Abstract
A rapid antigen test for the diagnosis of severe acute respiratory syndrome (SARS) is essential for control of this disease at the point of management. The nucleocapsid (N) protein of SARS-associated coronavirus (SARS-CoV) is abundantly expressed in infected-cell culture filtrate as demonstrable by Western blotting using convalescent-phase sera from patients with SARS. We used monoclonal antibodies specifically directed against N protein to establish a sensitive antigen capture sandwich enzyme-linked immunosorbent assay (ELISA) for the detection of SARS-CoV. The assay employed a mixture of three monoclonal antibodies for capture and rabbit polyclonal antibodies for detection of serum antigen in 32 cases of clinically probable SARS as defined by the World Health Organization during the epidemic in Guangzhou, China. Recombinant N protein was used as a standard to establish a detection sensitivity of approximated 50 pg/ml. The linear range of detection in clinical specimens was from 100 pg/ml to 3.2 ng/ml. Using a panel of sera collected at different points in time, the amount of circulating N antigen was found to peak 6 to 10 days after the onset of symptoms. The sensitivity of the assay was 84.6% in 13 serologically confirmed SARS patients with blood taken during the first 10 days after the onset of symptoms (11 of 13). The specificity of the assay was 98.5% in 1,272 healthy individuals (1,253 of 1,272). There was no cross-reaction with other human and animal coronaviruses in this assay. In conclusion, a sensitive and quantitative antigen capture ELISA was established for the early diagnosis and disease monitoring of SARS-CoV infection.
Severe acute respiratory syndrome (SARS) has infected more than 8,000 persons with 916 fatalities worldwide (http://www.who.int/csr/sars/country/en/country2003_08_15.pdf). Anovel coronavirus was consistently isolated from patients with clinical features fitting the World Health Organization case definition of SARS (6, 12, 16). Most of these patients did not have concomitant isolation of other pneumonic pathogens. Subsequently more than 90% of these patients had seroconversion against the SARS-associated coronavirus (SARS-CoV) without serological evidence of infection by other pneumonic agents (17). The Koch's postulates for causation were satisfied for this filterable agent, which alone can reproduce the clinical syndrome of acute respiratory distress syndrome and pathological changes of diffuse alveolar damage in monkeys (7). Again the SARS-CoV can be reisolated from these artificially inoculated monkeys, which subsequently seroconvert to the agent (13). Unfortunately, virus isolation by cell culture is slow and very inefficient (2) and there is no proven effective antiviral therapy. Isolation of suspected cases, contact tracing, and quarantine of contacts are the only means available for the control of this infection. A rapid and accurate diagnostic test at the point of management would be very important in case identification. Since the antibody response takes at least 10 to 28 days to develop after the onset of symptoms (17), detection of viral components is likely to be the best option for early detection. Complete genomic sequencing of many strains of SARS-CoV have provided information for the development of reverse transcription (RT)-PCR and cloning of major antigenic proteins (14). RT-PCR on nasopharyngeal specimens has a sensitivity of about 30% during the first few days after the onset of symptoms and the test has to be done in laboratories with expertise in molecular diagnostics (17). Antigen detection in serum by enzyme immunoassay (EIA) would be an important advance over RT-PCR in most health care settings since collection of nasopharyngeal aspirates may be associated with some risk to health care workers. In this paper we report on the development of an antigen detection test by using a batch of monoclonal antibodies specifically directed against the nucleocapsid (N) protein of SARS-CoV. The N antigen capture enzyme-linked immunosorbent assay (ELISA) was evaluated in a bank of sera collected from patients with SARS and healthy blood donors.
MATERIALS AND METHODS
Virus and cells.
Three SARS-CoV (HKU-39849, GZ01and BJ01) strains were isolated from fRhk-4 cell lines from Hong Kong Special Administrative Region, Guangdong Province, and Bejing, respectively (22). The SARS-CoVs were propagated in Vero E6 cell lines. Canine coronavirus 4 and avian infectious bronchitis virus 9 isolated from a dog and chicken, respectively, were kindly provided by the College of Veterinary Medicine, South China Agricultural University. They were propagated in Mardin Darby canine kidney (MDCK) cells. Human coronavirus strains 229E (ATCC VR740) and OC43 (ATCC VR759) were propagated in MRC-5 cells (ATCC CCL-171) and BS-C-1cells (ATCC CCL-26), respectively.
Cloning, expression, and purification of recombinant SARS-CoV N protein.
Reverse transcription and PCR were performed with a forward primer (5′-CGGGATCCCCATGTCTGATAATGGACCCCAATCAA-3′) and a reverse primer (5′-CGGAATTCTTATGCCTGAGTTGAATCAGCA-3′). The sequence coding nucleocapsid protein of SARS-CoV was amplified and ligated into the BamHI and EcoRI sites of the prokaryotic expression vector pGEX-5X-3 (Pharmacia Biotech, Uppsala, Sweden) in frame and downstream of the glutathione S-transferase (GST) coding sequence. The nucleoprotein inserts were sequenced to confirm the exactness of the N protein sequence and proper in-frame ligation. The complete sequence of SARS-CoV N protein cDNA has been published previously (14). The GST-N fusion protein was expressed and purified with a GST Gene Fusion System (Pharmacia Biotech) according to the manufacturer's instruction. The recombinant N fusion protein was detected by Western blot analysis with convalescent-phase sera at a dilution of 1 in 100 from serologically documented SARS patients as the primary antibody. As a positive control, the SARS-CoV infected-cell culture filtrate was concurrently run in the same assay. Horseradish peroxidase (HRP) labeled goat anti-human immunoglobulin G (IgG) was used as the secondary antibody. Aminoethyl carbazole Single Solution (Zymed Laboratories, Inc., So. San Francisco, Calif.) chromogen was used for signal detection. The concentration of purified recombinant N fusion protein was determined by the Coomassie Plus Protein Assay Reagent (Pierce Biotechnology, Rockford, Ill.).
Preparation of N protein specific monoclonal antibodies.
Six-week-old BALB/c mice (n = 10) were immunized with purified recombinant N protein mixed with an equal volume of monophosphoryl lipid A and trehalose dicorynomycolate (MPL+ TDM) Adjuvant (Sigma, St. Louis, Mo.). Splenocytes from the immunized mice were fused with NS-1 myeloma cells. The fused hybridoma cells were selected in RPMI 1640 medium supplemented with 10% fetal calf serum, hypoxanthine, aminopterin, and thymidine. Anti-N protein monoclonal antibody-producing hybridomas were selected by an ELISA with recombinant N protein and whole virus cell lysate as coating antigens, respectively, and the positive hybridomas were cloned at least twice by limiting dilution. Antibody subclasses were determined by ELISA using rabbit anti-mouse class-specific antibodies (Zymed Laboratories, Inc.). Monoclonal antibodies were purified by protein G column chromatography (Pharmacia Biotech).
Preparation of N protein specific antisera.
Rabbit antibodies to N protein were produced in New Zealand White rabbits. Rabbits (n = 4) were immunized with purified recombinant N protein mixed with an equal volume of complete Freund's adjuvant (Sigma) for the first injection and in Freund's incomplete Freund's adjuvant for the following three booster injections. Each injection contained 400 μg of recombinant N protein. Serum was collected 3 days after the final intravenous injection of 100 μg of recombinant N protein.
Capture ELISA assay for detection of the N antigen.
Wells of microtiter plates (Costar, Corning Incorporated, NY.) were coated with 100 μl of a mixture of 3 different anti-N protein monoclonal antibodies named as N10E4, N1E8, and N8E1 at 4°C for 14 to 16 h. Thereafter, the wells were washed twice with phosphate-buffered saline (PBS) at pH 7.4 with 0.05% Tween 20 (PBS-T) followed by incubation with a blocking reagent (2% sucrose, 0.5% casein-Na in PBS-T, pH 7.4) at 4°C for 14 to 16 h. After removing the blocking solution, the wells were vacuum dried and stored at 4°C. Human sera, or virus infected cell culture filtrate, or purified N protein diluted in PBS-T buffer were added to the wells (100 μl/well) and incubated for 1 h at 37°C. Sera were tested undiluted or at twofold dilutions during the assay. The wells were washed again and incubated for 1 h at 37°C with 100 μl (per well) of anti-N rabbit antiserum (diluted at 1/4,000 in PBS-T with 20% normal fetal serum). The wells were washed again and incubated for 1 h at 37°C with 100 μl (per well) of peroxidase-conjugated goat anti-rabbit IgG (Zymed) (diluted 1/5,000 in PBS-T with 20% normal goat serum). After three further washes, 100 μl of 3,3′,5,5′-tetramethylbenzidine solution (TMB) was added to each well. The color reaction was stopped after 15 min with 50 μl (per well) of 0.5 N sulfuric acid, and the plates were examined at 450 nm. The recombinant N protein concentrations in duplicates from 25 to 1,600 pg/ml were run on each plate, and the absorbance average readings were used to construct standard curves from which concentrations were evaluated. The experiments involving the use of sera from patients with SARS were performed in a biosafety level 2 laboratory.
Capture ELISA inhibition assay.
Sera that had positive reactions in the N antigen capture ELISA were confirmed in an inhibition assay, performed as described by Hunt et al. (9). Twenty microliters of convalescent-phase serum from a SARS patient with a titer of 1,280 by an indirect immunofluorescent assay (IFA) was mixed with 100 μl of each sample that was positive for N antigen, and incubated for 1 h at 37°C. Normal serum was used as the control antibody and was also mixed with each sample in a separate reaction mixture. These mixtures were retested in the capture ELISA by adding 100 μl of each mixture to a capture antibody-coated plate, and the assay was performed as above. The percentage of inhibition of antibody binding was calculated by the following equation: % inhibition = 1.00 − (A450 of sample + positive serum)/(A450 of sample + normal serum) × 100, if the % inhibition was greater than 50, test samples were considered to be confirmed as positive for N protein.
Clinical samples.
Normal sera were obtained from 1272 healthy blood donors. A total of 57 serum specimens were collected during the period from 3 to 90 days after the onset of symptoms from 32 patients with SARS in Nangfang Hospital of Guangzhou, China, between 15 February and 22 May 2003. All the patients were diagnosed according to the World Health Organization definition of SARS. 11 cases from whom acute and convalescent-phase sera were available were confirmed serologically by using an IFA for SARS-CoV antibody. Another 21 cases had only one serum sample, of which 11 did not have a detectable antibody response. All patients' sera were heat inactivated at 56°C 1 h before the experiments.
RESULTS
Selection and characterization of monoclonal antibodies for N antigen capture ELISA assay.
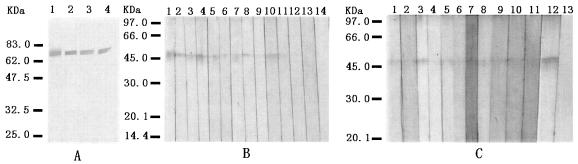
Hybridomas producing antibodies specific for N protein were successfully established from splenocytes following immunization with purified GST-N recombinant protein. Fusion of splenocytes from immunized mice with NS-1 myeloma cells produced more than 100 hybridoma lines, of which 9 cell lines were selected on the basis of their strong reactivity with the N protein in ELISA and Western blot assays. As shown in Fig. 1, monoclonal antibodies specifically bound to recombinant N protein and the virus culture filtrate on Western blot. The convalescent SARS patient serum produced only a single band with the infected-cell culture filtrate at exactly the same location of about 46-kDa as the assays with the monoclonal antibodies. The IFA showed that monoclonal antibodies strongly reacted with SARS-CoV infected Vero E6 cells, with no recognition of the other human coronaviruses and animal coronaviruses (data not shown). These results indicated that monoclonal antibodies to SARS-CoV N protein were specific for the SARS-CoV with no cross-reactivity with other coronaviruses. In order to establish a sensitive antigen capture ELISA for the SARS-CoV N protein, a rabbit polyclonal antiserum was used as a detector to evaluate the candidate monoclonal antibodies used for the antigen capture assay. Checkerboard analysis of a dilution series of the rabbit antiserum against single or combined monoclonal antibodies was performed. Using a standard concentration of purified recombinant N protein and a dilution series of the SARS-CoV infected Vero E6 cell culture filtrate, it was established that an optimum dilution of 1:4000 for the rabbit detection antiserum and a mixture of three monoclonal antibodies NE4A4, NE8A11, and NE1A17 as the capture monoclonal antibodies consistently yielded a strong signal in the N antigen capture assay. All three of the monoclonal antibodies were IgG1 isotype. Competitive assay demonstrated that the three monoclonal antibodies recognized different epitopes on N protein.
FIG. 1.
(A) Western blot analysis of monoclonal antibodies to recombinant N protein. Purified GST-N fusion protein was separated on the sodium dodecyl sulfate-10% polyacrylamide gel and transferred onto a nitrocellulose membrane. The strips were reacted with different anti-N protein monoclonal antibodies. Lanes 1 to 4: different monoclonal antibody clones (N1E8, N8E1, N10E4, and N10A4, respectively). (B and C) Western blot analysis of N protein in the culture filtrate of SARS-CoV. Culture filtrate from the Vero cells infected with SARS-CoV was concentrated and separated on the sodium dodecyl sulfate-10% polyacrylamide gel and transferred onto a nitrocellulose membrane. The strips were reacted with different anti-N protein monoclonal antibodies, control monoclonal antibodies and convalescent SARS patient sera, respectively. (B) Lanes 1 to 11, different monoclonal antibody clones (N1E8, N8E1, N10E4, N10A4, N14A3, N14E19, N14E7, N14E1, N14B6, N10E2, and N1A7, respectively); lanes 12 to 14, control monoclonal antibodies. (C) Lanes 1 to 12: 12 cases' sera; lane 13, normal serum control.
Sensitivity, specificity, and reproducibility of the assay.
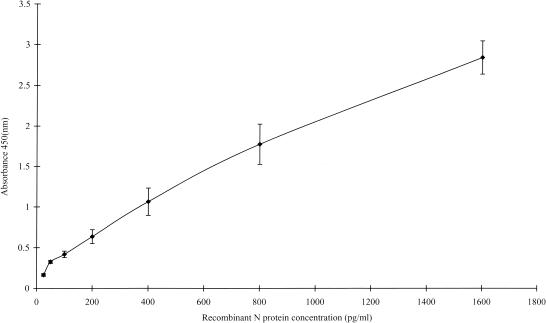
To evaluate the sensitivity of antigen capture assay, a serial dilution of the purified recombinant N protein was used to establish a standard curve for the N protein test (Fig. 2). Normal human sera were used as a diluent in order to determine the possible effect of serum on the sensitivity. Bovine serum albumin in human serum dilution was used to establish the baseline for the test at an optical density at 450 nm (OD450) of 0.061. The cutoff value was set to be 0.122, which is equal to twice the OD450 for bovine serum albumin. Using these criteria, the lower limit of detection of the recombinant protein with this capture ELISA is approximately 50 pg/ml.
FIG. 2.
Reproducibility of standard curve of N protein determine with a purified recombinant N protein. Various concentrations of N protein dilution were measured in duplicate by antigen capture ELISA. Results were mean values from three separate plates run under same conditions. Data points represent the means (error bars, standard deviations).
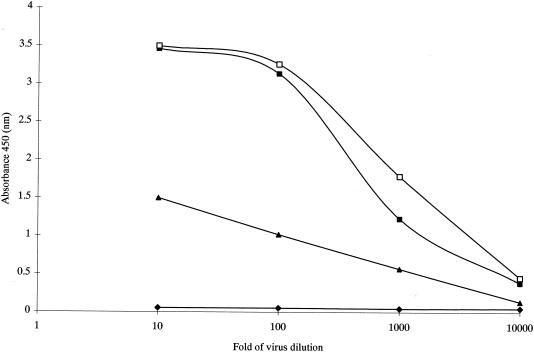
In order to examine the presence of N protein in the cell culture supernatant and the reactivity of N protein derived from SARS-CoV isolated from different areas in the capture assay, we tested culture filtrate harvested from Vero E6 cells infected by HKU-39849, GZ01 and BJ01 SARS-CoV stains, respectively. These viral supernatants were heat inactivated at 65°C overnight before assay. The results from a limiting-dilution experiment of a culture supernatant of virus are shown in Fig. 3. The high level of detection of N protein (approximately 10,000-fold) in the culture supernatant suggested that N protein is released as a soluble antigen in infected cell culture. This result raised the possibility that the N protein could be detected in serum specimens from infected patients.
FIG. 3.
Capture ELISA detection of N protein from Vero cells infected with SARS-CoV. Virus culture filtrate was isolated from Beijing (BJ01) (□), from HK (HKU-39849) (▪), from Guangdong (GZ01) (▴) (Vero cells culture filtrate, ♦).
To determine the specificity of the assay, culture filtrate obtained from other related viruses, human coronavirus strains 229E and OC43, canine coronavirus, avian infectious bronchitis virus, rhinovirus, adenovirus, influenza virus, and respiratory syncytial virus were analyzed by this assay. None of these virus-infected cell culture filtrates were detected in the N antigen capture assay (data not shown). Our initial findings indicate that this monoclonal antibody-based antigen capture assay may be specific for SARS-CoV and has no cross-reactivity with other related coronavirus.
To assess the reproducibility of the assay, we prepared an evaluation with three samples from virus culture filtrate diluted in serum at 1/100, 1,000, and 5,000, respectively, which were run many times within and between assays. The intra-assay variation was examined using 20 replicated tests of three samples within the same assay. The mean of OD values with standard deviation were 2.56 ± 0.16, 1.09 ± 0.08, and 0.46 ± 0.05, and the coefficients of variation (CV) were 6.3, 7.3, and 10.8%, respectively. We also used the same test samples to assess the reproducibility of interassay by repeating 10 tests from day to day. The mean of OD values with standard deviation were 2.37 ± 0.21, 1.15 ± 0.16 and 0.58 ± 0.07, the CV were 8.8, 13.9, and 12%, respectively. Standard curves produced on each plate using recombinant N protein gave good reproducibility (Fig. 2). Various concentration of recombinant N protein from 25 to 1,600 pg/ml were made by diluting buffer and were used to construct a standard curve for assessing the precision of the assay. Plate-to-plate variation of the standard curves was constructed. Plate-to-plate variation over the linear section of the curve assessed by calculating coefficients of variation within three condition plates used to construct standard curves showed a mean CV of 10.4% ± 3.8%.
Normal range of the assay.
To establish the normal range of this assay, serum specimens from 1272 healthy blood donors were analyzed. The mean of OD450 for these specimens as determined by the assay was 0.083, with a standard deviation of 0.03. The cutoff OD450 of the assay was calculated from 1,272 normal serum samples according to a Gaussian population distribution (5), for a 99% confidence interval, the cutoff was then defined as follows: mean of OD450 + 2.58 standard deviations = 0.083 + 2.56 × 0.03 = 0.160, which will cover the normal range within which 99% of the normal healthy population will fall. Thus, a total of 19 normal samples from 1,272 specimens were defined as low level false-positive with an OD value range from 0.16 to 0.28 (Table 1).
TABLE 1.
Distribution of OD value in sera from healthy blood donors
| A450 range | No. of serum samples | % of sera | Cumulative % |
|---|---|---|---|
| 0-0.01 | 0 | 0.00 | 0.00 |
| ≥0.01-0.02 | 0 | 0.00 | 0.00 |
| ≥0.02-0.03 | 2 | 0.16 | 0.16 |
| ≥0.03-0.04 | 39 | 3.07 | 3.22 |
| ≥0.04-0.05 | 149 | 11.71 | 14.94 |
| ≥0.05-0.06 | 148 | 11.64 | 26.57 |
| ≥0.06-0.07 | 120 | 9.43 | 36.01 |
| ≥0.07-0.08 | 120 | 9.43 | 45.44 |
| ≥0.08-0.09 | 188 | 14.78 | 60.22 |
| ≥0.09-0.1 | 182 | 14.31 | 74.53 |
| ≥0.1-0.11 | 135 | 10.61 | 85.14 |
| ≥0.11-0.12 | 63 | 4.95 | 90.09 |
| ≥0.12-0.13 | 48 | 3.77 | 93.87 |
| ≥0.13-0.14 | 30 | 2.36 | 96.23 |
| ≥0.14-0.15 | 13 | 1.02 | 97.25 |
| ≥0.15-0.16 | 16 | 1.26 | 98.51 |
| ≥0.16-0.17 | 7 | 0.55 | 99.06 |
| ≥0.17-0.18 | 5 | 0.39 | 99.45 |
| ≥0.18-0.19 | 1 | 0.08 | 99.53 |
| ≥0.19-0.2 | 2 | 0.16 | 99.69 |
| ≥0.2-0.28 | 4 | 0.31 | 100.00 |
Detection of N antigen in sera from patients with SARS.
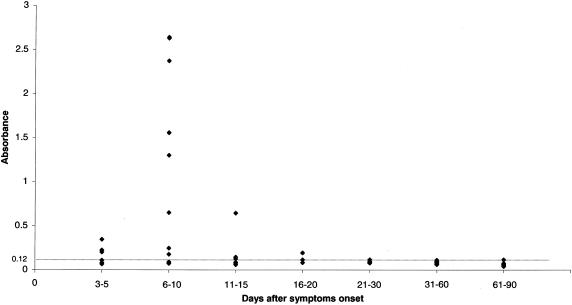
We analyzed a panel of sera from patients with clinically probable SARS with the N antigen capture ELISA. Sera collected up to day 10 after the onset of symptoms were considered as acute-phase sera. Convalescent-phase sera were collected beyond day 10. The sera were tested without dilution or at a 1/2 dilution. As sera from days 0 to 2 were not available, sera were tested from day 3 after the onset of symptoms. The percentage of N positive sera was 50% on days 3 to 5, peaked at 71% on days 6 to 10, and decreased to 50% on days 11 to 15. At day 19, 1 patient's serum was still found positive with an OD450 0.193. Serum N protein was never detected beyond day 20 (Table 2). During days 6 to 10, the ODs were highly variable from one individual to another (Fig. 4), with the OD varying between 0.177 and 2.64 with concentrations of N antigen in sera ranging from 100 pg/ml to 3.2 ng/ml by comparing the OD450 of the standard curve (Fig. 2).
TABLE 2.
N antigen detection in sera by a capture ELISA
| Days after onset of symptoms | No. of patients with positive sera/total no. of patients with SARS (%) |
|---|---|
| 0-2 | NA |
| 3-5 | 4/8 (50) |
| 6-10 | 10/14 (71) |
| 11-15 | 3/6 (50) |
| 16-20 | 1/3 (33) |
| 21-30 | 0/6 (0) |
| 31-60 | 0/15 (0) |
| 61-90 | 0/8 (0) |
FIG. 4.
N antigen detection in 57 serum specimens from 32 patients with SARS. Data represent the OD450 of sera tested at a dilution of 1/2. (dotted line) cutoff values.
Using the same bank of sera, we compared the sensitivity of the assay between the serologically confirmed SARS cases and clinical SARS cases from whom an acute-phase sera that was negative for SARS-CoV antibodies and convalescent-phase sera had not been obtained. We analyzed 11 cases who had seroconversion against SARS-CoV and 2 cases who had a acute-phase sera that were positive for SARS-CoV antibodies. Of the 11 who were test positive for N antigen by capture ELISA in the acute-phase sera, the N antigen positive rate in this group was 84.6%. On the other hand, the N antigen positive rate was 33% in 9 patients with negative antibody test in the acute-phase sera and whose convalescent-phase sera were not available.
The specificity of the positive reactions was confirmed by the inhibition assay. From a total of 18 N antigen positive sera, only 8 samples had sufficient residual volume to allow us to perform the N antigen capture inhibition assay, we found that the absorbance values in these reactions were reduced by more than 50% after blocking by SARS-CoV antibody positive serum (Table 3). This result indicated that the assay is highly specific for the N antigen.
TABLE 3.
Results of inhibition assay
| Sample no. |
A450 (nm)
|
% Inhibitionc | |
|---|---|---|---|
| Control seruma | Positive serumb | ||
| 1 | 0.97 | 0.239 | 76 |
| 2 | 0.196 | 0.087 | 66 |
| 3 | 0.253 | 0.092 | 64 |
| 4 | 1.728 | 0.229 | 87 |
| 5 | 1.907 | 0.132 | 94 |
| 6 | 0.434 | 0.119 | 73 |
| 7 | 2.209 | 0.356 | 84 |
| 8 | 0.422 | 0.08 | 81 |
N protein positive reaction serum (100 μl) was mixed with 20 μl of normal serum in each test.
N protein positive reaction serum (100 μl) was mixed with 20 μl of convalescent-phase serum from patients with SARS in each test.
Test samples were considered to be confirmed as positive for N protein if the percentage of inhibition was greater than 50.
DISCUSSION
One of the worst fears during the outbreak of SARS is the incorrect cohorting of non-SARS febrile pneumonic patients with those who are really suffering from SARS. This is likely to happen because of insufficient single rooms for isolation and insufficient nursing staff during a major outbreak. Thus, it is essential that a rapid and accurate diagnostic test is available at the point of patient care. Such a test would be important for any decisions on the use of antiviral treatment for cases or antiviral prophylaxis for the contacts in the management of SARS or in the setting of a clinical trial. At this stage in time, the RT-PCR assay for viral RNA in secretions or serum does not appear to be sensitive enough during the first 3 days after the onset of symptoms (21). There is also a long window period of 10 to 28 days if serum antibody is used as a diagnostic marker; however, seroconversion by neutralizing antibody or indirect immunofluorescent antibody against SARS-CoV infected cell lines is still the laboratory gold standard for retrospective case identification (2). A serum antigen assay will circumvent the need for nasopharyngeal aspirate collection and molecular diagnostic expertise.
Detection of circulating antigen has been successfully used in the diagnosis and monitoring of disease activity in human immunodeficiency virus (p24), hepatitis B virus (HBsAg, HBeAg), hepatitis C virus (core antigen), viral hemorrhagic fever (nucleoprotein), and cytomegalovirus (late pp65 antigen) infections (1, 3, 10, 15, 18, 19, 20). Mono-specific antibodies of either polyclonal or monoclonal nature have been used in a capture EIA format. In all of these clinical situations, antigen detection has shortened the window period by a positive test result preceding and later overlapping positive detection of antibody. It is therefore not unreasonable to develop a serum antigen capture test for SARS which has clinical evidence of multisystem involvement and laboratory evidence of circulating viral RNA detectable by RT-PCR.
In this study, we developed a capture ELISA for detection of N antigen. A batch of three monoclonal antibodies with high affinity and specificity for N protein was used for antigen capture and rabbit polyclonal antibodies were used for antigen detection in this assay. The capture ELISA appears to have a high degree of sensitivity. The limit of detection of the recombinant protein control was approximately 50 pg/ml. The analysis of produced N antigen in infected cell culture was up to 10,000-fold of culture filtrate. The high degree of sensitivity may be due to the fact that N protein is the predominant antigen produced in coronavirus-infected cells. Western blot analysis of the virus culture filtrate with convalescent patient serum in our data also shows that N protein is the abundant viral protein in the culture filtrate. These results further prove that N antigen is in the bloodstream of patients with SARS. The level of circulating N antigen in sera of SARS-CoV infected patients was estimated to range from 100 pg/ml to 3.2 ng/ml by calculation of OD450 within the liner part of the standard curve established by recombinant N protein. Using our N antigen ELISA, we also found the presence of N antigen during the early phase of the disease with antigen being detected in 50% of patients on days 3 to 5, and peaking on days 6 to 10 with 71% of patients being positive for antigen. It is understandable that the circulating N protein is maximally detected during the first 10 days of illness which coincides with our previous finding of the viral load in nasopharyngeal secretions peaking at day 10 with a subsequent decrease (17). Like most viruses, the nucleoprotein is well conserved and an ideal target for detection. This is a phosphoprotein with 397 amino acid residues that interact with the viral RNA genome to form the helical nucleocapsid. The amino acid homology of N protein of SARS-CoV with other known coronaviruses is low and varies from 27 to 37%. This strategy of detection of circulating antigen will eliminate the problem of cross-reactivity between the N protein of other human coronavirus 229E and OC43 because these two viruses do not cause systemic disease but rather just upper respiratory infections. The unique strategy of translation of coronaviruses by a series of subgenomic RNA starting at the individual open reading frames and terminating at the 3′ end results in abundant transcription and translation of the N protein. In terms of function, it was recently found that the amount of transcription factors binding to promoter sequences of c-Fos, ATF2, CREB-1, and FosB was increased by the expression of SARS-CoV N protein (8). Since these factors are related to the AP-1 signal transduction pathway, this suggests that SARS-CoV has encoded a strategy to regulate the cellular signaling process. Thus, the presence of circulating N protein in SARS patient may be related to its mode of expression in the viral replicative cycle. Previous studies from animal coronavirus have shown that the nucleocapsid (N) protein plays an important role in viral pathogenesis and replication (11). Such quantitation of a circulating antigen by a detection kit may facilitate both diagnosis and disease monitoring.
This is the first evaluation of an antigen capture assay that detects N antigen in SARS-CoV infected patient sera. The assay has proved to be highly specific and reproducible by precision performance and large normal population distribution analysis. The percentage of false positives is lower than 1.5%. Using seroconversion as the laboratory gold standard, the sensitivity is 84.6% in 13 cases who were confirmed positive by a detectable antibody response. There was no evidence of cross-reaction when tested with highly concentrated cell cultures of other related human and animal coronaviruses. The specificity of this ELISA was further proven by the blocking assay. It was shown that the SARS-CoV antibody positive serum significantly reduced the OD reading obtained from N antigen positive sera. The high sensitivity and specificity of the assay are likely to be due to the fact that a combination of three different but specific monoclonal antibodies is used for antigen capture, which may increase the reactivity of the assay system since both degraded and complete virions can be bound (4). One limitation of the present study is the relatively low number of patients because the study was initiated during the latter part of the SARS outbreak in Guangdong. However, these promising findings should be confirmed in a larger systemic field trial if SARS comes back in the coming winters.
Acknowledgments
This work was supported by the special programs of oppugning SARS from the Ministry of Science and Technology of the People's Republic of China and the Research Project of Guangdong Province for SARS Prevention and Treatment.
We thank Jin-lin Hou (Department of Infectious Diseases, Nanfang Hospital, First Military Medical University, Guangzhou, P. R. China) for supplying the sera from patients with SARS.
REFERENCES
- 1.Cano, H., M. J. Candela, M. L. Lozano, and V. Vicente. 2003. Application of a new enzyme-linked immunosorbent assay for detection of total hepatitis C virus core antigen in blood donors. Transfus. Med. 13:259-266. [DOI] [PubMed] [Google Scholar]
- 2.Chan, K. H., L. L. Poon, V. C. Cheng, Y. Guan, I. F. Hung, J. Kong, L. Y. Yam, W. H. Seto, K. Y. Yuen, and J. S. Peiris. 2004. Detection of SARS coronavirus in patients with suspected SARS. Emerg. Infect. Dis. 10:294-299. [DOI] [PMC free article] [PubMed]
- 3.Clement, F., P. Dewint, and G. Leroux-Roels. 2002. Evaluation of a new rapid test for the combined detection of hepatitis B virus surface antigen and hepatitis B virus e antigen. J. Clin. Microbiol. 40:4603-4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crouch, C. F., T. J. G. Raybould, and S. D. Acres. 1984. Monoclonal antibody capture enzyme-linked immunosorbent assay for detection of bovine enteric coronavirus. J. Clin. Microbiol. 19:388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deshpande, S. S. 1996. Enzyme immunoassay from concept to product development, 1st ed., p. 401-423. Kluwer Academic Publishers, New York, N.Y.
- 6.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 7.Fouchier, R. A., T. Kuiken, M. Schutten, G. van Amerongen, G. J. van Doornum, B. G. van den Hoogen, M. Peiris, W. Lim, K. Stohr, and A. D. Osterhaus. 2003. Aetiology: Koch's postulates fulfilled for SARS virus. Nature 423:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He, R., A. Leeson, A. Andonov, Y. Li, N. Bastien, J. Cao, C. Osiowy, F. Dobie, T. Cutts, M. Ballantine, and X. Li. 2003. Activation of AP-1 signal transduction pathway by SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 311:870-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunt, A. R., R. A. Hall, A. J. Kerst, R. S. Nasci, H. M. Savage, N. A. Panella, K. L. Gottfried, K. L. Burkhalter, and J. T. Roehrig. 2002. Detection of West Nile virus antigen in mosquitoes and avian tissues by a monoclonal antibody-based capture enzyme immunoassay. J. Clin. Microbiol. 40:2023-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Icardi, G., F. Ansaldi, B. M. Bruzzone, P. Durando, S. Lee, C. de Luigi, and P. Crovari. 2001. Novel approach to reduce the hepatitis C virus (HCV) window period: clinical evaluation of a new enzyme-linked immunosorbent assay for HCV core antigen. J. Clin. Microbiol. 39:3110-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.King, B., and D. A. Brian. 1982. Bovine coronavirus structural proteins. J. Virol. 42:700-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, L. J. Anderson, and SARS Working Group. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 13.Kuiken, T., R. A. Fouchier, M. Schutten, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, J. D. Laman, T. de Jong, G. van Doornum, W. Lim, A. E. Ling, P. K. Chan, J. S. Tam, M. C. Zambon, R. Gopal, C. Drosten, S. van der Werf, N. Escriou, J. C. Manuguerra, K. Stohr, J. S. Peiris, and A. D. Osterhaus. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 15.Mourton, C., B. Romestand, P. de Kinkelin, J. Jeffroy, R. Le Gouvello, and B. Pau. 1992. Highly sensitive immunoassay for direct diagnosis of viral hemorrhagic septicemia which uses antinucleocapsid monoclonal antibodies. J. Clin. Microbiol. 30:2338-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, K. Y. Yuen, and SARS Study Group. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peiris, J. S., C. M. Chu, V. C. Cheng, K. S. Chan, I. F. Hung, L. L. Poon, K. I. Law, B. S. Tang, T. Y. Hon, C. S. Chan, K. H. Chan, J. S. Ng, B. J. Zheng, W. L. Ng, R. W. Lai, Y. Guan, K. Y. Yuen, and HKU/UCH SARS Study Group. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.St George, K., M. J. Boyd, S. M. Lipson, D. Ferguson, G. F. Cartmell, L. H. Falk, C. R. Rinaldo, and M. L. Landry. 2000. A multisite trial comparing two cytomegalovirus (CMV) pp65 antigenemia test kits, biotest CMV brite and Bartels/Argene CMV antigenemia. J. Clin. Microbiol. 38:1430-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sutthent, R., N. Gaudart, K. Chokpaibulkit, N. Tanliang, C. Kanoksinsombath, and P. Chaisilwatana. 2003. p24 antigen detection assay modified with a booster step for diagnosis and monitoring of human immunodeficiency virus type 1 infection. J. Clin. Microbiol. 41:1016-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber, B., T. Dengler, A. Berger, H. W. Doerr, and H. Rabenau. 2003. Evaluation of two new automated assays for hepatitis B virus surface antigen (HBsAg) detection: IMMULITE HBsAg and IMMULITE 2000 HBsAg. J. Clin. Microbiol. 41:135-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yam, W. C., K. H. Chan, L. L. M. Poon, Y. Guan, K. Y. Yuen, W. H. Seto, and J. S. Peiris. 2003. Evaluation of reverse transcription-PCR assays for rapid diagnosis of severe acute respiratory syndrome associated with a novel coronavirus. J. Clin. Microbiol. 41:4521-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong, N. S., B. J. Zheng, Y. M. Li, L. L. Poon, Z. H. Xie, K. H. Chan, P. H. Li, S. Y. Tan, Q. Chang, J. P. Xie, X. Q. Liu, J. Xu, D. X. Li, K. Y. Yuen, J. S. Peiris, and Y. Guan. 2003. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet 362:1353-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]