Abstract
We investigated the utility of 500-bp 16S rRNA gene sequencing for identifying clinically significant species of aerobic actinomycetes. A total of 28 reference strains and 71 clinical isolates that included members of the genera Streptomyces, Gordonia, and Tsukamurella and 10 taxa of Nocardia were studied. Methods of nonsequencing analyses included growth and biochemical analysis, PCR-restriction enzyme analysis of the 439-bp Telenti fragment of the 65 hsp gene, susceptibility testing, and, for selected isolates, high-performance liquid chromatography. Many of the isolates were included in prior taxonomic studies. Sequencing of Nocardia species revealed that members of the group were generally most closely related to the American Type Culture Collection (ATCC) type strains. However, the sequences of Nocardia transvalensis, N. otitidiscaviarum, and N. nova isolates were highly variable; and it is likely that each of these species contains multiple species. We propose that these three species be designated complexes until they are more taxonomically defined. The sequences of several taxa did not match any recognized species. Among other aerobic actinomycetes, each group most closely resembled the associated reference strain, but with some divergence. The study demonstrates the ability of partial 16S rRNA gene sequencing to identify members of the aerobic actinomycetes, but the study also shows that a high degree of sequence divergence exists within many species and that many taxa within the Nocardia spp. are unnamed at present. A major unresolved issue is the type strain of N. asteroides, as the present one (ATCC 19247), chosen before the availability of molecular analysis, does not represent any of the common taxa associated with clinical nocardiosis.
Rapid and accurate identification of aerobic actinomycetes is becoming an increasingly important objective for clinical microbiology laboratories. These organisms, especially Nocardia spp., are more frequently isolated in the laboratory because of their potential to cause infection in the growing immunocompromised patient population. Once Nocardia spp. are isolated, rapid identification of Nocardia spp. is important for two reasons. First, some aerobic actinomycetes, such as Gordonia spp. and Streptomyces spp., are common in the environment and are more likely to be contaminants than true pathogens when they are isolated in the laboratory. This differs from Nocardia sp. isolates, which are often clinically significant. Therefore, rapid identification is important for determining the clinical relevance of an isolate. Second, these organisms differ in their susceptibilities to antimicrobials, and especially for Nocardia spp., species-level identification aids in predicting antimicrobial susceptibility (31). For example, Nocardia farcinica exhibits variable susceptibility to sulfamethoxazole and is resistant to extended-spectrum cephalosporins, such as cefotaxime and ceftriaxone, whereas N. nova is usually susceptible to these drugs (28).
Identification of aerobic actinomycetes by conventional biochemical assays requires expertise and time, and newer species such as N. nova can be difficult to separate with accuracy from other related species (28). These organisms are isolated infrequently enough that it is difficult to develop or maintain technical expertise among laboratory personnel. Since these bacteria are slowly growing, 2 to 4 weeks is required for genus-level identification and an additional 4 weeks or more is required for species-level identification. Alternative methods of identification, including high-performance liquid chromatography (HPLC) and molecular techniques, have been applied to this group of bacteria (3). HPLC is limited by the inability to determine a species-level identification. DNA amplification followed by PCR-restriction endonuclease analysis (PRA) of the 65 hsp gene has proven to be a more effective method of rapid identification (22).
Sequence-based identification is an alternative method of identifying clinical isolates that are either slowly growing or difficult to identify by biochemical profiling. Several reports have described the utility of sequencing a portion of the 16S rRNA gene for identifying clinical isolates of Mycobacterium spp. and other bacteria (for a review, see reference 17). We investigated the utility of 500-bp 16S rRNA gene sequence analysis for the identification of aerobic actinomycete isolates. Since the accuracy of sequence identification is directly dependent upon the sequence database that is queried, we evaluated two databases, the MicroSeq database (Applied Biosystems, Foster City, Calif.) and the GenBank database. The MicroSeq database is a commercial database that primarily consists of sequences of type strains. The GenBank database is a public database that contains a large number of sequences, including 16S rRNA sequences (2).
MATERIALS AND METHODS
Organisms.
A total of 99 isolates of aerobic actinomycetes from the Mycobacteria Nocardia Laboratory of the University of Texas Health Center at Tyler (UTHCT) were included in this study. These either were obtained directly from the American Type Culture Collection (ATCC; Manassas, Va.) (Table 1) or were taken from the frozen stocks in the UTHCT laboratory that were used as the sources for the strains submitted to ATCC. Several principles dictated what organisms were chosen for sequencing. If an ATCC strain representative of the group was available, especially the type strain, it was included. The remaining isolates were from clinical sources that were well characterized and included examples of any subgroups recognized within the member group. Clinical isolates were submitted for identification and/or susceptibility testing and were then frozen in tryptic soy broth plus 15% glycerol until needed. Most isolates had been used in prior taxonomic studies (22).
TABLE 1.
ATCC reference strains used in this study
| Prior classification | Isolate reference no. |
|---|---|
| Streptomyces spp. | |
| S. albus | ATCC 3004T |
| S. somaliensis | ATCC 33201T |
| S. griseus | ATCC 10137 |
| Gordonia spp. | |
| G. achiensis | ATCC 33611T |
| G. bronchialis | ATCC 25592T |
| G. sputi | ATCC 29627T |
| G. sputi | ATCC 33610 |
| Actinomadura spp. | ATCC 19425T |
| A. madurae | ATCC 13724 |
| A. madurae | ATCC 13724 |
| Tsukamurella spp. | |
| T. paurometabolum | ATCC 8368T |
| T. paurometabolum | ATCC 25938 |
| T. inchonensis | ATCC 700082T |
| Rhodococcus equi | ATCC 6939 |
| Nocardia spp. | |
| N. asteroides | ATCC 19247T |
| N. asteroides (type I) | ATCC 23824 |
| N. asteroides (type IV) | ATCC 49872 |
| N. asteroides (type IV) | ATCC 49873 |
| N. asteroides (type VI) | ATCC 14759 |
| N. nova | ATCC 33726T |
| N. nova | ATCC 33727 |
| N. farcinica | ATCC 3308 |
| N. farcinica | ATCC 3318T |
| N. brasiliensis | ATCC 19296T |
| N. transvalensis complex | ATCC 6865T |
| N. otitidiscaviarum | ATCC 14629T |
| N. pseudobrasiliensis | ATCC 51512T |
| N. paucivorans | ATCC BAA 278T |
| N. cyriacigeorgica | PSM 4484T |
Sixty-one isolates of Nocardia spp. belonging to 10 recognized species and multiple unnamed taxa were chosen for sequencing. Preliminary grouping of isolates in the N. asteroides complex was based on antimicrobial susceptibility patterns, which grouped the isolates into seven drug susceptibility types (29). Some of these types have been assigned to known species, some have been assigned to complexes, and some have no taxonomic status. Sequence analysis was performed with 41 isolates of the N. asteroides complex. The isolates included the N. asteroides type strain (ATCC 19247), three clinical isolates and one reference isolate (ATCC 23824) of N. asteroides with the type I drug susceptibility pattern, four clinical isolates of N. asteroides with the type II drug susceptibility pattern, and four clinical isolates and one reference strain (ATCC 14759) of N. asteroides with the type VI drug susceptibility pattern.
Fourteen isolates of the N. transvalensis complex (type IV drug susceptibility pattern) were studied. These included 11 clinical isolates and 3 reference strains (ATCC 6865T, ATCC 49872, and ATCC 49873).
Additionally, eight clinical isolates and two reference strains (ATCC 33726T and ATCC 33727) of N. nova (type III drug susceptibility pattern) and two clinical and two reference strains (ATCC 3308 and ATCC 3318T) of N. farcinica (type V drug susceptibility pattern) were selected for sequence analysis.
Nineteen additional Nocardia isolates belonging to six species other than those recognized within the N. asteroides complex were also selected for sequencing. The Nocardia species sequenced included three clinical strains and one reference strain (ATCC 19296T) of N. brasiliensis, two clinical strains and one reference strain (ATCC 51512T) of N. pseudobrasiliensis, nine clinical strains and one reference strain (ATCC 14629T) of N. otitidiscaviarum, one reference isolate of N. paucivorans (ATCC BAA-278T), and one reference strain of N. cyriacigeorgica (PSM 44484T).
Thirty-eight other aerobic actinomycetes were sequenced. These included four clinical isolates of Rhodococcus equi and one reference strain (ATCC 6939), five clinical isolates and three reference strains of Streptomyces spp. (Streptomyces albus ATCC 3004T, S. somaliensis ATCC 33201T, and S. griseus ATCC 10137T), and two reference strains of Actinomadura madurae (ATCC 19425T and ATCC 13724). Isolates of these genera were identified to the species level if they had typical colony morphologies and growth characteristics and if their PRA patterns matched those of the ATCC type strains for those species. Among the Gordonia spp., 11 clinical isolates and 4 reference strains (Gordonia bronchialis ATCC 25592T, G. sputi ATCC 29627T, G. sputi ATCC 33610, and G. aichiensis ATCC 33611T) were tested. For the genus Gordonia, the PRA patterns and carbohydrate utilization from mannitol, inositol, sorbitol, and citrate were used for species identification. If the results of both methods matched, an organism was given a species name. If they failed to match, the isolates were designated only to the genus level. Both G. aichiensis and G. sputi had the same PRA pattern, so they were separated only by carbohydrate utilization (unpublished data).
Five clinical strains of Tsukamurella spp., one reference strain of Tsukamurella paurometabola (ATCC 8368T), and two reference strains of T. inchonensis (ATCC 700082T, ATCC 25938) were studied. Isolates of Tsukamurella spp. were evaluated for carbohydrate utilization from mannitol, inositol, sorbitol, trehalose, galactose, rhamnose, xylose, and arabinose. They were also tested for acetamide utilization and growth at 45°C on tryptic soy agar. The PRA patterns of the Tsukamurella strains were compared to those of the five ATCC type strains. By use of these two techniques, all five species gave unique patterns by one or both of these tests.
PRA.
PRA of a 439-bp fragment of the 65 hsp gene (the Telenti fragment) was performed as described previously (26). The amplicon was digested with BstEII, HaeIII, MspI, and BsaHI; and the fragments were separated on 3% MetaPhor agarose gels (FMC Bioproducts, Rockland, Maine). Additional enzymes were used for selected taxa, including the N. transvalensis complex (34). The sizes of the fragments were estimated on a computerized Bio Image system (Millipore, Bedford, Mass.). The patterns were compared to those published previously (22, 23, 35) as well as unpublished in-house data derived from the ATCC type strain and clinical isolates.
Susceptibility testing.
Susceptibility testing was performed by the broth microdilution method in cation-adjusted Mueller-Hinton broth, as recommended by the recent NCCLS guidelines for Nocardia spp. and other aerobic actinomycetes (16). Quality control was performed by susceptibility testing with amoxicillin-clavulanic acid and Staphylococcus aureus ATCC 29213, Mycobacterium peregrinum ATCC 700686, and Enterococcus faecalis ATCC 29212.
Biochemical analysis.
Selected taxa were studied for growth and pigmentation, carbohydrate utilization, acetamide utilization, and growth at 45°C at 3 days on tryptic soy agar, as described previously (21, 28, 30, 34).
16S rDNA extraction, amplification, and sequencing.
DNA extracts of the bacteria were prepared by a mechanical disruption method, as described previously (18). The 5′ end of the 16S rRNA gene was amplified and sequenced by using reagents from the MicroSeq 500 16S rDNA Bacterial Sequencing kit (Applied Biosystems), as described previously (18, 25).
DNA sequence data analysis.
Sequence data were analyzed with MicroSeq software (version 1.40), as described previously (18). The unknown sequences were initially compared to all of the sequences in the MicroSeq database (version 0023b) by using the MicroSeq Basic Local Alignment Search Tool (BLAST) (1). At least 10 database sequences with the fewest sequence differences from the query sequence were compared to the query sequence for a second time, but with the Full Alignment Tool (33).
The sequences of the unknown isolates were also compared to sequences in the GenBank database by using the BLAST program available on the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov). The GenBank entry with the highest score from the search with the BLAST program was downloaded and trimmed so that the 3′ end did not extend beyond that of the region sequenced with the MicroSeq kit. The GenBank entry was compared to the unknown sequence by using the Full Alignment Tool of the MicroSeq software.
RESULTS
N. asteroides complex.
On the basis of the findings from a 1988 study (29), the N. asteroides complex was subclassified into six different drug susceptibility types. This phenotypic subclassification proved to have taxonomic significance. Subsequent studies resulted in the reclassification of isolates with drug susceptibility pattern III as N. nova and isolates with drug susceptibility pattern V as N. farcinica, and isolates with drug susceptibility pattern IV were reclassified as a subgroup of the N. transvalensis complex (28).
Thirteen of the sequenced isolates belong to the N. asteroides complex and were subclassified by drug susceptibility into three major unnamed groups (group types I, II, and VI). In general, the sequencing data for these three types indicate that isolates belonging to these three groups have 500-bp 16S rRNA gene sequences that are identical or very closely related. All four isolates classified as having the N. asteroides complex type I drug susceptibility pattern had similar biochemical patterns, including positivity for citrate utilization; similar drug susceptibility patterns; and identical PRA patterns (23). These four isolates also had identical 500-bp 16S rRNA sequences. The sequences did not clearly match any single sequence in the MicroSeq database, but in GenBank there was an exact match with the sequence from the newly described species N. abscessus (Table 2, sequence A1) (20). In fact, reference strain ATCC 23824, which is a drug susceptibility type I isolate by biochemical analysis, susceptibility studies, PRA, and 500-bp 16S rRNA gene sequence analysis, was one of the isolates characterized to define this new species. Thus, it appears that all of these isolates belong to the same species.
TABLE 2.
Database search results for Nocardia spp.
| Isolatea | No.b | Micro Seq database searchc
|
GenBank searchd
|
Sequencee | GenBank accession no.f | ||
|---|---|---|---|---|---|---|---|
| % Sequence difference (no. of bases) | Database entry | % Sequence difference | Database entryg (no. of bp)h | ||||
| N. asteroides ATCC 19247T | 1 | 0.00 | N. asteroides | 0.00 | N. asteroides Z36934 (477) | A28 | AY505492 |
| N. asteroides complex-type I drug pattern | 4 | 1.80 (4) | N. asteroides and N. brevicatena | 0.00 | N. abscessus AF430018 (477) | A1 | AY262328 |
| N. asteroides complex-type II drug pattern | 4 | 1.20 (1) | N. carnea | 0.00 | N. asteroides Z82231 (476) | A2 | AY262325 |
| 1.40 (2) | N. carnea | 0.00 | Nocardia sp. AF430022 (477) | A3 | AY262327 | ||
| 1.60 (1) | N. nova | 0.00 | N. paucivorans AF179865 (469) | A4 | AY262324 | ||
| N. nova (includes ATCC 33726T) | 10 | 0.00 (5)i | N. nova | 0.00 | N. nova AF4300028 (477) | A9 | AY262320 |
| 0.40 (1) | N. nova | 0.00 | N. nova AF430030 (477) | A10 | AY262319 | ||
| 0.60 (1) | N. nova | 0.21 | N. nova AF430030 (477) | A29 | AY505493 | ||
| 1.00 (1) | N. nova | 0.21 | N. veterana AF490540 (477) | A11 | AY262318 | ||
| 1.00 (2) | N. nova | 0.00 | N. veterana AF490540 (477) | A12 | AY262317 | ||
| N. farcinica (includes ATCC 3318T) | 4 | 0.00 (4)i | N. farcinica | 0.00 | N. otitidiscaviarum X80611 (487) | A8 | AY262321 |
| N. asteroides complex-type VI drug pattern | 5 | 1.60 (5) | N. nova and N. farcinica | 0.00 | N. cyriacigeorgica AF282889 (476) | A5 | AY262326 |
| N. cyriacigeorgica PSM 4484T | 1 | 1.60i | N. nova and N. farcinica | 0.00 | N. cyriacigeorgica AY244782 (488) | A5 | AY262326 |
| N. brasiliensis (includes ATCC 19296T) | 4 | 0.00 (3)i | N. brasiliensis | 0.00 | N. brasiliensis Z36935 (477) | A6 | AY262323 |
| 0.80 (1) | N. brasiliensis | 0.84 | N. brasiliensis Z36935 (477) | A7 | AY262322 | ||
| N. otitidiscaviarum (includes ATCC 14629T) | 10 | 0.00 (4)i | N. otitidiscaviarum | 0.85 | N. otitidiscaviarum M59056 (500) | A13 | AY262316 |
| 1.60 (1) | N. pseudobrasiliensis | 2.00 | Nocardia sp. AF421565 (499) | A14 | AY262315 | ||
| 1.80 (1) | N. pseudobrasiliensis | 0.58 | N. otitidiscaviarum Z82237 (476) | A15 | AY262314 | ||
| 2.20 (1) | N. pseudobrasiliensis | 0.16 | N. otitidiscaviarum Z82237 (476) | A16 | AY262313 | ||
| 2.30 (1) | N. pseudobrasiliensis | 0.26 | N. otitidiscaviarum Z82237 (476) | A17 | AY262312 | ||
| 2.30 (1) | N. pseudobrasiliensis and N. nova | 1.80 | N. crassostreae U92800 (500) | A18 | AY262311 | ||
| 2.30 (1) | N. nova | 0.42 | N. otitidiscaviarum Z82239 (476) | A19 | AY262310 | ||
| N. paucivorans ATCC BAA-278T | 1 | 1.60 (1)i | N. nova | 0.00 | N. paucivorans AF179865 (469) | A4 | AY262324 |
| N. pseudobrasiliensis (includes ATCC 51512T) | 3 | 0.00 (2) | N. pseudobrasiliensis | 0.00 | N. pseudobrasiliensis AF4300042 (477) | A20 | AY262309 |
| 1.20 (1) | N. pseudobrasiliensis | 0.00 | N. pseudobrasiliensis AB086862 (473) | A21 | AY262308 | ||
| N. transvalensis complex sensu stricto ATCC 6865T | 14 | 0.00 (1)i | N. transvalensis | 0.78 | N. transvalensis X80609 (482) | A22 | AY262307 |
| N. asteroides type IV drug pattern | 0.80 (6) | N. transvalensis | 0.21 | N. transvalensis AB084447 (486) | A23 | AY262306 | |
| New taxon 2 | 0.80 (1) | N. transvalensis | 0.21 | N. transvalensis AB084447 (486) | A24 | AY262305 | |
| New taxon 2 | 1.00 (3) | N. transvalensis | 0.00 | N. transvalensis AB084447 (487) | A25 | AY262304 | |
| New taxon 1 | 1.60 (1) | N. transvalensis | 0.42 | N. transvalensis Z82236 (476) | A26 | AY262303 | |
| New taxon 1 | 3.00 (2) | N. transvalensis | 0.00 | N. transvalensis Z82236 (476) | A27 | AY262302 | |
Identity by phenotyping, PRA, and drug susceptibility testing.
Number of isolates sequenced.
The entire MicroSeq database was searched by using the Full Tool of MicroSeq software (see Materials and Methods).
GenBank was searched by using the BLAST program available on the NI website of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) (see Materials and Methods).
A letter designation is given to each unique Nocardia sequence in this study.
GenBank accession number of each unique Nocardia sequence in this study.
The entry designates the name of the bacterium as it appears in the GenBank database and the GenBank accession number.
The number of base pairs (bp) indicates the number of bases from the GenBank entry, which corresponds to the region sequenced with the MicroSeq kit (see Materials and Methods).
Includes the ATCC type strain.
There was greater sequence diversity among the N. asteroides complex type II isolates. The four type II isolates had similar drug susceptibility patterns and biochemical profiles and exhibited two PRA patterns. Three of the four isolates that had the same PRA pattern also had very similar sequences, which were most closely related to that of the N. carnea type strain in the MicroSeq database. Sequence A2 was identical to a GenBank sequence identified as N. asteroides, and sequence A3 was identical to a GenBank sequence identified as a Nocardia sp. The isolate with the fourth type II drug susceptibility pattern (sequence A4), which has a PRA pattern different from those of the other three isolates in the group (i.e., the lack of a HinfI site) (23), also had a sequence that differed from those of the other isolates. This sequence most closely matched that of N. nova in the MicroSeq database but was identical to the sequence of a N. paucivorans isolate in the GenBank database as well as the sequence of the N. paucivorans type strain. The N. paucivorans type strain was studied, and it had the same drug susceptibility pattern and biochemical profile as the type II isolates. It also had the same PRA pattern as the fourth type II isolate (sequence A4).
The five N. asteroides complex, drug susceptibility pattern type VI isolates had similar susceptibilities, the same PRA patterns, and similar biochemical profiles except for variable results for citrate, rhamnose, and acetamide utilization. All isolates grew well on tryptic soy agar in 3 days at 45°C. All five of the N. asteroides complex, drug susceptibility pattern VI isolates had the same 500-bp 16S rRNA gene sequence. This sequence (Table 2, sequence A5) did not clearly match any single entry in the MicroSeq database (>1.5% discordance), but their sequences were an exact match to the sequence of a recently described species, N. cyriacigeorgica (37), which was in the GenBank database but not the MicroSeq database. The type strain of N. cyriacigeorgica was obtained; and its drug susceptibility pattern, biochemical profile, and PRA pattern are the same as those for these five type VI isolates.
N. farcinica.
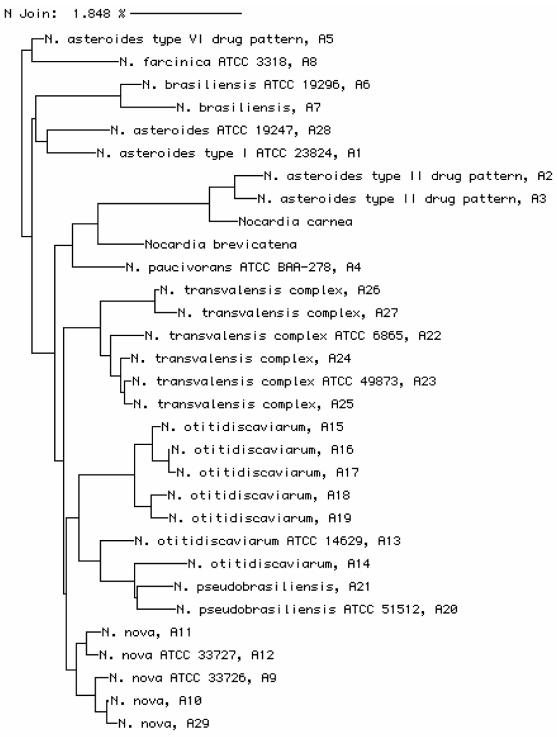
The two clinical isolates of N. farcinica and the two reference strains were the same by phenotypic analysis. They were positive for rhamnose and acetamide utilization, grew in 3 days at 45°C, and were resistant to tobramycin (30). The isolates also had identical results by 16S rRNA gene sequencing (Table 2, sequence A8). The GenBank database contains an N. farcinica sequence that exactly matches the sequences of the clinical and reference N. farcinica isolates (0.00% difference for a 477-base sequence of the N. farcinica strain with GenBank accession no. Z36936), yet a GenBank sequence identified as N. otitidiscaviarum received the highest BLAST score. In fact, the N. otitidiscaviarum sequence in GenBank (GenBank accession no. X80611) was also an exact match to the sequences of the clinical N. farcinica isolates. However, according to a phylogenetic analysis based on the 500-bp 16S rRNA gene sequences (Fig. 1), these two species are not closely related. This suggests that the sequence with GenBank accession no. X80611 is misidentified.
FIG. 1.
Phylogenetic tree of all unique Nocardia sequences generated in this study. The tree was created by the neighbor-joining method (24). Sequences without letter designations are sequences of type strains from the MicroSeq library.
N. nova.
The nine N. nova isolates gave typical biochemical profiles, including a positive 14-day arylsulfatase utilization test; typical drug susceptibility patterns; and typical mycolic acids by HPLC analysis. According to their PRA patterns, the isolates were identical with the four enzymes tested (BstEII, MspI, HinfI, and BsaHI), as previously described for the species (28). By fatty acid analysis, however, ATCC 33726T and ATCC 33727 are known to belong to different groups, designated groups A and B (15). DNA sequencing of the N. nova isolates produced five unique sequences. The sequences of four clinical isolates were an exact match for the N. nova type strain ATCC 33726 sequence (Table 2, sequence A9). The sequences of one clinical isolate and strain ATCC 33727 differed by five bases from this sequence (sequence A12). The sequences of three other clinical isolates differed from the sequence of the type strain by two, three, and five bases, respectively (sequences A10, A29, and A11, respectively). Thus, the 10 N. nova isolates which appeared to differ phenotypically only in their fatty acid profiles (15) differed from type strain ATCC 33726 by 0 to 5 bases by partial 16S rRNA gene sequencing. N. nova sequences A9, A10, and A29 most closely matched the N. nova sequences in the GenBank database. However, sequences A11 and A12 differed by 1 base or were identical to a recently described species, N. veterana (8).
N. transvalensis complex.
The N. transvalensis complex isolates have been divided into four subgroups by biochemical testing, HPLC, susceptibility testing, and PRA (34). N. transvalensis sensu stricto is the group that includes type strain ATCC 6865. Isolates in this group are positive for hypoxanthine, citrate, trehalose, mannitol, galactose, erythritol, adonitol, inositol, and sorbitol utilization. The second group is referred to as N. transvalensis new taxon 1 and differs from the sensu stricto species in being negative for inositol utilization and having a different PRA pattern. The third group is referred to as N. transvalensis new taxon 2 and differs from the sensu stricto group by being negative for inositol, mannitol, and sorbitol utilization and having a unique PRA pattern. The fourth group represents the former N. asteroides complex drug susceptibility type IV and differs from the sensu stricto group by being negative for inositol, mannitol, sorbitol, adonitol, and erythritol utilization and having a unique PRA pattern. It includes strains ATCC 49872 and ATCC 49873. Sequencing of the 16S rRNA gene confirmed the phenotypic and PRA heterogeneity of N. transvalensis and validated its present description as a “complex.” As in prior studies (20), by sequencing, members of each of the four subgroups were more closely related to each other than to members of the other three subgroups. Among the isolates in new taxon 1, two isolates had the same sequence and one isolate differed by 2 bases. They differed by 10 and 8 bases, respectively, from ATCC 6865T (sensu stricto). Among the isolates in new taxon 2, three of the four isolates had the same sequence and differed by 5 bases from ATCC 6865T. The six isolates of N. asteroides drug susceptibility group IV all had the same 500-base rRNA gene sequences and differed from ATCC 6865T by 4 bases. They differed from three of the four new taxon 2 isolates, however, by only a single base. Thus, the four phenotypic groups presently clustered within the N. transvalensis complex differed by 4 to 10 bases from N. transvalensis ATCC 6865T by 500-bp 16S rRNA gene sequencing.
N. otitidiscaviarum.
Ten N. otitidiscaviarum isolates were studied. Their drug susceptibility patterns and biochemical profiles were similar, but by PRA with the three routine restriction enzymes (BstEII, MspI, and HinfI) (23), three patterns were seen by restriction fragment polymorphism (RFLP) analysis. One pattern was actually a failure to generate an amplicon with the standard primers for two isolates. This RFLP pattern was previously observed with 36% of the N. otitidiscaviarum isolates (23). Four patterns were seen by use of a fourth enzyme (BsaHI). As noted previously (22, 23), these patterns are unique for this species. Sequencing identified three groups. Three clinical isolates had the same sequence as the type strain, ATCC 14629 (Table 2, sequence A13). These sequences were most closely related to the N. otitidiscaviarum sequences in the MicroSeq and GenBank databases. The remaining six isolates each had a unique sequence. Five of these sequences (sequences A15 to A19) are very similar to each other (0.5 to 6 base differences among the group) but differ from the sequence of ATCC 14629T by 12.5 to 16.5 bases. These five sequences cluster together in a dendrogram generated by 500-bp 16S rRNA gene sequencing (Fig. 1). All of these sequences were most closely related to the N. pseudobrasiliensis sequence in the MicroSeq database; however, all but one sequence matched an N. otitidiscaviarum sequence in the GenBank database. The remaining sequence, sequence A14, differed from the ATCC 14629T sequence by 9 bases and differed from the cluster of five sequences by 14.5 to 16 bases. Sequence A14 actually clustered with the N. pseudobrasiliensis sequences on the dendrogram (Fig. 1). Neither database contained an N. otitidiscaviarum sequence that most closely matched sequence A14.
We observed several sites of heterogeneity among the sequences of the N. otitidiscaviarum isolates. In other words, single isolates contained multiple 16S rRNA gene copies that differed by 1 or more bases. These sites of heterogeneity were detected in the sequence electropherogram data as overlapping peaks, and these differences are calculated as a fraction of a base difference (e.g., a sequence with a base designated Y [T or C] for one isolate is only 0.5 base different from the sequence of an isolate that consistently has a T at the same position but that is otherwise identical). In addition to these sites of heterogeneity, we found a large degree of sequence diversity among these isolates, the degree of which suggests that some of these isolates may represent new species.
N. brasiliensis and N. pseudobrasiliensis.
The sequences of all isolates identified by phenotypic studies and PRA as N. brasiliensis and N. pseudobrasiliensis most closely matched that of the type strain of the same species in the MicroSeq database. However, in some cases there was significant sequence diversity between the sequences of the N. brasiliensis and N. pseudobrasiliensis isolates and the sequence of the type strain (i.e., differences of 0.80 and 1.20%, respectively). A search of the GenBank database produced similar results for the N. brasiliensis, N. pseudobrasilienis, and N. transvalensis complex isolates.
Identification of Actinomadura, Gordonia, Rhodococcus, Streptomyces, and Tsukamurella isolates.
Three of the 26 species that constitute the genus Actinomadura are considered clinically relevant: A. madurae, A. pelletieri, and A. latina. The MicroSeq database contains four sequences of Actinomadura species; however, none of them are clinically significant species. Two isolates with a phenotypic identification of A. madurae were sequenced in this study. The closest MicroSeq database entries for these two sequences were the same genus but different species, A. brunnae and A. yumaensis (Table 3). A search of the GenBank database yielded sequences that were more similar to the sequences of the test isolates than did a search of the MicroSeq database. However, for one isolate, the sequence match in the GenBank database was of a different identified as an unclassified member of the Pseudonocardiaceae family. The GenBank database A. madurae sequence entry, and this entry generated a high ranking score for both queries with the BLAST program, but this entry did not have the highest ranking score because of poor sequence quality at the 5′ end of the 16S rRNA gene.
TABLE 3.
Database search results for Actinomadura spp., Gordona spp., Rhodococcus spp., Streptomyces spp., and Tsukamurella spp.
| Isolatea | No.b | MicroSeq database searchc
|
GenBank searchd
|
Sequencee | GenBank accession no.f | ||
|---|---|---|---|---|---|---|---|
| % Sequence difference (no. of bases) | Database entry | % Sequence difference | Database entryg (no. of bph) | ||||
| A. madurae (includes ATCC 19425T) | 2 | 2.59 (1) | A. brunnea | 1.25 | Pseudonocardiaceae strain AF223350 (480) | B1 | AY262333 |
| 3.39 (1) | A. yumaensis | 1.48 | A. livida AF163116 (472) | B2 | AY262334 | ||
| G. aichiensis (includes ATCC 33611T) | 2 | 0.00 (2)i | G. aichiensis | 0.60 | Gordona sp. AF150493 (501) | C1 | AY262332 |
| G. bronchialis (includes ATCC 25592T) | 5 | 0.00 (5)i | G. bronchialis | 0.80 | G. desulfuricans AF101417 (500) | C2 | AY262331 |
| G. sputi (includes ATCC 29627T) | 5 | 0.00 (1)i | G. sputi | 0.80 | Gordona sp. AF150493 (501) | C3 | AY262330 |
| 0.10 (3) | G. sputi | C4 | AY262329 | ||||
| 0.00 (1) | G. aichiensis | C1 | AY262332 | ||||
| Gordona spp. | 3 | 0.00 (2) | G. bronchialis | C2 | AY262331 | ||
| 0.00 (1) | G. sputi | C3 | AY262330 | ||||
| R. equi | 5 | 0.00 (2) | R. equi, C. hoagii | 0.00 | R. equi X80614 (490) | D1 | AY262301 |
| 0.20 (3) | R. equi, C. hoagii | D2 | AY262300 | ||||
| S. albus ATCC 3004T | 1 | 0.10i | S. albus albus | E1 | AY262299 | ||
| S. somaliensis ATCC 33201T | 1 | 0.00i | S. somaliensis | 0.00 | S. somaliensis AJ007403 (500) | E2 | AY262297 |
| S. griseus | 1 | 0.40 | S. anulatus | E3 | AY262298 | ||
| Streptomyces spp. | 5 | 0.10 (1) | S. albus albus | E4 | AY262296 | ||
| 0.30 (1) | S. albus albus | 1.90 | S. albus X53163 (473) | E5 | AY262295 | ||
| 0.50 (1) | S. albus albus | E6 | AY262294 | ||||
| 0.80 (1) | S. anulatus | 0.95 | Streptomyces sp. AF112177 (500) | E7 | AY262293 | ||
| 1.10 (1) | S. violaceoruber | 0.00 | Streptomyces sp. AF223892 (501) | E8 | AY262292 | ||
| T. paurometabola ATCC 8368T | 1 | 0.00 (1)i | T. paurometabola | 0.20 | T. paurometabola AF283280 (499) | F1 | AY262291 |
| T. pulmonis | 1 | 0.00 (1) | T. pulmonis | 0.60 | T. inchonensis AF283281 T. strandjordae AF283283 (499) | F2 | AY262290 |
| T. tyrosinosolovens | 4 | 0.20 (1) | T. tyrosinosolovens | 0.60 | T. inchonensis AF283281, T. strandjordae AF283283 (499) | F3 | AY262289 |
| 0.40 (1) | T. tyrosinosolovens | F4 | AY262288 | ||||
| 0.40 (1) | T. inchonensis or T. paurometabola | 0.20 | T. inchonensis AF283281, T. strandjordae AF283283 (499) | F5 | AY262287 | ||
| 1.00 (1) | T. inchonensis or T. paurmetabola or T. pulmonis | 0.60 | T. inchonensis AF283281, T. strandjordae AF283283 (499) | F6 | AY262286 | ||
| T. inchonensis (includes ATCC 700082T) | 2 | 0.00 (2)i | T. inchonensis | 0.00 | T. inchonensis AF283281, T. strandjordae AF283283 (499) | F7 | AY262285 |
Identity determined by phenotyping and PRA.
Number of isolates sequenced.
The entire MicroSeq database was searched by using the Full Tool of MicroSeq software (see Materials and Methods).
GenBank was searched by using the BLAST program available on the website of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov) (see Materials and Methods).
A letter designation is given to each unique Nocardia sequence in this study.
GenBank accession number of each unique Nocardia sequence in this study.
The entry designates the name of the bacterium as it appears in the GenBank database and the GenBank accession number.
The number of base pairs (bp) indicates the number of bases from the GenBank entry, which corresponds to the region sequenced with the MicroSeq kit (see Materials and Methods).
Includes the ATCC type strain.
All isolates of Gordonia spp., Rodococcus spp., Streptomyces spp., and Tsukamurella spp. were correctly identified to the genus level when the sequences in either the MicroSeq or the GenBank database were searched. In general, the MicroSeq database provided more species information for these isolates than did the GenBank database (Table 3).
Among the isolates in the genus Gordonia, PRA patterns and carbohydrate utilization identified five G. bronchialis isolates, five G. sputi isolates, two G. aichiensis isolates, and three Gordonia species isolates. Sequencing confirmed the species identifications for all five G. bronchialis isolates, four of five of the G. sputi isolates, and both G. aichiensis isolates. Among the three isolates identified phenotypically as Gordonia species, two were G. bronchialis isolates and one was a G. sputi isolate by sequencing.
One discrepancy between phenotypic identification and sequence identification was noted for the isolates in the genus Gordonia. An isolate identified as G. sputi by carbohydrate utilization studies (positive for mannitol and sorbitol utilization, negative for inositol utilization) had a sequence that was identical to that of G. aichiensis. The 500-bp rRNA gene sequence difference between the G. sputi type strain and the G. aichiensis type strain was 2.5 bases (the 0.5-base difference exists because G. sputi has a G-T polymorphism at a location where the G. aichiensis sequence contains a G). The sequence similarity of the entire 16S rRNA genes of these two species is 99.7%. This is in contrast to the results of DNA-DNA relatedness studies, which indicate only 40% relatedness (14). These data suggest that 16S rRNA gene sequencing may not be a definitive method for distinguishing between G. sputi and G. aichiensis. However, sequencing does aid in the identification of isolates with indeterminate phenotypic or PRA results.
All five R. equi isolates were accurately identified by use of the sequences in both databases. The MicroSeq database also contains the sequence of the Corynebacterium hoagii type strain, and this sequence is identical to that of the R. equi type strain, which is also in the MicroSeq database. C. hoagii and R. equi belong to the same species and the name C. hoagii is no longer used. Therefore, the C. hoagii sequence should be removed from the MicroSeq database.
Sequence-based identification correlated with HPLC-based identification for isolates of the genus Streptomyces. The organisms in the genus Streptomyces were identified to the species level if their PRA patterns matched those of one of the ATCC reference strains. Strains that failed to match were identified only as Streptomyces species. By these methods, the three reference strains each had unique PRA patterns, two clinical isolates had the pattern of S. albus, and three clinical isolates were unique.
Until recently, T. paurametabolum was the only species in the genus Tsukamurella. The genus now consists of five species, and the sequences of all five type strains are in the MicroSeq database (36). We sequenced eight isolates of Tsukamurella identified as T. paurometabola (one isolate), T. pulmonis (one isolate), T. tyrosinosolovens (four isolates), or T. inchonensis (two isolates) by a combination of phenotypic methods and PRA. The sequence-based identifications obtained by using the MicroSeq database were consistent with the phenotyping- and PRA-based identifications for 6 of the isolates. The only perfect matches were for one clinical strain identified as T. pulmonis, the T. inchonensis type strain (ATCC 700082), and ATCC 25938, which was previously named T. paurametabolum but which was recently reported to be a misnamed T. inchonensis isolate (12). The sequences of two T. tyrosinosolovens isolates, isolates F-5 and F-6 (Table 3), were equally different from more than one database sequence; and in both cases the T. tyrosinosolovens database entry was not among the closest-matching sequences. These discrepancies could be explained by previous studies, which demonstrated that the entire 16S rRNA gene sequences of Tsukamurella spp. have 99.4% or greater similarity. Despite this similarity in the 16S rRNA sequence, there are significant differences in DNA-DNA relatedness by hybridization studies, and these species are reported to be distinguishable by phenotypic tests (36). Therefore, 500-bp 16S rRNA gene sequence identification is useful for distinguishing Tsukamurella spp. from isolates of other genera, but this identification method has a limited ability to provide a species-level identification.
DISCUSSION
In this study, we evaluated partial 16S rRNA gene sequences and compared them to the profiles obtained by biochemical analysis, HPLC, fatty acid analysis, drug susceptibility testing, and PRA of the 65 hsp Telenti sequence for identification of isolates of the aerobic actinomycetes. In general, we found that sequence differences correspond to the differences obtained by traditional identification methods. For example, isolates with the N. asteroides complex type I drug susceptibility pattern had the same biochemical pattern and unique PRA patterns, and all had the same 16S rRNA gene sequence. This sequence differs from the sequences of all other Nocardia species by at least 9 bases (1.8%). Figure 1 shows a phylogenetic tree based on the 500-bp 16S rRNA gene sequences of Nocardia sp. isolates. This tree demonstrates that this sequence provides sufficient information to distinguish between species or complexes. This finding is consistent with those in a recent report by Roth et al. (20), who compared the full 16S rRNA gene sequences (1,500 bp) of 74 Nocardia isolates, and a report by Cloud et al. (5), who compared the 500-bp 16S rRNA gene sequences of 28 reference Nocardia strains.
For the most part, partial 16S rRNA gene sequencing provided sufficient information to identify R. equi isolates, to provide genus-level identifications of Actinomadura spp., and to distinguish among species of Gordonia and Streptomyces. However, the sequencing of the 500-bp fragment demonstrated a limited ability to distinguish among species that share significant 16S rRNA gene sequence similarities yet that differ from each other by overall DNA similarity and phenotypic attributes. Most notably, this was the case for Tsukamurella spp. Since Tsukamurella spp. are uncommon clinical isolates and little is known regarding the differences in clinical significance or antibiotic susceptibilities among the various species, genus-level identification is usually sufficient for the clinical management of patients.
The primary obstacle to obtaining an accurate sequence-based identification for this group of bacteria is the lack of an adequate database. Both the MicroSeq database and the GenBank database demonstrated limitations. The MicroSeq database is highly accurate because it only contains the sequences of reference isolates. However, not all species examined in this study were represented in the database; and the sequences of many species in Nocardia taxon, which are unnamed at present, are also not represented in the database. In addition, this database contains only one entry per species, which does not take into account the occurrence of intraspecies variability. The GenBank database contains significantly more sequences, but there were also problems generating an identification with the sequences in GenBank. These problems were threefold. First, the database does not contain sequences for many of the species evaluated in this study. Second, the species assignments for the sequences in the database may be incorrect, such as the sequence with GenBank accession no. X80611, which is reported to belong to N. otitidiscaviarum but which is identical to the sequence of N. farcinica. Finally, some of the sequences are of poor quality or of insufficient length (e.g., the 5′ end of the sequence is missing) and as a result fail to generate sufficient similarity scores during the comparison. Similar problems with the use of these databases were noted when the sequences in the databases were applied to the identification of Mycobacterium spp. (27). Use of the RIDOM database is promising for the identification of bacteria by 16S rRNA gene sequencing. This database is available to the public, and it contains sequences from well-defined isolates of medically relevant bacteria. At present, the database contains the sequences of a limited number of genera, but Nocardia sequences were recently added (2). In addition to using these databases, clinical laboratories may wish to build their own databases with sequences that are generated in-house from reference or well-characterized isolates. All of the sequences generated from this study were deposited in GenBank (Tables 2 and 3).
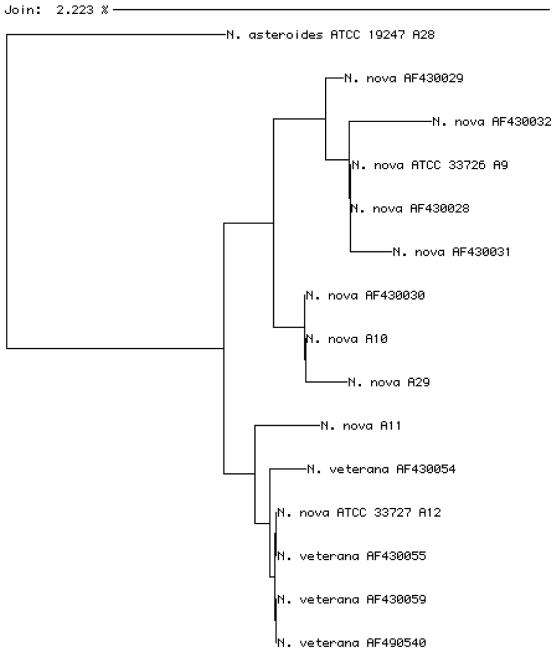
A major observation of this study is the heterogeneity of the 16S rRNA gene sequences within individual Nocardia species, especially N. nova, N. transvalensis, and N. otitidiscaviarum. McNabb et al. (15) recently reported on the results of a large study in which they characterized the fatty acid profiles of 39 type strains and 529 clinical or reference strains of pathogenic aerobic actinomycetes using a commercial system (Microbial Identification System; Microbial Identification Inc., Newark, Del.). They identified four subgroups within N. nova, with the two major subgroups represented by ATCC 33726T and ATCC 33727, respectively. We identified five sequences within N. nova by partial 16S sequencing. These sequences clustered into two major groups (Fig. 1), and these groups are represented by the same two reference strains. These two groups differed from each other by 5 bases (1.0%) within the first 500 bases. A study by Roth et al. (20) comparing the 16S rRNA gene sequences of Nocardia species also demonstrated sequence heterogeneity among N. nova isolates. A recently described species, N. veterana, has significant sequence similarity to N. nova isolates (8). In addition, N. veterana and N. nova are indistinguishable by PRA, biochemical analysis, and drug susceptibility testing (19). A comparison of partial N. nova sequences from this study and the GenBank database, along with N. veterana sequences from the GenBank database, demonstrates two primary clusters (Fig. 2). As noted before, these sequences cluster around N. nova ATCC 33726T and N. nova ATCC 33727. It is interesting that three of the four N. veterana isolates have sequences identical to the N. nova ATCC 33727 sequence. Since the characterization of N. veterana did not include this N. nova reference isolate, additional studies are necessary to distinguish the relationship between these two species. Three other sequences were identified, and preliminary collaborative studies by complete 16S rRNA gene sequencing suggest that these sequences represent additional unnamed taxa (Patricia S. Conville, unpublished observations). We propose that the clinical identities of isolates within the N. nova-N. veterana group be reported as N. nova complex until the taxonomy of this group is better established.
FIG. 2.
Phlogenetic tree of N. nova and N. veterana sequences. The tree was created by the neighbor-joining method (24). Where appropriate, the ATCC number, GenBank accession number, or letter designation given to each unique Nocardia sequence from this study (Table 1) is provided.
McNabb et al. (15) also assessed the fatty acid profiles of 13 N. transvalensis clinical isolates and found that none matched that of ATCC 6865T and that, with one exception, were so dissimilar that they were unable to group them. Four major phenotypic groups were seen in the present study of 13 N. transvalensis isolates, which gave six different sequences, with no sequence matching that of ATCC 6865T. This molecular heterogeneity supports the continued reporting of isolates in this group as N. transvalensis complex (as recommended previously [34]) until the taxonomy of this group is better defined.
McNabb et al. (15) also analyzed 20 strains of N. otitidiscaviarum and found them to be heterogeneous. By using three reference strains, including ATCC 14629T, as representatives of three groups, only six isolates (30%) could be clustered in these groups. In the present study with 10 N. otitidiscaviarum strains, four patterns were identified by PRA, and the 10 isolates gave seven different sequences, with the sequences of only 3 of the 9 clinical isolates (33%) matching that of ATCC 14629T. Given the molecular heterogeneity of N. otitidiscaviarum, it seems certain that there are multiple other species represented within it. On this basis we propose that clinical N. otitidiscaviarum isolates be called members of the N. otitidiscaviarum complex (as recommended for N. nova, N. transvalensis, and N. asteroides) until the taxonomy of this group is more clearly defined.
In contrast to the results of fatty acid analysis, which demonstrated marked intraspecies variations for the three species mentioned above, the results for isolates of N. farcinica and the predominant group within N. asteroides complex, the group with the type VI drug susceptibility pattern (29) or group B of McNabb et al. (15), showed that the isolates are highly homogeneous. These two taxa also gave homogeneous sequencing results, with only a single 16S rRNA gene sequence detected for each group. Thus, the results of the study of McNabb et al. (15), based on fatty acid profiles, strongly support the heterogeneity of N. nova, N. otitidiscaviarum, and N. transvalensis identified by 16S rRNA gene sequencing in the present study, as well as the homogeneity of other groups, such as N. farcinica and the N. asteroides type VI group.
The greatest taxonomic issue raised by this study is which taxa should represent the real N. asteroides. Gordon and Mihm (7) selected ATCC 19247T as the representative N. asteroides strain and discarded the previously used type strain. Their choice was based strictly on biochemical testing, as no molecular techniques, HPLC, or drug susceptibility tests were yet available. Unfortunately, isolates within the N. asteroides complex are largely asaccharolytic and are positive for the utilization of few biochemicals, making it difficult to select among strains. It is the major reason why isolates of the N. nova complex, N. farcinica, N. abscessus, N. paucivorans, and isolates of the N. transvalensis complex were not recognized and were misidentified in the early years (prior to the 1990s) as N. asteroides. Unfortunately, what appeared to Gordon and Mihm (7) as a typical strain phenotypically is anything but typical by more modern methods of strain comparison. Recent evaluations by fatty acid analysis (15) and, now, 16S rRNA gene sequencing have shown that the strain chosen by Gordon and Mihm (ATCC 19247T) is not representative of N. asteroides but represents a rare subgroup almost never seen clinically. The present study clearly helps to define this. The group within the N. asteroides complex closest taxonomically to ATCC 19247T is the N. asteroides complex group with the type I drug susceptibility pattern, and the sequences of the two groups differ by 9 bp (1.8%) in the first 500 bp. Previous studies have shown that the largest group within the N. asteroides complex that has not been designated as another species (i.e., other than N. nova, N. farcinica, and members of the N. transvalensis complex) has the type VI drug susceptibility pattern. Members of this group represent about 60% of the clinical isolates within the N. asteroides complex in the United States (29) and are represented by reference strain ATCC 14759.
Recently, Yassin et al. (37) described a single Nocardia strain with a 16S rRNA gene sequence that differed from those of other reference strains. Those investigators named this strain N. cyriacigeorgica. Unfortunately, this strain was not compared to the other recognized but unnamed taxonomic groups within the N. asteroides complex. On the basis of the findings of the present study, it appears that N. cyriacigeorgica may be the same as the major unnamed group of isolates (i.e., type VI) within the N. asteroides complex. It is our opinion that Gordon and Mihm (7) and others intended for the type strain of Gordon and Mihm to represent the clinical isolates responsible for most cases of nocardiosis. In our opinion, Gordon and Mihm chose the wrong type strain. However, to accept other names for all of the subgroups within N. asteroides is to doom the long-standing name to obscurity, since it will no longer represent the group that it was intended to represent.
If further studies that include sequencing of the complete 16S rRNA gene and, it is hoped, DNA-DNA homology studies confirm that isolates with the type VI drug susceptibility pattern and N. cyriacigeorgica are the same taxon, two options seem to be available. One is to not accept N. cyriacigeorgica as the name for the type VI taxonomic group but to continue to call that group N. asteroides. A new type strain should be chosen, and we propose ATCC 14759 as a reasonable choice as a replacement for ATCC 19247T. A second option would be to retain the new name N. cyriacigeorgica as well as N. asteriodes (the latter would refer to those isolates represented by ATCC 19247). Since ATCC 19247T is indistinguishable from type VI N. cyriacigeorica isolates according to their drug susceptibility patterns, biochemical utilization patterns, and 65 hsp PRA patterns, one could continue to call members of these two taxa members of the N. asteroides complex when they are identified by nonsequencing methods in the clinical laboratory. Either option would retain isolates with the name N. asteroides as a major cause of clinical nocardiosis. However, if N. cyriacigeorgica is not the same as the type VI group, we propose that the type VI group be named N. asteroides, with selection of a new type strain. A review by taxonomic committees may be required to reach a decision on this issue.
Several taxonomic groups of Nocardia likely represent new species. As mentioned above, N. asteroides drug susceptibility pattern type I isolates probably belong to the newly described species N. abscessus. Other groups that may represent new species include N. asteroides drug susceptibility pattern type II isolates, three of the subgroups within the N. transvalensis complex, multiple strains of N. otitidiscaviarum, and, possibly, the present N. asteroides type strain, ATCC 19247. Complete 16S rRNA gene sequencing and DNA-DNA homology studies are ongoing to try to help resolve some of these issues.
A large number of new species of Nocardia not addressed in this study have recently been described primarily on the basis of differences in their 16S rRNA gene sequences. These include N. bejingensis (32), N. pseudovaccini (13), N. ignorata (38), N. uniformis (11), N. crassostreae (6), N. salmonicida (10), N. africana (9), and N. flavorosea (4). The present study identified one clinical strain of N. paucivorans. It had a unique PRA pattern among the N. asteroides complex isolates with the type II drug susceptibility pattern. This pattern and the 16S sequence matched those of the present reference strain of N. paucivorans, ATCC BAA-278. This is the only clinical isolate of this new species that we have encountered, suggesting that it is very rare in the United States. Examples of the other species within the clinical Nocardia isolates were not identified.
Sequence-based identification is becoming an increasingly important identification tool. Although the present cost of performing such tests limits its application in most clinical laboratories, it is a useful alternative for the identification of bacterial isolates that are slowly growing or for which specialized identification techniques are required. Our results indicate that sequencing of the 500-bp 16S rRNA gene provides enough information for the species-level identification of most aerobic actinomycetes. However, the accurate assignment of a species name with a sequence will depend upon the continued generation of sequences from well-characterized isolates and the deposition of these sequences in databases that are available to the public, as well as additional taxonomic studies to resolve the appropriate classification of presently unnamed species.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2004. GenBank: update. Database issue. Nucleic Acids Res. 32:D23-D26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler, W. R., J. O. Kilburn, and G. P. Kubica. 1987. High-performance liquid chromatography analysis of mycolic acids as an aid in laboratory identification of Rhodococcus and Nocardia species. J. Clin. Microbiol. 25:2126-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chun, J., C. N. Seong, K. S. Bae, K. J. Lee, S. O. Kang, M. Goodfellow, and Y. C. Hah. 1998. Nocardia flavorosea sp. nov. Int. J. Syst. Bacteriol. 48(Pt 3):901-905. [DOI] [PubMed] [Google Scholar]
- 5.Cloud, J. L., P. S. Conville, A. Croft, D. Harmsen, F. G. Witebsky, and K. C. Carroll. 2004. Evaluation of partial 16S ribosomal DNA sequencing for identification of Nocardia species by using the MicroSeq 500 system with an expanded database. J. Clin. Microbiol. 42:578-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman, C. S., B. L. Beaman, J. Chun, M. Goodfellow, A. Gee, and R. P. Hedrick. 1998. Nocardia crassostreae sp. nov., the causal agent of nocardiosis in Pacific oysters. Int. J. Syst. Bacteriol. 48(Pt 1):237-246. [DOI] [PubMed] [Google Scholar]
- 7.Gordon, R. E., and J. M. Mihm. 1959. A comparison of Nocardia asteroides and Nocardia brasiliensis. J. Gen. Microbiol. 20:129-135. [DOI] [PubMed] [Google Scholar]
- 8.Gurtler, V., R. Smith, B. C. Mayall, G. Potter-Reinemann, E. Stackebrandt, and R. M. Kroppenstedt. 2001. Nocardia veterana sp. nov., isolated from human bronchial lavage. Int. J. Syst. Evol. Microbiol. 51:933-936. [DOI] [PubMed] [Google Scholar]
- 9.Hamid, M. E., L. Maldonado, G. S. Sharaf Eldin, M. F. Mohamed, N. S. Saeed, and M. Goodfellow. 2001. Nocardia africana sp. nov., a new pathogen isolated from patients with pulmonary infections. J. Clin. Microbiol. 39:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isik, K., J. Chun, Y. C. Hah, and M. Goodfellow. 1999. Nocardia salmonicida nom. rev., a fish pathogen. Int. J. Syst. Bacteriol. 49(Pt 2):833-837. [DOI] [PubMed] [Google Scholar]
- 11.Isik, K., J. Chun, Y. C. Hah, and M. Goodfellow. 1999. Nocardia uniformis nom. rev. Int. J. Syst. Bacteriol. 49(Pt 3):1227-1230. [DOI] [PubMed] [Google Scholar]
- 12.Kattar, M. M., B. T. Cookson, L. D. Carlson, S. K. Stiglich, M. A. Schwartz, T. T. Nguyen, R. Daza, C. K. Wallis, S. L. Yarfitz, and M. B. Coyle. 2001. Tsukamurella strandjordae sp. nov., a proposed new species causing sepsis. J. Clin. Microbiol. 39:1467-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, K. K., A. Roth, S. Andrees, S. T. Lee, and R. M. Kroppenstedt. 2002. Nocardia pseudovaccinii sp. nov. Int. J. Syst. Evol. Microbiol. 52:1825-1829. [DOI] [PubMed] [Google Scholar]
- 14.Klatte, S., F. A. Rainey, and R. M. Kroppenstedt. 1994. Transfer of Rhodococcus aichiensis Tsukamura 1982 and Nocardia amarae Lechevalier and Lechevalier 1974 to the genus Gordona as Gordona aichiensis comb. nov. and Gordona amarae comb. nov. Int. J. Syst. Bacteriol. 44:769-773. [DOI] [PubMed] [Google Scholar]
- 15.McNabb, A., R. Shuttleworth, R. Behme, and W. D. Colby. 1997. Fatty acid characterization of rapidly growing pathogenic aerobic actinomycetes as a means of identification. J. Clin. Microbiol. 35:1361-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. 2003. Susceptibility testing of mycobateria, nocardiae, and other aerobic actinomycetes; M24-A. National Committee for Clinical Laboratory Standards, Wayne, Pa. [PubMed]
- 17.Patel, J. B. 2001. 16S rRNA gene sequencing for bacterial pathogen identification in the clinical laboratory. Mol. Diagn. 6:313-321. [DOI] [PubMed] [Google Scholar]
- 18.Patel, J. B., D. G. Leonard, X. Pan, J. M. Musser, R. E. Berman, and I. Nachamkin. 2000. Sequence-based identification of Mycobacterium species using the MicroSeq 500 16S rDNA bacterial identification system. J. Clin. Microbiol. 38:246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pottumarthy, S., A. P. Limaye, J. L. Prentice, Y. B. Houze, S. R. Swanzy, and B. T. Cookson. 2003. Nocardia veterana, a new emerging pathogen. J. Clin. Microbiol. 41:1705-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth, A., S. Andrees, R. M. Kroppenstedt, D. Harmsen, and H. Mauch. 2003. Phylogeny of the genus Nocardia based on reassessed 16S rRNA gene sequences reveals underspeciation and division of strains classified as Nocardia asteroides into three established species and two unnamed taxons. J. Clin. Microbiol. 41:851-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruimy, R., P. Riegel, A. Carlotti, P. Boiron, G. Bernardin, H. Monteil, R. J. Wallace, Jr., and R. Christen. 1996. Nocardia pseudobrasiliensis sp. nov., a new species of Nocardia which groups bacterial strains previously identified as Nocardia brasiliensis and associated with invasive diseases. Int. J. Syst. Bacteriol. 46:259-264. [DOI] [PubMed] [Google Scholar]
- 22.Steingrube, V. A., B. A. Brown, J. L. Gibson, R. W. Wilson, J. Brown, Z. Blacklock, K. Jost, S. Locke, R. F. Ulrich, and R. J. Wallace, Jr. 1995. DNA amplification and restriction endonuclease analysis for differentiation of 12 species and taxa of Nocardia, including recognition of four new taxa within the Nocardia asteroides complex. J. Clin. Microbiol. 33:3096-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steingrube, V. A., R. W. Wilson, B. A. Brown, K. C. Jost, Jr., Z. Blacklock, J. L. Gibson, and R. J. Wallace, Jr. 1997. Rapid identification of clinically significant species and taxa of aerobic actinomycetes, including Actinomadura, Gordona, Nocardia, Rhodococcus, Streptomyces, and Tsukamurella isolates, by DNA amplification and restriction endonuclease analysis. J. Clin. Microbiol. 35:817-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Studier, J. A., and K. J. Keppler. 1988. A note on the neighbor-joining algorithm of Saitou and Nei. Mol. Biol. Evol. 5:729-731. [DOI] [PubMed] [Google Scholar]
- 25.Tang, Y. W., N. M. Ellis, M. K. Hopkins, D. H. Smith, D. E. Dodge, and D. H. Persing. 1998. Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli. J. Clin. Microbiol. 36:3674-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Bottger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turenne, C. Y., L. Tschetter, J. Wolfe, and A. Kabani. 2001. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J. Clin. Microbiol. 39:3637-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallace, R. J., Jr., B. A. Brown, M. Tsukamura, J. M. Brown, and G. O. Onyi. 1991. Clinical and laboratory features of Nocardia nova. J. Clin. Microbiol. 29:2407-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace, R. J., Jr., L. C. Steele, G. Sumter, and J. M. Smith. 1988. Antimicrobial susceptibility patterns of Nocardia asteroides. Antimicrob. Agents Chemother. 32:1776-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallace, R. J., Jr., M. Tsukamura, B. A. Brown, J. Brown, V. A. Steingrube, Y. S. Zhang, and D. R. Nash. 1990. Cefotaxime-resistant Nocardia asteroides strains are isolates of the controversial species Nocardia farcinica. J. Clin. Microbiol. 28:2726-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallace, R. J., Jr., K. Wiss, R. Curvey, P. H. Vance, and J. Steadham. 1983. Differences among Nocardia spp. in susceptibility to aminoglycosides and beta-lactam antibiotics and their potential use in taxonomy. Antimicrob. Agents Chemother. 23:19-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, L., Y. Zhang, Z. Lu, Y. Shi, Z. Liu, L. Maldonado, and M. Goodfellow. 2001. Nocardia beijingensis sp. nov., a novel isolate from soil. Int. J. Syst. Evol. Microbiol. 51:1783-1788. [DOI] [PubMed] [Google Scholar]
- 33.Waterman, M. S. 1995. Introduction to computational biology, p. 201-202. Chapman & Hall, London, England.
- 34.Wilson, R. W., V. A. Steingrube, B. A. Brown, Z. Blacklock, K. C. Jost, Jr., A. McNabb, W. D. Colby, J. R. Biehle, J. L. Gibson, and R. J. Wallace, Jr. 1997. Recognition of a Nocardia transvalensis complex by resistance to aminoglycosides, including amikacin, and PCR-restriction fragment length polymorphism analysis. J. Clin. Microbiol. 35:2235-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson, R. W., V. A. Steingrube, B. A. Brown, and R. J. Wallace, Jr. 1998. Clinical application of PCR-restriction enzyme pattern analysis for rapid identification of aerobic actinomycete isolates. J. Clin. Microbiol. 36:148-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yassin, A. F., F. A. Rainey, J. Burghardt, H. Brzezinka, S. Schmitt, P. Seifert, O. Zimmermann, H. Mauch, D. Gierth, I. Lux, and K. P. Schaal. 1997. Tsukamurella tyrosinosolvens sp. nov. Int. J. Syst. Bacteriol. 47:607-614. [DOI] [PubMed] [Google Scholar]
- 37.Yassin, A. F., F. A. Rainey, and U. Steiner. 2001. Nocardia cyriacigeorgici sp. nov. Int. J. Syst. Evol. Microbiol. 51:1419-1423. [DOI] [PubMed] [Google Scholar]
- 38.Yassin, A. F., F. A. Rainey, and U. Steiner. 2001. Nocardia ignorata sp. nov. Int. J. Syst. Evol. Microbiol. 51:2127-2131. [DOI] [PubMed] [Google Scholar]