Diagn. Lab. Immunol. 4:715-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, R. I., and R. S. Campbell. 1982. Immunological studies on equine phycomycosis. Aust. Vet. J. 58:227-231. [DOI] [PubMed] [Google Scholar]
- 37.Möller, E. M., G. Bahnweg, H. Sandermann, and H. H. Geiger. 1992. A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res. 20:6115-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park, C. S., M. Kakinuma, and A. Hideomi. 2002. Detection and quantitative analysis of zoospores of Pythium porphyrae, causative organism of red rot disease in Porphyra, by competitive PCR. J. Appl. Phycol. 13:433-441. [Google Scholar]
- 39.Patterson, D. J. 1989. Stramenopiles, chromophytes from a protistan perspective, p. 357-379. In J. C. Green, B. S. C. Leadbeater, and E. L. Diver (ed.), The chromophyte algae, problems, and perspectives. Clarendon Press, Oxford, England.
- 40.Pracharktam, R., P. Changtrakool, B. Sathapatayavongs, P. Jayanetra, and L. Ajello. 1991. Immunodiffusion test for diagnosis and monitoring of human pythiosis insidiosi. J. Clin. Microbiol. 29:2661-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reis, J. L., Jr., E. C. de Carvalho, R. H. Nogueira, L. S. Lemos, and L. Mendoza. 2003. Disseminated pythiosis in three horses. Vet. Microbiol. 96:289-295. [DOI] [PubMed] [Google Scholar]
- 42.Riethmüller, A., H. Voglmayr, M. Göker, M. Weib, and F. Oberwinkler. 2002. Phylogenetic relationships of the downy mildews (Peronosporales) and related groups based on nuclear large subunit ribosomal DNA sequences. Mycologia 94:834-849. [DOI] [PubMed] [Google Scholar]
- 43.Rippon, J. W. 1998. Zygomycosis. p. 681-713. In M. Wonsiewicz (ed.), Medical mycology: the pathogenic fungi and the pathogenic actinomycetes, 3rd ed. W. B. Saunders Co., Philadelphia, Pa.
- 44.Sandhu, G. S., B. C. Kline, L. Stockman, and G. D. Roberts. 1995. Molecular probes for diagnosis of fungal infections. J. Clin. Microbiol. 33:2913-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandhu, G. S., R. A. Aleff, B. C. Kline, and C. da Silva Lacaz. 1997. Molecular detection and identification of Paracoccidioides brasiliensis. J. Clin. Microbiol. 35:1894-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schurko, A. M., L. Mendoza, A. W. A. M. de Cock, and G. R. Klassen. 2003. Evidence for geographic clusters: molecular genetic differences among strains of Pythium insidiosum from Asia, Australia, and the Americas are explored. Mycologia 95:200-208. [PubMed] [Google Scholar]
- 47.Schurko, A. M., L. Mendoza, C. A. Lévesque, N. L. Désaulniers, A. W. A. M. de Cock, and G. R. Klassen. 2003. A molecular phylogeny of Pythium insidiosum. Mycol. Res. 107:537-544. [DOI] [PubMed] [Google Scholar]
- 48.Shenep, J. L., B. K. English, L. Kaufman, T. A. Pearson, J. W. Thompson, R. A. Kaufman, F. Frisch, and M. G. Rinaldi. 1998. Successful medical therapy for deeply invasive facial infection due to Pythium insidiosum in a child. Clin. Infect. Dis. 27:1388-1393. [DOI] [PubMed] [Google Scholar]
- 49.Southern, E. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503. [DOI] [PubMed] [Google Scholar]
- 50.Thianprasit, M., A. Chaiprasert, and P. Imwidthaya. 1996. Human pythiosis. Curr. Top. Med. Mycol. 7:43-54. [PubMed] [Google Scholar]
- 51.Thitithanyanont, A., L. Mendoza, A. Chuansumrit, R. Pracharktam, J. Laothamatas, B. Sathapatayavongs, S. Lolekha, and L. Ajello. 1998. Use of an immunotherapeutic vaccine to treat a life-threatening human arteritic infection caused by Pythium insidiosum. Clin. Infect. Dis. 27:1394-1400. [DOI] [PubMed] [Google Scholar]
- 52.Thomas, R. C., and D. T. Lewis. 1998. Pythiosis in dogs and cats. Compendium 20:63-75. [Google Scholar]
- 53.Van der Plaats-Niterink, A. J. 1981. Monograph of the genus Pythium. Studies Mycol. 21:1-242. [Google Scholar]
- 54.Yao, C. L., C. W. Magill, and R. A. Frederiksen. 1991. Use of an A-T-Rich DNA clone for identification and detection of Peronosclerospora sorghi. Appl. Environ. Microbiol. 57:2027-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Znajda, N. R., A. M. Grooters, and R. Marsella. 2002. PCR-based detection of Pythium and Lagenidium DNA in frozen and ethanol-fixed animal tissues. Vet. Dermatol. 13:187-194. [DOI] [PubMed] [Google Scholar]