Abstract
Enterococci possess capsular carbohydrate antigens that are targets of opsonic antibodies. These antigens may be used to develop alternative options for the treatment and prevention of enterococcal infections. The present study was done to analyze the diversity of capsular polysaccharides in Enterococcus faecalis. Four type-specific sera were used in an enzyme-linked immunosorbent assay format to detect polysaccharide antigen extracted from bacterial cell walls. A total of 55% of a collection of 29 E. faecalis strains could be grouped into one of four serogroups. Additional analysis of the strains by opsonophagocytic assays revealed agreement between the results of the two methods for 72% of the isolates. An additional four strains could be assigned to a serogroup on the basis of opsonic killing by sera with antibodies against the four prototypes strains, provisionally named CPS-A to CPS-D. The results of the two methods disagreed for one strain (4%). When the results of both methods were combined, 66% of the strains could be classified. One strain had to be assigned to two serogroups. The assignments to the four serogroups were confirmed by analysis of the genetic organization of the biosynthetic capsular polysaccharide (cps) locus. All strains grouped into serotypes CPS-A and CPS-B possessed only the cpsA and cpsB genes, while all strains grouped into serogroups CPS-C and CPS-D possessed an additional eight or nine genes. Our results suggest the existence of a limited number of E. faecalis capsule serotypes, and we provisionally propose four serotypes, named CPS-A to CPS-D, and the respective prototype strains for these families.
Enterococci are one of the leading causes of infections in hospitalized patients and the third most common cause of nosocomial bloodstream infections in the United States (6-8, 20). The number of strains resistant to virtually all clinically available antibiotics is increasing (20). Recent studies demonstrate that up to 25% of enterococcal hospital isolates are resistant to glycopeptide antibiotics (1, 20), the mainstay of the therapeutic armory. Resistant enterococcal strains cause significant morbidity and mortality, especially among immunocompromised patients.
Huebner et al. (16) recently identified a capsular polysaccharide in an Enterococcus faecalis strain and a vancomycin-resistant E. faecium strain that is the target of opsonic antibodies. Compositional and structural analyses revealed a teichoic acid-like molecule. Antibodies raised against this antigen protected mice in an experimental model of systemic enterococcal infection (15), thus offering new options for treatment and prevention. In 2002, Hancock and colleagues (10, 12) identified a genetic locus involved in the biosynthesis of a cell wall polysaccharide of E. faecalis strain FA2-2. This strain reacts with a type-specific antiserum raised against Maekawa E. faecalis strain type 2 (19). The corresponding cps locus consists of 11 open reading frames.
Some clues to the diversity of E. faecalis capsular polysaccharides can be gleaned from previous attempts to establish serotyping systems (19, 24), although it is not known at present how many structurally different capsular polysaccharides exist. Sharpe and Shattok (24) used HCl extracts of bacteria to raise rabbit sera with antibodies against surface polysaccharides. Their serotyping scheme is likely based on polysaccharide antigens, but considerable taxonomic changes subsequent to development of the scheme (19) limited its applicability. Maekawa et al. (19) used formaldehyde-killed bacteria to immunize rabbits against 21 prototype strains. Their serogroups are probably based not only on polysaccharide antigens but on a number of different surface antigens as well.
With an interest in the ultimate development of immunotherapeutics, we undertook a study to reexamine the diversity of E. faecalis capsules using strains identified by modern taxonomy and to compare the serological diversity observed with the organization of the genes in the recently identified E. faecalis capsule locus (10).
(This work was presented in part at the 101st General Meeting of the American Society for Microbiology, Orlando, Fla., 2001.)
MATERIALS AND METHODS
Strains, culture, and sera.
The bacterial strains and sera used in these studies are described in Tables 1 and 2 (10, 15-17, 19, 21). E. faecalis 12030 is the only E. faecalis strain for which structural information on the capsular polysaccharide exists (16, 27). Strain E. faecalis 12107 (15) is a clinical isolate from the United States, and the 21 prototype E. faecalis strains previously described by Maekawa et al. (19) are of E. faecalis types 2 and 5. HG101 is an insertion mutant of laboratory strain FA2-2 that is unable to produce the capsular polysaccharide (10). All E. faecalis strains were grown without agitation at 37°C either in Todd-Hewitt broth (Becton Dickinson, Sparks, Md.) or, for the capsular polysaccharide-specific enzyme-linked immunosorbent assay (CPS-ELISA), in Columbia broth (Difco Laboratories, Detroit, Mich.) with the addition of 0.5% glucose. Erythromycin (Sigma Chemical Co., St. Louis, Mo.) was added at a concentration of 10 μg/ml for mutant strain HG101.
TABLE 1.
E. faecalis strains used in the present study
| Strain | Origin (reference or source) |
|---|---|
| 12030 | Clinical isolate (16) |
| 12107 | Clinical isolate (15) |
| type 1-21 | Prototype Japanese strains from Maekawa et al. (19) |
| FA2-2 | Laboratory strain |
| HG101 | cpsI mutant of FA2-2 (10) |
| OG1-RF | Laboratory strain (ATCC 47077) |
| V583 | Vancomycin-resistant clinical strain (ATCC 700802) |
| MMH 594 | Epidemic strain (17) |
| R19.001 | Epidemic strain from Children's Hospital, Boston, Mass. (21) |
| 68114 | Clinical isolate |
TABLE 2.
Sera and serum absorptions used in the present study
| Antigen against which immune rabbit serum was directed | E. faecalis strain(s) used for absorption in CPS-ELISA | Homologous and heterologous strains used for absorption in opsonophago- cytic assay |
|---|---|---|
| Strain 12030 polysaccharide | Type 20 | 12030,a 12107b |
| Strain 12107 polysaccharide | 68114 | 12107,a 12030b |
| Type 2 whole bacteria (heat and proteinase K treated) | 12107 | Type 2,a 12030b and 12107b |
| Type 5 whole bacteria (heat and proteinase K treated) | 12030, 12107, type 2 | Type 5,a 12030b and 12107b |
Homologous strain.
Heterologous strain.
Sera with antibodies against the purified capsular polysaccharides of strains E. faecalis 12030 (15) and E. faecalis 12107 (16) were prepared as described elsewhere (15). In addition, sera with antibodies against two of the prototype strains described by Maekawa et al. (19) (E. faecalis Maekawa type 2 and E. faecalis Maekawa type 5) were prepared by using heat-killed and proteinase K-treated whole bacterial cells. New Zealand White rabbits were injected with two doses of 100 μg of purified polysaccharide antigen subcutaneously and three doses of 10 μg of purified polysaccharide antigen intravenously as described previously (15). Immunization with whole bacterial cells (i.e., E. faecalis type 2 and type 5) was done by using doses of 3 × 108 bacteria injected intravenously three times a week for 5 weeks. The bacterial cells were heat killed (70°C for 1 h) and digested with proteinase K (0.1 mg/ml at 56°C for 1 h; Invitrogen, Carlsbad, Calif.) prior to intravenous injection. Rabbits were bled 5 weeks after the last injection and periodically for 20 weeks without boosting. The resulting four groups of serum samples, representing four serologically distinct families, were used in a newly developed capsular polysaccharide-specific ELISA (CPS-ELISA) and in opsonophagocytic assays.
CPS-ELISA.
Crude polysaccharide extracts were prepared from the enterococcal strains. Bacteria were grown overnight at 37°C to stationary phase in 50 ml of Columbia broth supplemented with 0.5% glucose. The cell pellet was suspended in 500 μl of 10% trichloroacetic acid (TCA) and incubated on a rotor rack at 4°C for 18 to 24 h (a modification of the procedure of Heckels and Virji [13]). The suspension was then centrifuged at 12,000 rpm for 10 min (Hettich Micro 20 instrument), and the supernatant was precipitated with 1.25 ml of ethanol overnight at −20°C. After centrifugation (10 min at 12,000 rpm) the supernatant was discarded and the pellet was dissolved in 1 ml of distilled H2O. Microtiter plates (96 wells; Immulon 2HB; Dynex Technologies, Chantilly, Va.) were coated overnight with 80 μl of antigen solution along with 20 μl of 5× sensitizing buffer (180 mg of NaH2PO4 and 1.7 g of NaHPO4 per 100 ml) at 4°C. The plates were washed three times with phosphate-buffered saline (PBS), blocked with 3% skim milk in PBS for 1 h, and again washed three times with PBS. The primary antibody was applied in dilutions of 1:100 and 1:500, and the plates were incubated for 60 min at 37°C. In order to eliminate cross-reactivity and to improve specificity, all four prototype groups of sera were absorbed with either one or a combination of three different heterologous E. faecalis strains (Table 2). All serum samples were absorbed at a 1:20 dilution with 2 × 109 bacteria per ml at 4°C for 60 min on a rotor rack. This procedure was done three times for every serum sample. After an additional washing step, the secondary antibody (goat anti-rabbit alkaline phosphatase-conjugated immunoglobulin G antibody; ICN Biomedicals, Aurora, Ohio) was applied at a dilution of 1:1,000 and the mixture was incubated for 60 min at 37°C. After the plates were washed with PBS, the detection reagent p-nitrophenyl-phosphate (Sigma Chemical Co.) was added. The plates were read at 405 nm after 15, 30, 60, and 90 min in an EL 309 ELISA reader (Bio-Tek Instruments, Winooski, Vt.). Each strain was tested at least in duplicate. Both positive and negative controls were included on all plates.
Characterization of TCA extracts.
For quantitative measurement of the protein contents of TCA extracts, a modified dye-binding assay of Bradford (9) was used. For quantitative estimation of the carbohydrate contents of TCA extracts, the phenol-sulfuric acid method was performed (9).
Opsonophagocytic assays.
Opsonophagocytic assays with all enterococcal strains were performed as described elsewhere (16). Each bacterial strain was tested with the four groups of unabsorbed prototype sera. In addition, the sera were absorbed either with the homologous, immunizing strain or with single or multiple combinations of heterologous prototype strains (Table 2). Samples without serum and samples with normal rabbit serum (NRS) were used as controls, and each experiment was performed at least in duplicate. The percent killing was calculated by comparing the inoculum (viable counts at time zero [T0]) with the colony counts obtained after incubation for 90 min at 37°C (T90) on a rotor rack by use of the following formula: [(mean CFU at T0 − mean CFU at T90)/(mean CFU at T0)] × 100.
Immunoelectron microscopy.
Immunoelectron microscopy was performed with the four prototype strains, E. faecalis 12030, 12107, type 2, and type 5, as described elsewhere (16). The four groups of unabsorbed sera (data not shown) and all four groups of sera absorbed either with the homologous strain or with each of the three heterologous strains were used as primary antibodies.
PCR analysis of cps locus.
The primers and PCR conditions used to amplify the 11 genes of the cps locus have been described elsewhere (10).
Restriction fragment length polymorphism (RFLP) analysis of the cps locus.
Southern blot hybridization analysis of chromosomal DNA digested with different restriction enzymes (Invitrogen) was performed as described elsewhere (10).
RESULTS
Generation of capsule-specific antisera.
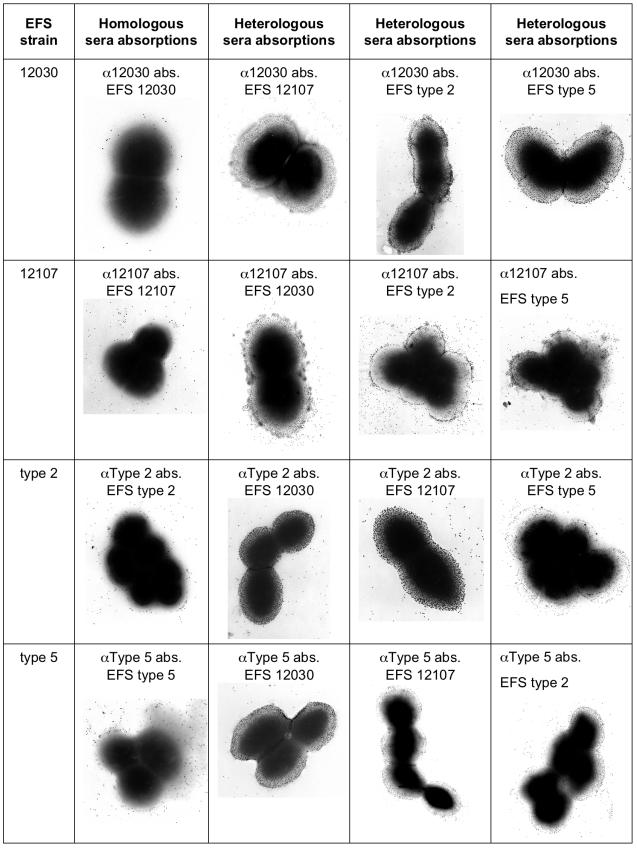
Four groups of rabbit sera with antibodies against enterococcal surface polysaccharide antigens were prepared to establish a system for the serotyping of E. faecalis. Each of the four groups of sera reacted specifically with the homologous strain (Fig. 1), outlining a capsule-like structure in all cases. Absorption of the respective serum group with the corresponding homologous strain abolished the visualization of the capsule-like structure (Fig. 1, second column), whereas absorption of the four groups of sera with any of the three heterologous strains had no effect on the ability to visualize the capsule (Fig. 1, columns three to five). This indicates that all four groups of sera reacted with antigens present on the surface of the bacterial cells and that each type of serum bound specifically to its prototype strain. Only low levels of binding of gold-labeled protein A were observed when NRS was used as a negative control (data not shown).
FIG. 1.
Immunoelectron microscopy of the four prototype E. faecalis (EFS) strains labeled with the prototype serum absorbed with either the homologous strain or the three heterologous strains.
Type-specific reactivity of antisera.
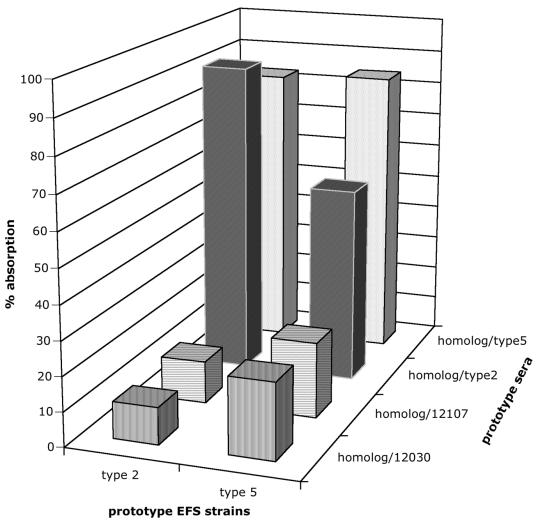
Three of the groups of absorbed prototype sera (i.e., α12030, α12107, and αType 5; Table 2) reacted only with the corresponding prototype strain by CPS-ELISA (Table 3). The type 2-specific serum reacted with both E. faecalis type 2 and E. faecalis type 5 antigens, indicating cross-reactivity between the two strains (Table 3). This cross-reactivity was further substantiated by the testing of the two strains in an absorption CPS-ELISA (Fig. 2). The two groups of sera were absorbed with the respective homologous strains as well as with each of the three heterologous strains. Binding of both groups of sera was reduced by absorption with the homologous strain (84% reduction for the type 5-specific serum, 91% reduction for the type 2-specific serum). Absorption of the two groups of sera with the respective heterologous strain, E. faecalis type 2 or E. faecalis type 5, led to a higher degree of reduction (57% reduction for the type 5-specific serum, 83% reduction for the type 2-specific serum) than absorption with the heterologous strains, E. faecalis 12030 or E. faecalis 12107 (levels of reduction, between 11 and 22%). Despite the cross-reactivity of the type 2-specific serum between E. faecalis type 2 and E. faecalis type 5, these two strains were distinguished in the CPS-ELISA. Type 2-related strains showed high levels of reactivity (as determined by measurement of the optical density at 405 nm [OD405]) with the type 2-specific serum but low levels of reactivity with the type 5-specific serum. In contrast, type 5-related strains reacted with both types of sera, with the strains having higher levels of reactivity with type 5-specific serum than type 2-specific serum (134 and 100%, respectively; Table 3).
TABLE 3.
Results of CPS-ELISA with TCA extracts of 14 E. faecalis strains
| Proposed serogroup | E. faecalis strain | OD405a
|
|||
|---|---|---|---|---|---|
| α12030 | α12107 | αType 2 | αType 5 | ||
| CPS-A | 12030 | 2.56 | 0.772 | 0.456 | 0.324 |
| Type 7 | 1.855 | 0.495 | 0.459 | 0.253 | |
| CPS-B | 12107 | 0.232 | 2.529 | 0.501 | 0.356 |
| Type 1 | 0.275 | 2.232 | 0.843 | 0.306 | |
| OG1-RF | 0.319 | 2.263 | 0.489 | 0.285 | |
| CPS-C | Type 2 | 0.586 | 0.384 | 2.444 | 0.243 |
| FA2-2 | 0.469 | 0.546 | 1.86 | 0.818 | |
| Type 12 | 0.254 | 0.306 | 2.029 | 0.218 | |
| R19.001 | 0.288 | 0.642 | 2.402 | 0.33 | |
| V583 | 0.42 | 0.718 | 2.476 | 0.514 | |
| CPS-D | Type 5 | 0.222 | 0.482 | 1.128 | 1.512 |
| Type 18 | 0.523 | 0.482 | 0.976 | 1.305 | |
| CPS-C/D | MMH 594 | 0.184 | 0.337 | 2.388 | 2.396 |
| NTb | Type 9 | 0.347 | 0.775 | 1.4 | 0.515 |
The OD405s of the four prototype groups of sera for the respective prototype strain are shown in boldface and italic. Boldface alone indicates the assignment of the strain to one of the four prototype strain types.
NT, nontypeable.
FIG. 2.
Absorption CPS-ELISA of E. faecalis (EFS) type 2 and type 5 prototype strains with the two corresponding prototype sera. The two types of sera were absorbed either with the respective homologous strain or with each of the three heterologous strains. The percent absorption values reflect the ratio of the amount of absorbed sera to the amount of unabsorbed sera. Both types of sera could be absorbed with the homologous strain (84 and 91%, respectively). Absorption of both types of sera with heterologous strains E. faecalis type 2 and E. faecalis type 5, respectively, led to a higher degree of absorption (57 and 83%, respectively) than absorption with the heterologous strains E. faecalis 12030 and E. faecalis 12107 (range, 11 to 22%).
Diversity of binding of antisera to capsular polysaccharides in the CPS-ELISA.
The assignment of a test strain to a prototype strain was made if the OD405 was at least 70% of the value obtained with the corresponding prototype strain (Table 3). This threshold was chosen on the basis of the assumption that the reactivity of a related strain with one of the prototype groups of sera should exceed the cross-reactivity of the other three heterologous prototype strains with the respective groups of sera. The following formula was used to obtain a threshold value: sum of the OD405s of all four groups of sera reacting with all three heterologous strains divided by the sum of the OD405s of all four groups of sera reacting with the respective homologous strains. A value of 62.9% was obtained by this formula. To decrease the error margin, a 70% cutoff was finally chosen as the assignment criterion.
A panel of 29 different E. faecalis strains was tested by the CPS-ELISA. By using the criteria described above, 55% (i.e., 16 of 29) of the strains tested could be classified into one of the four serotypes. One strain (E. faecalis MMH 594) reacted with two groups of sera (i.e., type 2- and type 5-specific sera), and thus, 41% (12 of 29) of the strains could not be classified unambiguously by the CPS-ELISA.
Diversity of capsular polysaccharides in opsonophagocytic killing.
To confirm the findings obtained by CPS-ELISA, all strains were analyzed by opsonophagocytic assays that tested for a clinically relevant biological function, i.e., the opsonic killing activity of the sera. To assess the specificity of the killing, the sera were absorbed with the respective homologous strain as well as with different heterologous strains (Table 2). Isolates were assigned to one of the four serotypes if (i) opsonic killing by a prototype serum was greater than 70% and (ii) the killing was abolished by more than 50% by absorption with the homologous strain but not to the same degree by absorption with the heterologous strains.
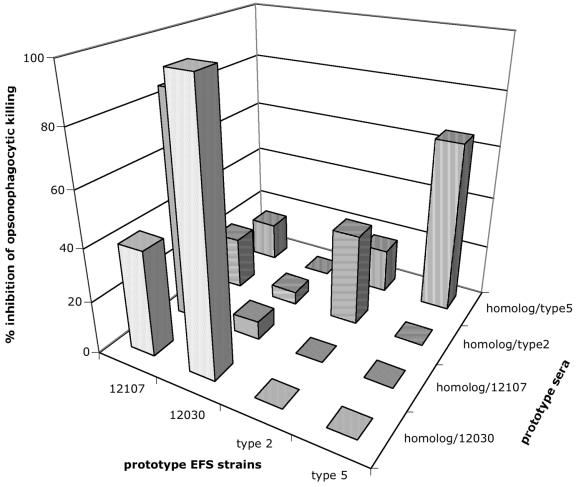
Again, the three prototypes strains, E. faecalis 12030, E. faecalis 12107, and E. faecalis type 5, were most effectively killed and, according to the assignment criterion, were also specifically killed by their homologous sera (Fig. 3). Type 2-specific serum killed both E. faecalis type 2 and E. faecalis type 5, supporting the findings of cross-reactivity between these two strains. Ten of the 29 (34%) E. faecalis strains could be unambiguously assigned to one of the four serotypes. For a further 24% (7 of 29) of the strains, the opsonic killing activity was not able to distinguish between type 2- or type 5-related strains. One strain (Maekawa strain type 7) was killed to the same degree by both strain 12030- and strain 12107-specific sera. The overall agreement between the CPS-ELISA and the opsonophagocytic assay was 72% (21 of 29 strains). Seven (24%) strains that were nontypeable by one assay could be assigned to a serogroup by the other test. Only one strain (Maekawa strain type 6 strain) gave conflicting results by the two assays.
FIG. 3.
Phagocytic inhibition assay of the four prototype groups of sera against the four prototype E. faecalis (EFS) strains. Each prototype serum was tested against its homologous prototype E. faecalis strain by absorption with the respective homologous strain and with the three heterologous strains. The percent inhibition values reflect the ratio of the amount of absorbed sera to the amount of unabsorbed sera. The killing activities of the four types of sera were most effectively inhibited by absorption with the homologous strain (100% for E. faecalis 12030, 84% for E. faecalis 12107, 66% for E. faecalis type 5). Type 2-specific sera could not be absorbed to the same degree (absorption, 34%). Inhibition of killing activity by heterologous strains was greatly reduced for all four prototype sera, with cross-reactivity between strains E. faecalis 12030 and E. faecalis 12107.
When the results of the two assays are taken together, 66% (19 of 29) of the E. faecalis strains could be assigned to one of the four serogroups. Two additional strains were grouped into two serogroups (types 2 and 5; Table 4).
TABLE 4.
Proposal for a serotyping system for E. faecalis based on capsular polysaccharides and their relationship with the cps locusa
| Proposed serogroup | E. faecalis strain | Homology by:
|
RFLP pattern of CPS locus | |
|---|---|---|---|---|
| CPS-ELISA | Opsonophago- cytic assay | |||
| CPS-A | 12030 | 12030 | 12030 | I |
| Type 3 | 12030 | I | ||
| Type 7 | 12030 | 12030 and 12107 | I | |
| CPS-B | 12107 | 12107 | 12107 | I |
| OG1-RF | 12107 | 12107 | I | |
| Type 1 | 12107 | I | ||
| Type 10 | 12107 | I | ||
| Type 16 | 12107 | I | ||
| Type 20 | 12107 | I | ||
| CPS-C | Type 2 | Type 2 | Type 2 | III |
| V583 | Type 2 | Type 2 and type 5 | III | |
| FA2-2 | Type 2 | Type 2 and type 5 | III | |
| R19.001 | Type 2 | Type 2 | NDb | |
| Type 12 | Type 2 | Type 2 and type 5 | II | |
| Type 14 | Type 2 | Type 2 and type 5 | II | |
| Type 21 | Type 2 | II | ||
| CPS-D | Type 5 | Type 5 | Type 2 and type 5 | II |
| 68114 | Type 5 | Type 2 and type 5 | ND | |
| Type 18 | Type 5 | Type 2 and type 5 | II | |
| CPS-C and CPS-D | MMH 594 | Type 2 and type 5 | III | |
| Type 6 | Type 5 | Type 2 | II | |
| NTc | Type 4 | I | ||
| Type 8 | III | |||
| Type 9 | I | |||
| Type 11 | III | |||
| Type 13 | I | |||
| Type 15 | I | |||
| Type 17 | I | |||
| Type 19 | I | |||
E. faecalis strains were assigned to one of the four capsular polysaccharide serotypes (CPS-A to CPS-D) if the definition criteria for one of the two diagnostic assays were met or if one assay eliminated cross-reactivity between two serotypes in the second assay. Boldface indicates the prototype strain.
ND, not determined.
NT, nontypeable.
Genetic diversity of capsular polysaccharides.
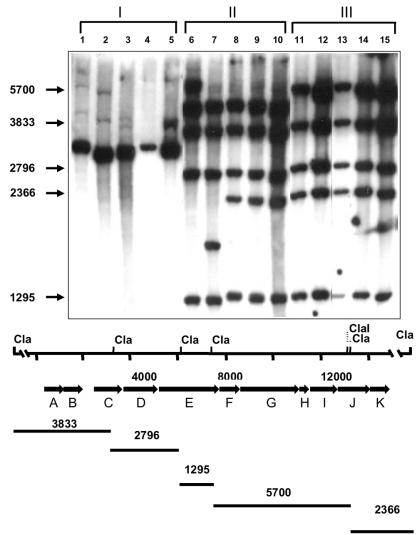
The assignment of a given strain to a serogroup was correlated with the genetic organization of the cps locus, shown by Hancock and Gilmore (10) to be involved in the biosynthesis of capsular polysaccharides. Representative strains of the four serogroups were chosen, and PCR was used to test for the presence of the 11 cps genes (Table 5). Two different patterns could be shown: E. faecalis strains of the strain 12030- and 12107-related groups consistently possessed only the first two genes of the cps locus (i.e., cpsA and cpsB), while all strains of the type 2- and type 5-related groups had the cpsA to cpsE and the cpsG to cpsK genes. The cpsF gene was consistently missing from the type 5-related strains and was variably present in the type 2-related strains. No specific pattern could be observed for the nontypeable strains. The results of the PCR were verified by Southern blot hybridization analysis. Hybridization analysis of DNA digested with ClaI, using the 14.8-kb cpsA to cpsK fragment amplified from strain V583 as a probe, demonstrated various degrees of conservation among the isolates examined. Strains belonging to the E. faecalis 12030 and E. faecalis 12107 groups all showed an RFLP class I pattern, with only a single hybridization band, the size of which corresponds to those of the cpsA and cpsB genes, as well as a conserved region downstream of cpsK (Fig. 4). E. faecalis type 5 strains yielded an RFLP class II pattern and were more closely related to class III strains than to the other classes of strains, with the class II and class III strains appearing to differ by only a single ClaI restriction fragment.
TABLE 5.
PCR analysis of the cps locus in a representative collection of E. faecalis strains
| Proposed serogroup | E. faecalis strain | Result for gene:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cpsA | cpsB | cpsC | cpsD | cpsE | cpsF | cpsG | cpsH | cpsI | cpsJ | cpsK | ||
| CPS-A | 12030 | + | + | − | − | − | − | − | − | − | − | − |
| Type 7 | + | + | − | − | − | − | − | − | − | − | − | |
| CPS-B | 12107 | + | + | − | − | − | − | − | − | − | − | − |
| Type 1 | + | + | − | − | − | − | − | − | − | − | − | |
| OG1-RF | + | + | − | − | − | − | − | − | − | − | − | |
| CPS-C | Type 2 | + | + | + | + | + | + | + | + | + | + | + |
| FA2-2 | + | + | + | + | + | + | + | + | + | + | + | |
| Type 12 | + | + | + | + | + | − | + | + | + | + | + | |
| R19.001 | + | + | + | + | + | − | + | + | + | + | + | |
| CPS-D | Type 5 | + | + | + | + | + | − | + | + | + | + | + |
| Type 18 | + | + | + | + | + | − | + | + | + | + | + | |
| CPS-C and CPS-D | MMH 594 | + | + | + | + | + | + | + | + | + | + | + |
| NTa | Type 9 | + | + | − | − | − | − | − | − | − | − | − |
NT, nontypeable.
+, present; −, absent.
FIG. 4.
Southern blot analysis of capsular polysaccharide determinants from various E. faecalis strains. DNA was restricted with ClaI and probed with the radiolabeled fragment of the cpsA to cpsK genes from E. faecalis strain V583. Lanes 1 to 5, strains with RFLP pattern I; lanes 6 to 10, strains with RFLP pattern II; lanes 11 to 15, strains with RFLP pattern III. Lane 1, E. faecalis 12107; lane 2, E. faecalis 12030; lane 3, E. faecalis type 4; lane 4, E. faecalis type 1; lane 5, E. faecalis type 7; lane 6, E. faecalis type 6; lane 7, E. faecalis type 5; lane 8, E. faecalis type 18; lane 9, E. faecalis type 14; lane 10, E. faecalis type 12; lane 11, E. faecalis type 8, lane 12, E. faecalis type 2; lane 13, E. faecalis MMH 594; lane 14, E. faecalis V583; lane 15, E. faecalis FA2-2. The sizes (in base pairs) of the E. faecalis V583 sequence fragments hybridizing to the probe are shown at the left. The ClaI restriction map for the E. faecalis V583 capsular polysaccharide determinant is shown below. The molecular sizes (in base pairs) of the regions spanned by the ClaI sites in E. faecalis V583 are listed below the map.
The difference between RFLP class II and III strains is likely attributable to the absence of the cpsF gene in the class II strains. The 5.7-kb ClaI fragment, which spans the 3′ end of cpsE through the 5′ end of cpsJ and which is present in class III strains, is absent from class II strains. However, a conserved 4.7-kb ClaI fragment from class II strains hybridizes to the probe (Fig. 4). An apparent ClaI polymorphism was observed for class II strains. With the exception of the 2.4-kb ClaI fragment, E. faecalis type 5 and type 6 strains have ClaI restriction patterns identical to those of E. faecalis types 12, 14, and 18. In place of the 2.4-kb fragment, E. faecalis type 5 has a slightly smaller ClaI fragment of approximately 1.8 kb, whereas E. faecalis type 6 has a much larger ClaI fragment of 6.0 kb (Fig. 4). E. faecalis class III strains previously identified as being reactive with type 2-specific serum (10), namely, E. faecalis FA2-2, E. faecalis V583, and E. faecalis MMH 594, showed identical hybridization patterns with the E. faecalis type 2 prototype strain (Fig. 4).
Capsule-specific reactivity of antisera.
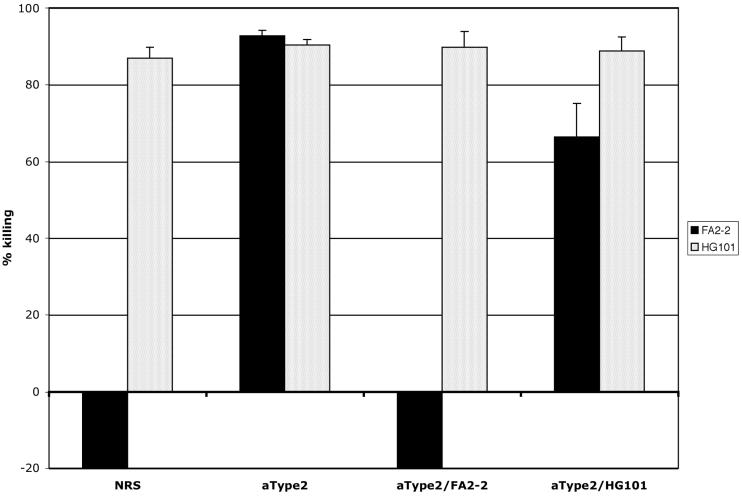
To confirm the hypothesis that the typing sera reacted with capsular antigens encoded by the cps locus, a CPS-ELISA (data not shown) and an opsonophagocytic assay were performed with the acapsular mutant HG101 and its wild-type (WT) capsular strain, E. faecalis FA2-2 (Fig. 5). E. faecalis FA2-2 reacts specifically with type 2-specific serum (10). NRS was used as a negative control. The WT strain was resistant to killing by NRS, whereas the capsule-negative mutant was highly susceptible to killing by NRS. Type 2-specific serum killed both strains efficiently. Absorption of the serum with the WT strain abolished the killing activity against the WT strain but not the mutant, and absorption of the serum with the mutant had little or no effect on the killing activity of the type 2-specific serum. These data support the hypothesis that the activity of the type 2-specific serum is directed against the capsule produced at least in part by enzymes encoded by the cps locus.
FIG. 5.
Comparison of E. faecalis strains FA2-2 and HG101 in the opsonophagocytic assay with NRS and the type 2-specific sera. Negative killing reflects growth during the 90-min incubation period of the assay.
DISCUSSION
An enterococcal surface antigen that is the target of opsonic antibodies has previously been isolated from E. faecalis 12030 and characterized (16). Compositional and structural analyses of this antigen revealed a teichoic acid composed of a repeating unit of α-d-glucopyranosyl-(1-2)-α-d-glucopyranose (kojibose) attached to a glycerol diphosphate (16, 27). Passive and active immunotherapies reduced the bacterial counts in a mouse model of bacteremia when the animals were challenged with the homologous strain. This serum also protected mice when they were challenged with a vancomycin-resistant E. faecium strain (15). However, it is not known how many structurally and immunologically different capsular polysaccharides that may be used as vaccine candidates exist in enterococci.
Earlier studies by Sharpe (23), Sharpe and Shattok (24), and Maekawa et al. (18, 19) attempted to establish systems for the serotyping of enterococci. The latter scheme was based on 6 of Sharpe's original prototype strains, 2 strains from the Centers for Disease Control and Prevention, and 13 clinical isolates from Japan (19). In order to relate our present findings to those of previous investigators, we included the strain collection of Maekawa et al. (19) in the present effort, since these isolates represent E. faecalis strains with a high degree of antigenic diversity (although the diversity is not necessarily based solely on carbohydrate antigens). Maekawa’s collection also included the strains from the study of Sharpe (23) and that of Sharpe and Shattok (24).
A CPS-ELISA was developed on the basis of the preferential extraction of cell wall teichoic acids. In the assay, we used sera that were absorbed with different heterologous strains (Table 2). The four prototype strains used in this study were chosen for the following reasons: the capsular polysaccharide structure of E. faecalis 12030 has been elucidated (16, 27). The opsonic killing activities of antibodies against the purified capsular polysaccharide from this strain were tested against a collection of clinical isolates, and it was found that 33% of the strains were killed by this serum (16). One of these strains (E. faecalis 12107) was not killed by this serum, and preliminary structural analysis of capsular polysaccharide material from this strain indicated that its cell wall teichoic acid structure may be different from that of E. faecalis 12030 (unpublished data).
In studies performed by Maekawa and Habadera (18), 30% of urinary tract isolates were of the type 2 serotype. Studies performed contemporaneously by Hancock and Gilmore (10) indicated that the cps locus in the 21 Maekawa prototype strains showed three different patterns, with E. faecalis 12030 and E. faecalis 12107 being indistinguishable by this method. However, two additional patterns were observed, and E. faecalis type 2 and E. faecalis type 5 were arbitrarily chosen as representatives of the remaining two RFLP patterns, respectively.
The four types of sera used in this study were all directed against capsular polysaccharides and reacted specifically with the respective prototype strain, independently of the method used to raise the sera. Immunoelectron microscopy of the four prototype strains reacting with the respective sera clearly showed that the capsule is the target of the antibodies (Fig. 1). However, E. faecalis type 2 and E. faecalis type 5 showed certain degrees of cross-reactivity. Heat inactivation and proteinase K treatment of whole bacterial cells elicited antibodies directed against capsular structures, a finding which is confirmed by the opsonophagocytic assay with WT E. faecalis strain FA2-2 and its capsule-negative mutant, HG101. The WT strain was killed only by the type 2-specific serum, indicating that the serum contained opsonic antibodies directed against the capsule. In contrast, the capsule-negative mutant was already sensitive to killing by preexisting natural antibodies in NRS, which were not necessarily directed against the capsule. A similar phenomenon has been observed previously (2, 3, 15, 16).
In order to increase the specificity of the reactivities of the four types of sera against capsular polysaccharides, we used TCA extraction to isolate crude polysaccharide antigens from the bacterial cells in the CPS-ELISA. We found that the protein levels in TCA extracts were below the detection limit of the modified Bradford assay used. Treatment of TCA extracts with sodium periodate reduced the carbohydrate content to 18% of that of pure TCA extracts and reduced the reactivity with the type-specific serum to 8% (data not shown), indicating that the majority of the antibody binding activity was to a carbohydrate antigen. In contrast, treatment of the TCA extract with proteinase K had no effect on the reactivities of the antisera (98% of that of pure TCA extracts). From these data we conclude that the TCA extracts primarily consisted of carbohydrate antigens, with the majority presumably being capsular polysaccharide antigens. This assumption was further supported by the observation that type 2-specific serum revealed a strong reactivity with E. faecalis FA2-2 (OD405, 1.860) but a strongly reduced reactivity with capsule-negative mutant HG101 (OD405, 0.526).
Using the criteria mentioned above to assign an isolate to one of the four serogroups, we were unequivocally able to group 55% of the strains tested. However, one of the serum types (i.e., the type 2-specific serum) showed a high degree of cross-reactivity with a different prototype strain (i.e., E. faecalis type 5). This may be explained by chemical and/or structural similarities of the different polysaccharides, a situation not uncommon for polysaccharide antigens from other bacteria (5, 25). Alternatively, a given strain may be able to produce two different variant forms of carbohydrate antigen, depending on the conditions used.
To correlate the data obtained by CPS-ELISA with the biological response (i.e., opsonic killing) relevant to the intended use of the antigens as the basis of a vaccine, we compared the results obtained by CPS-ELISA with those obtained by an opsonophagocytic assay. While the majority of strains reacted identically in the opsonophagocytic assay, we also observed some differences. The high degree of cross-reactivity between E. faecalis type 2 and E. faecalis type 5 in the opsonophagocytic assay points toward the possibility that these two antigens have close structural similarity. Taking into account this cross-reactivity, as well as the number of strains that were not typeable by both methods, the agreement between the two assays was 72%. Four strains could be grouped only by the opsonophagocytic assay, perhaps because the amount of specific polysaccharide on the surface of the bacterial cell was too small to be detected by the CPS-ELISA. This amount of antigen may still provide enough antigenic epitopes for opsonization, leading to killing in the opsonophagocytic assay. One of the two strains typeable only by CPS-ELISA (Maekawa strain type 20) was highly resistant to killing by any of the sera, and factors other than capsule production that contribute to serum resistance may be responsible for this effect. The other strain, E. faecalis type 1, was highly sensitive to killing by all four types of sera, including NRS (data not shown), suggesting that this strain was unable to resist non-antibody-mediated opsonic killing, even though it has a capsule.
To date there is no proven link between the structure of capsular polysaccharide and a genetic locus responsible for the biosynthesis of the capsular polysaccharide. E. faecalis 12030 is the only strain for which structural analysis of the capsular polysaccharide has been performed (16, 27). The cps locus is known to be involved in the biosynthesis of capsular polysaccharides (10), but so far only a compositional analysis of the respective antigen exists. The homology of one of the enzymes within this gene locus with a glycerophosphotransferase involved in the synthesis of teichoic acids in Bacillus subtilis and the presence of quantitative amounts of phosphate indicate that the locus described by Hancock and Gilmore (10) may be responsible for the production of the capsular polysaccharide described by Huebner and colleagues (16, 27). Three different organizations of the gene locus have been observed: E. faecalis 12030 and E. faecalis 12107 showed an RFLP class I pattern, with the presence of only two open reading frames, i.e., cpsA and cpsB. Both of these genes may be essential in E. faecalis, and they are present in all strains tested so far, independent of their serotype.
Hancock and Gilmore (11) and Xu et al. (28-30) have described a different locus involved in the synthesis of an integral enterococcal cell wall polysaccharide. While Huebner and colleagues (16, 27) were able to isolate and initially characterize a similar tetraheteroglycan material from the cell wall of our prototype strain, E. faecalis 12030, this antigen is distinct from the teichoic acid-like capsular polysaccharides, and the preparation of this tetraheteroglycan shows no immunoreactivity with serum raised against the capsular polysaccharides examined here (data not shown).
E. faecalis type 2- and E. faecalis type 5-related strains, which we refer to as the CPS-C and the CPS-D serogroups, respectively, possessed at least 10 of the 11 genes of the cps locus. While strains of the CPS-D serogroup did not contain the cpsF gene, its presence was variable in CPS-C strains. At present, it is unclear why RFLP class II strains represent both CPS-C and CPS-D strains. Some class II strains (i.e., E. faecalis types 5 and 18) react with type 5-specific serum in the CPS-ELISA, whereas others (i.e., E. faecalis types 12 and 14) react with type 2-specific serum. E. faecalis strain MMH 594, which was previously shown to react with type 2-specific serum (10), produced a hybridization pattern identical to that of RFLP class III strains; but it is now known that it cross-reacts with type 5-specific serum, which suggests that it may modulate expression of its cps genes, possibly leading to antigenic variation through the expression of multiple serotypes. Interestingly, E. faecalis type 8 produced the same hybridization profile as E. faecalis type 2 (Fig. 4) but was nontypeable with type 2-specific serum. It remains unclear how E. faecalis types 2, 8, and 11 (data not shown) can yield identical restriction patterns for the cps determinant yet still be serologically distinct. It is possible that E. faecalis types 8 and 11 have lost the ability to produce the capsular serotype, thus exposing alternative typing antigens. Alterations in promoter sequences or the presence of regulatory elements could modulate the extent to which the polysaccharide is expressed. Alternatively, the product(s) of polysaccharide biosynthetic genes not found in the cps determinant could alter the capsular structure through the placement of additional carbohydrate residues on the capsular polysaccharide.
The presence of the genes in the cps locus does not completely explain the serologic specificity observed. Structural analysis of defined knockout mutants may be able to determine whether the difference between CPS-C and CPS-D can be explained by the expression of the cpsF gene. The additional genes that are probably necessary for the production of the CPS-A and CPS-B polysaccharides need to be identified and sequenced.
It appears that E. faecalis capsular polysaccharides have evolved from two distinct families. The first family comprises lineages containing all (RFLP class III) or most (RFLP class II) of the cpsC to cpsK determinant, whereas the second family consists of a third lineage that lacks this determinant. The importance of the capsular polysaccharide to the biology of the organism would suggest that at some time in the distant past the second family acquired the ability to make a capsule, independent of the cps determinant. A closer examination of the Southern blot analysis (Fig. 4) reveals additional faint bands for DNA from strains of the second family, suggesting that the evolution of capsular polysaccharide diversity in this family may have paralleled that in the first family to allow some cross hybridization with related genes located elsewhere on the chromosome. This observation is consistent with the evolution of capsular polysaccharide determinants in other organisms (22). In Staphylococcus aureus, the capsular polysaccharides of serotypes 5 and 8 are similar and their biosynthetic genes occur at the same point on the S. aureus genomic map, whereas the determinant for type 1 capsule biosynthesis occurs elsewhere on the chromosome. The limited homology between these genetic pathways is detectable only by hybridization under low-stringency conditions, even though the structures of the type 1, 5, and 8 capsular polysaccharides are similar (22).
From the present data we conclude that a limited number of enterococcal serogroups exist. The four serogroups described here represent a major portion (about 60%) of clinically relevant E. faecalis isolates. The methods described here can be used to identify strains with serologically different polysaccharides. Ongoing studies aimed at identifying additional serogroups have so far not given an indication of any serologically diverse polysaccharide capsules (data not shown). The presence of strains that do not react with our antisera may be due to the absence of a polysaccharide capsule in these strains.
In contrast to the studies of Sharpe and Shattok (24), we investigated the serologic diversity of enterococcal polysaccharides not for epidemiologic analysis but for better evaluation of the feasibility of immunotherapy regimens based on capsular polysaccharide antigens. Serotyping of bacteria has been widely used as a tool in bacterial epidemiology. Powerful new molecular typing methods have replaced this technique. Methods such as pulsed-field gel electrophoresis (26) have better discriminatory power, and randomly amplified polymorphic DNA analysis (4) and multilocus sequence typing (14) are also able to show phylogenetic and evolutionary relationships between strains. Studies are under way to elucidate the possible relationships between serogroups and the evolutionary lineages of enterococcal strains of different geographical origins.
Compositional and structural analyses of the polysaccharides are under way to confirm the serogroups described here. These data will facilitate the comprehensive establishment of a system for the serotyping of enterococci and will help to determine the relevant serotypes to be included in a broadly active enterococcal polysaccharide vaccine. The limited number of serotypes found so far suggests that the development of an enterococcal vaccine will be feasible.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (grant AI5066701 to J.H.) and an Infectious Disease Fellowship by the Walter-Marget Foundation (to M.H.)
The studies described in this report were reviewed and approved by the Institutional Animal Care and Use Committee at Harvard University.
REFERENCES
- 1.Anonymous. 2000. National Nosocomial Infections Surveillance (NNIS) system report, data summary from January 1992-April 2000, issued June 2000. Am. J. Infect. Control 28:429-448. [DOI] [PubMed] [Google Scholar]
- 2.Arduino, R. C., K. Jacques-Palaz, B. E. Murray, and R. M. Rakita. 1994. Resistance of Enterococcus faecium to neutrophil-mediated phagocytosis. Infect. Immun. 62:5587-5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arduino, R. C., B. E. Murray, and R. M. Rakita. 1994. Roles of antibodies and complement in phagocytic killing of enterococci. Infect. Immun. 62:987-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbier, N., P. Saulnier, E. Chachaty, S. Dumontier, and A. Andremont. 1996. Random amplified polymorphic DNA typing versus pulsed-field gel electrophoresis for epidemiological typing of vancomycin-resistant enterococci. J. Clin. Microbiol. 34:1096-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beynon, L. M., J. C. Richards, and M. B. Perry. 1997. Identification of the common antigenic determinant shared by Streptococcus pneumoniae serotype 35A and 20 capsular polysaccharides—structural analysis of the Streptococcus pneumoniae serotype 35A capsular polysaccharide. Eur. J. Biochem. 250:163-167. [DOI] [PubMed] [Google Scholar]
- 6.Cetinkaya, Y., P. Falk, and C. G. Mayhall. 2000. Vancomycin-resistant enterococci. Clin. Microbiol. Rev. 13:686-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diekema, D. J., S. L. Coffman, S. A. Marshall, M. L. Beach, K. V. Rolston, and R. N. Jones. 1999. Comparison of activities of broad-spectrum beta-lactam compounds against 1,128 gram-positive cocci recently isolated in cancer treatment centers. Antimicrob. Agents Chemother. 43:940-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edmond, M. B., S. E. Wallace, D. K. McClish, M. A. Pfaller, R. N. Jones, and R. P. Wenzel. 1999. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 29:239-244. [DOI] [PubMed] [Google Scholar]
- 9.Hancock, I., and I. Poxton. 1988. Appendix 1: general methods, p. 269-286. In I. Hancock and I. Poxton (ed.), Bacterial cell surface techniques. John Wiley & Sons, Chichester, United Kingdom.
- 10.Hancock, L. E., and M. S. Gilmore. 2002. The capsular polysaccharide of Enterococcus faecalis and its relationship to other polysaccharides in the cell wall. Proc. Natl. Acad. Sci. USA 99:1574-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hancock, L. E., and M. S. Gilmore. 1997. Identification of a highly conserved lipopolysaccharide (LPS) modification operon in Enterococcus faecalis. Adv. Exp. Med. Biol. 418:1049-1050. [DOI] [PubMed] [Google Scholar]
- 12.Hancock, L. E., B. D. Shepard, and M. S. Gilmore. 2003. Molecular analysis of the Enterococcus faecalis serotype 2 polysaccharide determinant. J. Bacteriol. 185:4393-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heckels, J. E., and M. Virji. 1988. Separation and purification of surface components, p. xv and 329. In I. Hancock and I. Poxton (ed.), Bacterial cell surface techniques. John Wiley & Sons, Chichester, United Kingdom.
- 14.Homan, W. L., D. Tribe, S. Poznanski, M. Li, G. Hogg, E. Spalburg, J. D. Van Embden, and R. J. Willems. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40:1963-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huebner, J., A. Quaas, W. A. Krueger, D. A. Goldmann, and G. B. Pier. 2000. Prophylactic and therapeutic efficacy of antibodies to a capsular polysaccharide shared among vancomycin-sensitive and -resistant enterococci. Infect. Immun. 68:4631-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huebner, J., Y. Wang, W. A. Krueger, L. C. Madoff, G. Martirosian, S. Boisot, D. A. Goldmann, D. L. Kasper, A. O. Tzianabos, and G. B. Pier. 1999. Isolation and chemical characterization of a capsular polysaccharide antigen shared by clinical isolates of Enterococcus faecalis and vancomycin-resistant Enterococcus faecium. Infect. Immun. 67:1213-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huycke, M. M., C. A. Spiegel, and M. S. Gilmore. 1991. Bacteremia caused by hemolytic, high-level gentamicin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 35:1626-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maekawa, S., and S. Habadera. 1996. Comparative distribution of the serotypes of Enterococcus faecalis isolated from the urine of patients with urinary tract infections and the feces of healthy persons as determined by the slide agglutination reaction. Kansenshogaku Zasshi 70:168-174. [DOI] [PubMed] [Google Scholar]
- 19.Maekawa, S., M. Yoshioka, and Y. Kumamoto. 1992. Proposal of a new scheme for the serological typing of Enterococcus faecalis strains. Microbiol. Immunol. 36:671-681. [DOI] [PubMed] [Google Scholar]
- 20.Murray, B. E. 2000. Vancomycin-resistant enterococcal infections. N. Engl. J. Med. 342:710-721. [DOI] [PubMed] [Google Scholar]
- 21.Rhinehart, E., N. E. Smith, C. Wennersten, E. Gorss, J. Freeman, G. M. Eliopoulos, R. C. Moellering, Jr., and D. A. Goldmann. 1990. Rapid dissemination of beta-lactamase-producing, aminoglycoside-resistant Enterococcus faecalis among patients and staff on an infant-toddler surgical ward. N. Engl. J. Med. 323:1814-1818. [DOI] [PubMed] [Google Scholar]
- 22.Sau, S., and C. Y. Lee. 1996. Cloning of type 8 capsule genes and analysis of gene clusters for the production of different capsular polysaccharides in Staphylococcus aureus. J. Bacteriol. 178:2118-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharpe, M. 1964. Serological types of Streptococcus faecalis and its varieties and their cell wall type antigen. J. Gen. Microbiol. 36:151-160. [DOI] [PubMed] [Google Scholar]
- 24.Sharpe, M., and P. Shattok. 1952. The serological typing of group D streptococci associated with outbreaks of neonatal diarrhoea. J. Gen. Microbiol. 6:150-165. [DOI] [PubMed] [Google Scholar]
- 25.Szu, S., C. J. Lee, D. Carlo, and J. Henrichsen. 1981. Immunochemical characterization of cross-reactivity of pneumococcal group 9 capsular polysaccharide types 9N, 9A, 9L, and 9V. Infect. Immun. 31:371-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomayko, J. F., and B. E. Murray. 1995. Analysis of Enterococcus faecalis isolates from intercontinental sources by multilocus enzyme electrophoresis and pulsed-field gel electrophoresis. J. Clin. Microbiol. 33:2903-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, Y., J. Huebner, A. O. Tzianabos, G. Martirosian, D. L. Kasper, and G. B. Pier. 1999. Structure of an antigenic teichoic acid shared by clinical isolates of Enterococcus faecalis and vancomycin-resistant Enterococcus faecium. Carbohydr. Res. 316:155-160. [DOI] [PubMed] [Google Scholar]
- 28.Xu, Y., L. Jiang, B. E. Murray, and G. M. Weinstock. 1997. Enterococcus faecalis antigens in human infections. Infect. Immun. 65:4207-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu, Y., B. E. Murray, and G. M. Weinstock. 1998. A cluster of genes involved in polysaccharide biosynthesis from Enterococcus faecalis OG1RF. Infect. Immun. 66:4313-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu, Y., K. V. Singh, X. Qin, B. E. Murray, and G. M. Weinstock. 2000. Analysis of a gene cluster of Enterococcus faecalis involved in polysaccharide biosynthesis. Infect. Immun. 68:815-823. [DOI] [PMC free article] [PubMed] [Google Scholar]