Abstract
A recent screening program in Melbourne, Victoria, Australia, has shown that Chlamydia trachomatis is an important infection among men who frequent male-only saunas. To evaluate the C. trachomatis isolates circulating in local saunas, the C. trachomatis-positive samples collected during the program underwent amplification and sequencing of the omp1 gene, and the corresponding serovars were deduced. Forty-seven C. trachomatis-positive samples collected (from October 2001 to September 2002) from 39 men were evaluated. The deduced serovars found, in descending order of prevalence, were D, G, and J; and serovars B, E, F, and H were each found in single samples. The seven different serovars identified in the study sample indicate that local saunas are a reservoir of multiple C. trachomatis strains, possibly maintained by the introduction of new patrons or regular patrons who have been exposed to C. trachomatis elsewhere. No significant genetic variants were found, as most variable positions were silent and were detected only in single samples.
Bacterial sexually transmitted infections (STIs) and transmission risk behaviors among men who have sex with men (MSM) are an important public health concern in Australia and other industrialized countries (6, 10, 13, 24). In Melbourne, Victoria, Australia, two recent reports have highlighted rectal chlamydia infection as an important STI among MSM (20, 22). An audit of tests conducted at the Melbourne Sexual Health Centre between August 2001 and July 2002 showed that 15% of MSM tested positive for rectal chlamydia (22), and a recent screening program in Melbourne male-only saunas showed that 6% of MSM tested positive for rectal chlamydia (20). The Chlamydia trachomatis-positive samples from the Melbourne sauna program offer a good opportunity to identify the C. trachomatis types circulating in this population at present.
C. trachomatis serovars can be differentiated by sequencing the omp1 gene, which codes for the major outer membrane protein (MOMP) (1, 5, 8, 15-17, 25, 31). Preliminary evidence suggests that MOMP is a porin that assembles as β barrels containing conserved domains (CDs) of transmembrane β strands and periplasmic loops and four variable domains (VDs) located on the outer loops (14, 28). Serovars exhibit MOMP sequence heterogeneity that is mainly localized to the VDs (27, 36). Variability in the MOMP sequence is presumably a result of host selection and bacterial adaptation (3, 4, 19), although recent analysis of nucleotide and amino acid substitutions for omp1 and MOMP, respectively, showed a lack of evolutionary pattern with respect to disease or tissue tropism (29). The findings from studies of the clinical manifestations of disease and the association of those manifestations with specific serovars (2, 7, 12, 17, 23, 26, 34), geographic clustering (5, 8, 11), and the amount of variation within genotypes and the changes in the frequencies of the genotypes circulating over time (16, 17, 25, 30, 35) have been mixed. Other studies have highlighted the importance of MOMP diversity in strains isolated from sex workers, whose sexual network was described as a core group creating selective immune pressure on circulating strains (3, 4). Overall, of the serovars usually associated with urogenital infection (serovars D to K), serovars D, E, and F are the most common (1, 5, 8, 11, 12, 15, 17, 23, 30, 31).
The aims of the present study were to characterize the C. trachomatis strains detected in samples collected during a sauna screening program (20) by sequencing the omp1 gene and determining the corresponding serovars. Determination of the serovars and the omp1 variability of the C. trachomatis strains detected in these samples may give insight into the C. trachomatis strains circulating among MSM who frequent saunas.
MATERIALS AND METHODS
Clinical samples.
A sample of 47 C. trachomatis-positive specimens collected from 39 men who participated in a screening program in male-only saunas in Melbourne, Victoria, Australia (20), was used. These 47 samples comprised 34 anal swab specimens, 10 urine samples, and 3 throat swab specimens. Two anal swab specimens were collected from each of five men, and two urine samples were collected from each of two men; i.e., the samples were collected at the initial consultation in the sauna and repeat samples were then collected (for confirmation before antibiotic treatment). C. trachomatis was detected at two anatomical sites in one man: from an anal swab specimen and a urine sample.
Isolation of DNA and omp1-specific PCR.
DNA extraction and omp1-specific PCR were performed as described previously (20, 21). Three omp1-specific PCRs were used: PCR VD1-4 with primers P1 and OMP2 to amplify a region of the gene containing all four variable domains (17), nested PCR VD1/2 with primers P1 and CT6 (5) to amplify a region containing VD1 and VD2, and nested PCR VD3/4 with primers CT6 (sense) and OMP2 to amplify a region containing VD3 and VD4. Another nested PCR was performed to ensure sufficient overlap and fidelity after sequencing. This PCR used primers NL-f (5′-TGG GAT CGY TTT GAT GTA [G or A][G or A]-3′) and NL-r (5′-CCA ATG TA[G or A] GGA GTG AAC AT-3′) to amplify a region of omp1 that included VD2 and VD3, with the reaction and cycle conditions being the same as those described for PCRs VD1/2 and VD3/4.
The PCR products were purified by use of the QIAquick PCR Purification kit (Qiagen, Valencia, Calif.), according to the instructions of the manufacturer. Purified amplicons underwent gel electrophoresis and were quantitated by comparison with a molecular weight marker by using the Gene Genius Gel Documentation system (Syngene, Cambridge, United Kingdom).
DNA sequencing.
Approximately 65 ng per kb of each of the PCR products was added to the sequencing reaction mixtures prepared with a CEQ Dye Terminator Cycle Sequencing Quick Start kit (Beckman Coulter Inc., Fullerton, Calif.), according to the instructions of the manufacturer. The reaction mixtures contained one of the primers used in the PCR (primer P1, OMP2, CT6, NL-f, or NL-r) at a concentration of 1.6 μM, and thermal cycling was conducted according to the instructions of the manufacturer. Unincorporated dye terminators and salts were removed from postreaction products according to the instructions of the manufacturer (Beckman Coulter Inc.). The sequences of the omp1 PCR products were determined with an automated capillary sequencer (CEQ 8000; Beckman Coulter Inc.).
omp1 sequence analysis.
The partial sequences obtained for each sample were assembled to form a contiguous gene fragment sequence containing all four VDs of the omp1 gene. The sequences of the omp1 gene fragments sequenced were first compared to known omp1 nucleotide sequences in the GenBank database by use of the BLAST search algorithm (http://www.ncbi.nlm.nih.gov.blast). The deduced amino acid sequences of the omp1 gene fragments were then aligned with reference sequences and reported variants in GenBank by use of the alignment program ClustalW (32). The sequences of 288 amino acids of the coding region were compared. The alignment was manually checked and corrected according to known sequence motifs and differences between serovars (36). Genetic distances were calculated with the Jones-Taylor-Thornton model, the Dayhoffs PAM 001 matrix, and the Kimura formula in the program PRODIST in the Phylogeny Inference Package (PHYLIP) (9). Phylogenetic dendrograms were constructed from the PRODIST program output by a least-squares method with the FITCH program in PHYLIP, and the NEIGHBOR program in PHYLIP was used to construct dendrograms by the neighbor-joining method. C. pneumoniae and C. psittaci strains were used as the outgroup to root the dendrograms, as these strains have previously been identified as the outgroup with respect to C. trachomatis strains (37). Bootstrap analysis was performed by resampling the protein sequence data 1,000 times, calculating evolutionary distances by use of the Jones-Taylor-Thornton model, constructing 1,000 dendrograms by the neighbor-joining method with the NEIGHBOR program in PHYLIP with jumbled sequence addition, and producing a consensus dendrogram. The ClustalW and PHYLIP programs were implemented with the Institut Pasteur server (http://bioweb.pasteur.fr).
RESULTS
Sequence analysis of the C. trachomatis detected in samples from 39 men showed that the most prevalent omp1 sequences corresponded to serovar D (53.8%), followed by serovars G (25.6%) and J (10.2%); and strains of serovars B, E, H, and F were each found in single samples (Table 1). For seven men the same deduced serovar was detected in repeat samples. No mixed infections with more than one serovar were observed in this sample of men.
TABLE 1.
omp1 genotype distribution of C. trachomatis-positive samples from 39 men in male-only saunas in Melbourne, Australia
| Genotype | No. of clinical samples in which the omp1 genotype was detecteda
|
||
|---|---|---|---|
| Anal swab | Urine | Throat swab | |
| B | 1 | 0 | 0 |
| D | 16 | 2 | 3 |
| E | 1 | 0 | 0 |
| F | 1 | 0 | 0 |
| Gb | 6 | 5 | 0 |
| H | 0 | 1 | 0 |
| J | 4 | 0 | 0 |
| Total | 29 | 8 | 3 |
Repeat samples from the same individual were excluded. These include five anal swab specimens (genotypes D [n = 2], F [n = 1], G [n = 1], and J [n = 1]) and two urine samples (genotypes D [n = 1] and G [n = 1]). The genotypes from the repeat samples were identical to those from the samples collected initially.
One man was positive for C. trachomatis at two anatomical sites. The same genotype (genotype G) was detected in anal swab and urine samples.
Contiguous omp1 gene fragment sequences were obtained from 36 samples, and these sequences were analyzed for nucleotide differences. Of the 36 samples, we observed 11 different omp1 nucleotide sequences (Table 2). Twenty-seven samples had omp1 sequences identical to sequences in GenBank, and the other nine samples had nucleotide variations. More variable positions were detected within conserved regions (n = 8) than in surface-exposed VD2 and VD4 (n = 4). Multiple-sequence alignment of the sequences from all 36 samples showed a total of 12 different variable nucleotide positions, of which 4 resulted in amino acid replacements.
TABLE 2.
omp1 sequences in C. trachomatis-positive samples from men in male-only saunas in Melbourne, Australia
| Sauna omp1 sequence (GenBank accession no.) | No. of samplesa | Most similar GenBank sequence (GenBank accession no.) | Sequence variation
|
||
|---|---|---|---|---|---|
| No. of variant positions | Nucleotide and position (domain)b | Amino acid change | |||
| B complex | |||||
| B (AY464143) | 1 | B/IU-1226 (AF063208) | 2 | T→C 18 (CD1)c | Silent |
| G→A 475 (CD3) | V→I | ||||
| D (AY464160-75) | 16 | D/1b-IS5643 (AF414949) | 0 | ||
| D (AY464176-7) | 2 | D/1b-IS5643 (AF414949) | 1 | C→T 743 (CD4) | Silent |
| D (AY464178) | 1 | D/1b-IS5643 (AF414949) | 1 | T→C 821 (VD4)d | Silent |
| E (AY464144) | 1 | E/Bour (X52557) | 1 | C→A 666 (CD3) | Silent |
| Intermediate group | |||||
| F (AY464145) | 1 | F/IU-1607 (AF063213) | 1 | G→A 669 (CD4) | Silent |
| G (AY464150-7) | 8 | G/1a-IS4481 (AF414956)e | 0 | ||
| G (AY464158) | 1 | G/1a-IS4481 (AF414956) | 2 | C→A 558 (CD3) | Silent |
| G→C 559 (CD3) | E→Q | ||||
| G (AY464159) | 1 | G/1a-IS4481 (AF414956) | 1 | A→T 843 (VD4) | Silent |
| C complex | |||||
| J (AY464147-9) | 3 | J/1a-IS255 (AF414962)f | 0 | ||
| H (AY464146) | 1 | H/1a-IS1075 (AF414959) | 3 | T→A 450 (VD2) | D→E |
| A→G 457 (VD2) | T→A | ||||
| C→T 753 (CD4) | Silent | ||||
A total of 36 samples were tested.
Position of the nucleotide variation(s) according to the omp1 nucleotide sequence of the most similar GenBank sequence.
This nucleotide difference is also observed for the sequences of B/Apache-2 (AF063194), B/TW-5/OT (M17342), B/Jali-20 (M33636), and B/Alfa/95 (U80075) (GenBank accession numbers are given in parentheses).
This nucleotide difference is also observed for the sequence of Da/TW-448 (GenBank accession no. X62921).
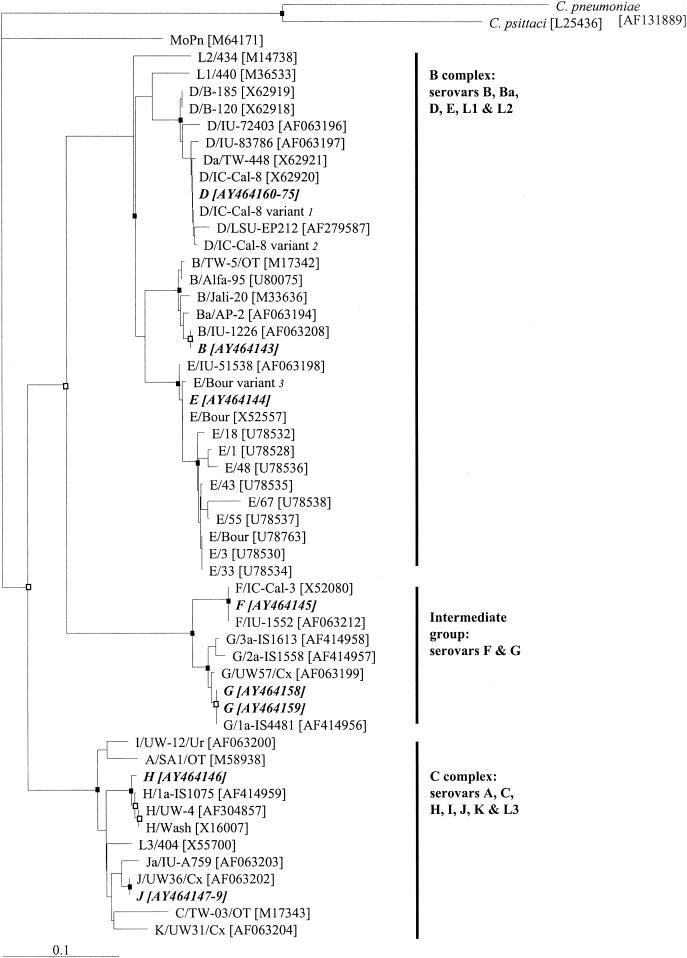
Overall, eight different phenotypes were identified from the deduced amino acid sequences. Phylogenetic analysis of the deduced amino acid sequences and GenBank sequences are shown in Fig. 1. Figure 1 is similar to previously described dendrograms constructed with human C. trachomatis sequences and the mouse pneumonitis strain of C. trachomatis (MoPn), in which the human and mouse strains formed separate clusters and in which the human sequences segregated into three major clusters for the B-complex, C-complex, and intermediate-group serovars (17, 29, 37). The branching pattern of this neighbor-joining dendrogram also agreed with those in dendrograms constructed by different distance methods with the PROTDIST program in PHYLIP (data not shown) and the consensus neighbor-joining dendrogram. We observed strongly supported coherent clusters of serovars D, B, E, F, G, and H, while serovars with only one or two representative sequences were not observed to form coherent clusters by bootstrap analysis of the data set. The differences in branching order between Fig. 1, the dendrograms that we generated by other methods with the PROTDIST program, and previously published dendrograms were mostly observed at nodes with low bootstrap values, indicating a low level of confidence.
FIG. 1.
Dendrogram illustrating relationships of deduced amino acid sequences of C. trachomaits omp1 genes recovered from men in Melbourne male-only saunas. The dendrogram was generated by use of the Jones-Taylor-Thornton model and the neighbor-joining method with the PROTDIST program in PHYLIP (9). Each branch terminus contains the names and the GenBank accession numbers (in brackets) of the MOMP sequences. The deduced MOMP sequences from this study are shown in boldface italicized text and are assigned the same letter for the corresponding serovar. The different clusters of C. trachomatis serovars (18, 33, 37) are indicated on the right. C. pneumoniae and C. psittaci strains were used as the outgroup to root the dendrogram. Bootstrap values were obtained from a consensus tree based on 1,000 randomly generated data sets with jumbled sequence addition. Nodes recovered in 90% or more of the 1,000 bootstrap dendrograms are indicated by a closed box, those recovered in 70 to 89% of the 1,000 bootstrap dendrograms are indicated by open boxes, and those recovered in less than 70% of the 1,000 bootstrap dendrograms have no symbol. Bar, 0.10 inferred amino acid changes per position. Notes: 1, the D/IC-Cal-8 variant was described by Jurstrand et al. (17), with a difference of R→H at position 72 in comparison to the MOMP sequence of D/IC-Cal-8 (GenBank accession no. X62920); 2, the D/IC-Cal-8 variant was described by Jurstrand et al. (17), with a difference of A→T at position 326 in comparison to the MOMP sequence of D/IC-Cal-8 (GenBank accession no. X62920); 3, the E/Bour variant was described by Jurstrand et al. (17), with a difference of A→T at position 333 in comparison to the MOMP sequence of E/Bour (GenBank accession no. X52557).
DISCUSSION
The C. trachomatis omp1 genes detected in samples from men in male-only saunas most frequently corresponded to those of serovars D and G. Also relatively common was omp1 serovar J, whereas the remaining serovars, serovars B, E, F, and H, were observed in single samples only. We found minor omp1 sequence variation within the different genotypes in our samples, and most of these genetic variants were detected only in single samples and did not result in amino acid changes.
The deduced serovar distribution was different from most distributions reported for C. trachomatis from developed countries, where serovar E was identified as the most prevalent (5, 8, 12, 16, 17, 23, 25, 30, 31). Although most studies have reported that serovar is D common (1, 5, 12, 15, 16, 23) and Geisler et al. (11) reported that most chlamydia infections among MSM were caused by serovars G (47.9%) and D (29.6%), the predominance of type D (53.8%) in Melbourne MSM suggests that the exposures of this population are partially independent from those in other centers in the world. The proportion of men at each sauna testing positive was not significantly different (20), and we did not observe geographic clustering of serovars with the saunas (data not shown). In comparison to the types detected in the samples from MSM, the preliminary findings in our diagnostic laboratory have shown that type E is most frequently detected by omp1 sequencing in C. trachomatis-positive urogenital samples from women (unpublished data). Geisler et al. (11) also found that the serovar distribution of C. trachomatis infections among MSM differed from the C. trachomatis causing urogenital infections among men and women in the same community. It is likely that different serovars circulate in heterosexual versus MSM networks. Future work could investigate the serovar distributions in other MSM and heterosexual populations in Australia.
Most of the C. trachomatis omp1 genes detected in anal swab specimens corresponded to B-complex serovars (serovars B and D), and most of the C. trachomatis serovars detected in urine specimens corresponded to intermediate-group serovars (serovars F and G). The phylogenetic analyses and a review of studies conducted by Stothard et al. (29) did not support an association of a particular serovar with disease or tissue tropism. By consideration of the findings presented in that report, it is unlikely that MOMP tissue tropism has resulted in the different serovar distributions in anal swab and urine specimens. This pattern of serovar distribution in anal swab and urine specimens may be explained by a confounding effect of other ligands responsible for directing the colonization of various tissues. Another explanation may be the differences in the efficiency of C. trachomatis transmission during a single exposure. For example, a difference in transmission efficiency between urethra-rectum and rectum-urethra may explain the different serovar distributions in anal swab and urine specimens.
The omp1 variability within genotypes was minor, as has been observed in other studies (17, 25). The three most prevalent omp1 sequences were identical to GenBank sequences, and the majority of variants were evolutionarily neutral, with no amino acid changes. This is highlighted in the phylogenetic dendrogram based on the amino acid sequences (Fig. 1), which shows that the sequences in samples from men at the sauna are closely related or identical to GenBank sequences. We observed most variation within CDs, and Pedersen et al. (25) reported a similar proportion of variable CD positions. All except two of the CD nucleotide variations were silent (Table 2). One variation in CD3 resulted in the amino acid replacement V→I, which probably caused little antigenic, functional, or conformational change to MOMP, as both amino acids have similar properties and CD3 is not predicted to be surface exposed (14, 28). The other mutation in CD3 resulted in the amino acid replacement E→Q, and this change may also have only a minor consequence if this amino acid contributes to a hydrophilic area of the predicted porin-like ion channel. We observed four nucleotide variations within VDs (Table 2); but only two resulted in amino acid changes in VD2, which may have been advantageous for the pathogen to escape immune pressure directed at omp1. All except one of the observed omp1 variants were detected only in single samples, and we believe that it is unlikely that these nucleotide variations have originated in vitro by PCR amplification or that they represent sequencing artifacts. The omp1-specific PCR was performed directly with clinical samples; and for samples with omp1 variations, the variable nucleotides were observed in all amplified gene fragments (sense and antisense sequences, overlapping sequences, or sequences from repeat amplification). Overall, we found that the most successful phenotypes (as measured by prevalence in the population) do not appear to be highly variable. This could indicate a lack of immune pressure on these strains among sexual networks in local saunas, or it may indicate that a less vigorous immune response is triggered by these strains and that MOMP conservation is important.
In conclusion, seven of the eight serovars usually associated with urogenital infections were identified in our study sample, and this indicates that local saunas are a reservoir of multiple C. trachomatis strains. This suggests that MSM who frequent saunas do not represent a closed community or an exclusive core group, as our study showed that MSM were exposed to multiple C. trachomatis urogenital serovars; three serovars were predominant (serovars D, G, and J), and other serovars were observed only in single samples. The number of circulating serovars in this population may be maintained by the introduction of new sauna patrons or regular patrons who have been exposed to different C. trachomatis strains elsewhere. The existence of a smaller subgroup of individuals who pass the predominant serovars among each other is also a plausible interpretation of our findings, especially when the predominance of serovar D is considered.
Acknowledgments
We thank Matthew Stevens (Royal Women's Hospital, Melbourne) for laboratory support. We gratefully acknowledge the research team involved in the collection of the C. trachomatis-positive samples used in this study.
This project was approved by the Victorian Department of Human Services Human Research Ethics Committee, Melbourne, Victoria, Australia. This work was supported by funding from the Public Health Research Projects 2002-03 (Communicable Diseases), Victorian Department of Human Services. Anthony Smith is supported by a senior research fellowship from the Victorian Health Promotion Foundation, Melbourne, Victoria, Australia.
This work was performed at Department of Microbiology & Infectious Diseases, Royal Women's Hospital, Women's and Children's Health, Melbourne, Victoria, Australia.
REFERENCES
- 1.Bandea, C. I., K. Kubota, T. M. Brown, P. H. Kilmarx, V. Bhullar, S. Yanpaisarn, P. Chaisilwattana, W. Siriwasin, and C. M. Black. 2001. Typing of Chlamydia trachomatis strains from urine samples by amplification and sequencing the major outer membrane protein gene (omp1). Sex. Transm. Infect. 77:419-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boisvert, J. F., L. A. Koutsky, R. J. Suchland, and W. E. Stamm. 1999. Clinical features of Chlamydia trachomatis rectal infection by serovar among homosexually active men. Sex. Transm. Dis. 26:392-398. [DOI] [PubMed] [Google Scholar]
- 3.Brunham, R., C. Yang, I. Maclean, J. Kimani, G. Maitha, and F. Plummer. 1994. Chlamydia trachomatis from individuals in a sexually transmitted core group exhibit frequent sequence variation in the major outer membrane protein (omp1) gene. J. Clin. Investig. 94:458-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunham, R. C., J. Kimani, J. Bwayo, G. Maitha, I. Maclean, C. Yang, C. Shen, S. Roman, N. J. Nagelkerke, M. Cheang, and F. A. Plummer. 1996. The epidemiology of Chlamydia trachomatis within a sexually transmitted diseases core group. J. Infect. Dis. 173:950-956. [DOI] [PubMed] [Google Scholar]
- 5.Cabral, T., A. M. Jolly, and J. L. Wylie. 2003. Chlamydia trachomatis omp1 genotypic diversity and concordance with sexual network data. J. Infect. Dis. 187:279-286. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 1999. Resurgent bacterial sexually transmitted disease among men who have sex with men—King County, Washington, 1997-1999. Morb. Mortal. Wkly. Rep. 48:773-777. [PubMed] [Google Scholar]
- 7.Dean, D., E. Oudens, G. Bolan, N. Padian, and J. Schachter. 1995. Major outer membrane protein variants of Chlamydia trachomatis are associated with severe upper genital tract infections and histopathology in San Francisco. J. Infect. Dis. 172:1013-1022. [DOI] [PubMed] [Google Scholar]
- 8.Falk, L., M. Lindberg, M. Jurstrand, A. Backman, P. Olcen, and H. Fredlund. 2003. Genotyping of Chlamydia trachomatis would improve contact tracing. Sex. Transm. Dis. 30:205-210. [DOI] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package), version 3.5c. Department of Genetics, University of Washington, Seattle. Accessed 1 September 2003. [Online.] http://bioweb.pasteur.fr.
- 10.Fox, K. K., C. del Rio, K. K. Holmes, E. W. Hook III, F. N. Judson, J. S. Knapp, G. W. Procop, S. A. Wang, W. L. H. Whittington, and W. C. Levine. 2001. Gonorrhea in the HIV era: a reversal in trends among men who have sex with men. Am. J. Public Health 91:959-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geisler, W. M., W. L. Whittington, R. J. Suchland, and W. E. Stamm. 2002. Epidemiology of anorectal chlamydia and gonococcal infections among men having sex with men in Seattle: utilizing serovar and auxotype strain typing. Sex. Transm. Dis. 29:189-195. [DOI] [PubMed] [Google Scholar]
- 12.Geisler, W. M., R. J. Suchland, W. L. Whittington, and W. E. Stamm. 2003. The relationship of serovar to clinical manifestations of urogenital Chlamydia trachomatis infection. Sex. Transm. Infect. 30:160-165. [DOI] [PubMed] [Google Scholar]
- 13.Handsfield, H. H. 2002. posting date. Conference report: highlights from the 2002 STD Conference, March 4-7, 2002, San Diego, California. Medscape Infect. Dis. 4(1). [Online.]
- 14.Hughes, E. S., K. M. Shaw, and R. H. Ashley. 2001. Mutagenesis and functional reconstitution of chlamydial major outer membrane proteins: VS4 domains are not required for pore formation but modify channel function. Infect. Immun. 69:1671-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikehata, M., K. Numazaki, and S. Chiba. 2000. Analysis of Chlamydia trachomatis serovars in endocervical specimens derived from pregnant Japanese women. FEMS Immunol. Med. Microbiol. 27:35-41. [DOI] [PubMed] [Google Scholar]
- 16.Jonsdottir, K., M. Kristjansson, J. Hjaltalin Olafsson, and O. Steingrimsson. 2003. The molecular epidemiology of genital Chlamydia trachomatis in the greater Reykjavik area, Iceland. Sex. Transm. Dis. 30:249-256. [DOI] [PubMed] [Google Scholar]
- 17.Jurstrand, M., L. Falk, H. Fredlund, M. Lindberg, P. Olcen, S. Andersson, K. Persson, J. Albert, and A. Backman. 2001. Characterization of Chlamydia trachomatis omp1 genotypes among sexually transmitted disease patients in Sweden. J. Clin. Microbiol. 39:3915-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lampe, M. F., R. J. Suchland, and W. E. Stamm. 1993. Nucleotide sequence of the variable domains within the major outer membrane protein gene from serovariants of Chlamydia trachomatis. Infect. Immun. 61:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lampe, M. F., K. G. Wong, L. M. Kuehl, and W. E. Stamm. 1997. Chlamydia trachomatis major outer membrane protein variants escape neutralization by both monoclonal antibodies and human immune sera. Infect. Immun. 65:317-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lister, N. A., A. Smith, S. N. Tabrizi, P. Hayes, N. A. Medland, S. Garland, and C. K. Fairley. 2003. Screening for Neisseria gonorrhoeae and Chlamydia trachomatis in men who have sex with men at male-only saunas. Sex. Transm. Dis. 30:886-889. [DOI] [PubMed] [Google Scholar]
- 21.Lister, N. A., S. Tabrizi, C. K. Fairley, and S. Garland. 2004. Validation of Roche COBAS AMPLICOR assay for detection of Chlamydia trachomatis in rectal and pharyngeal specimens by an omp1 PCR assay. J. Clin. Microbiol. 42:239-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lister, N. A., A. Smith, T. Read, and C. K. Fairley. 2004. Testing men who have sex with men for Neisseria gonorrhoeae and Chlamydia trachomatis at an STD clinic in Mebourne prior to the introduction of guidelines. Sex. Health 1:47-50. [DOI] [PubMed]
- 23.Morre, S. A., L. Rozendaal, I. G. van Valkengoed, A. J. Boeke, P. C. van Voorst Vader, J. Schirm, S. de Blok, J. A. van Den Hoek, G. J. van Doornum, C. J. Meijer, and A. J. van den Brule. 2000. Urogenital Chlamydia trachomatis serovars in men and women with a symptomatic or asymptomatic infection: an association with clinical manifestations? J. Clin. Microbiol. 38:2292-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Centre in HIV Epidemiology and Clinical Research. 2003. HIV/AIDS, viral hepatitis and sexually transmissible infections. In Australia annual surveillance report 2003. National Centre in HIV Epidemiology and Clinical Research, The University of New South Wales, Sydney, New South Wales, Australia. Accessed 31 October 2003. [Online.] http://www.med.unsw.edu.au/nchecr.
- 25.Pedersen, L. N., H. O. Kjaer, J. K. Moller, T. F. Orntoft, and L. Ostergaard. 2000. High-resolution genotyping of Chlamydia trachomatis from recurrent urogenital infections. J. Clin. Microbiol. 38:3068-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Persson, K., and S. Osser. 1993. Lack of evidence of a relationship between genital symptoms, cervicitis and salpingitis and different serovars of Chlamydia trachomatis. Eur. J. Clin. Microbiol. Infect. Dis. 12:195-199. [DOI] [PubMed] [Google Scholar]
- 27.Poole, E., and I. Lamont. 1992. Chlamydia trachomatis serovar differentiation by direct sequence analysis of the variable segment 4 region of the major outer membrane protein gene. Infect. Immun. 60:1089-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez-Maranon, M. J., R. M. Bush, E. M. Peterson, T. Schirmer, and L. M. de la Maza. 2002. Prediction of the membrane-spanning beta-strands of the major outer membrane protein of Chlamydia. Protein Sci. 11:1854-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stothard, D. R., G. Boguslawski, and R. B. Jones. 1998. Phylogenetic analyses of the Chlamydia trachomatis major outer membrane protein and examination of potential pathogenic determinants. Infect. Immun. 66:3618-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suchland, R. J., L. O. Eckert, S. E. Hawes, and W. E. Stamm. 2003. Longitudinal assessment of infecting serovars of Chlamydia trachomatis in Seattle public health clinics: 1988-1996. Sex. Transm. Dis. 30:357-361. [DOI] [PubMed] [Google Scholar]
- 31.Sylvan, S. P., G. Von Krogh, A. Tiveljung, B. M. Siwerth, L. Henriksson, L. Noren, A. K. Asp, and L. Grillner. 2002. Screening and genotyping of genital Chlamydia trachomaits in urine specimens from male and female clients of youth-health centres in Stockholm County. Sex. Transm. Dis. 29:379-386. [DOI] [PubMed] [Google Scholar]
- 32.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, S. P., C. C. Kuo, R. C. Barnes, R. S. Stephens, and J. T. Grayston. 1985. Immunotyping of Chlamydia trachomatis with monoclonal antibodies. J. Infect. Dis. 152:791-800. [DOI] [PubMed] [Google Scholar]
- 34.Workowski, K. A., C. E. Stevens, R. J. Suchland, K. K. Holmes, D. A. Eschenbach, M. B. Pettinger, and W. E. Stamm. 1994. Clinical manifestations of genital infection due to Chlamydia trachomatis in women: differences related to serovar. Clin. Infect. Dis. 19:756-760. [DOI] [PubMed] [Google Scholar]
- 35.Yang, C. L., I. Maclean, and R. C. Brunham. 1993. DNA sequence polymorphism of the Chlamydia trachomatis omp1 gene. J. Infect. Dis. 168:1125-1130. [DOI] [PubMed] [Google Scholar]
- 36.Yuan, Y., Y. X. Zhang, N. G. Watkins, and H. D. Caldwell. 1989. Nucleotide and deduced amino acid sequences for the four variable domains of the major outer membrane proteins of the 15 Chlamydia trachomatis serovars. Infect. Immun. 57:1040-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, Y., J. G. Fox, Y. Ho, L. Zhang, H. F. Stills, Jr., and T. F. Smith. 1993. Comparison of the major outer-membrane protein (MOMP) gene of mouse pneumonitis (MoPn) and hamster SFPD strains of Chlamydia trachomatis with other Chlamydia strains. Mol. Biol. Evol. 10:1327-1342. [DOI] [PubMed] [Google Scholar]