Abstract
The shortcomings of the licensed polysaccharide-based pneumococcal vaccine are driving efforts toward development of a protein-based vaccine that is serotype independent and effective in all age groups. An opsonophagocytic killing assay (OPKA) is used to evaluate the antibody response against polysaccharide-based pneumococcal vaccines. However, the OPKA is not reliable for noncapsular antigens. Thus, there is a need to develop an in vitro surrogate for protection for protein vaccine candidates like pneumococcal surface antigen A (PspA). PspA is a serologically variable cell surface virulence factor. Based on its sequence, PspA has been classified into families 1 (clade 1 and 2), 2 (clades 3, 4 and 5), and 3 (clade 6). Here, we report the characterization of 18 IgG anti-PspA monoclonal antibodies (anti-PspAhkR36A MAbs) generated from mice immunized with heat-killed strain R36A (clade 2). An enzyme-linked immunosorbent assay (ELISA)-based analysis of the reactivity of the MAbs with recombinant PspAs from the 6 clades indicated that they were family 1 specific. This was confirmed by flow cytometry using a hyperimmune serum generated against PspA from R36A. Eight MAbs that bind at least one clade 1- and clade 2-expressing strain were evaluated for complement deposition, bactericidal activity, and passive protection. The anti-PspAhkR36A MAb-dependent deposition of complement on pneumococci showed a positive correlation with passive protection against strain WU2 (r = 0.8783, P = 0.0041). All of our protective MAbs showed bactericidal activity; however, not all MAbs that exhibited bactericidal activity conferred protection in vivo. The protective MAbs described here can be used to identify conserved protection eliciting B cell epitopes for engineering a superior PspA-based vaccine.
INTRODUCTION
The bacterial pathogen Streptococcus pneumoniae (pneumococcus) is responsible for causing pneumonia, septicemia, meningitis, and otitis media in humans (1). According to the estimate made by the World Health Organization in 2005, 1.6 million individuals die of diseases caused by S. pneumoniae every year, and most of these deaths occur in developing countries (2). In the year 2000, it was estimated that pneumococcal disease was responsible for about 800,000 deaths of children <5 years of age (3). The currently available pneumococcal polysaccharide vaccine is not effective in children <2 years of age. Pneumococcal conjugate vaccines, however, overcome this limitation and are effective in children but have limited serotype coverage (4). The development of antibiotic resistance and the emergence of nonvaccine serotypes pose difficulties in the management of pneumococcal infections. Efforts are being made globally to develop a protein-based pneumococcal vaccine that confers serotype-independent protection in all age groups (5–7).
A polysaccharide capsule envelops S. pneumoniae, and it serves as the major virulence factor by shielding pneumococci from immune attack. The unencapsulated strains of S. pneumoniae are known to be avirulent or highly attenuated. In addition to the capsule, several surface-associated proteins have been demonstrated to be involved in pneumococcal virulence, and one such protein is pneumococcal surface protein A (PspA) (8). A well-established opsonophagocytic killing assay (OPKA) is available for evaluating the pneumococcal polysaccharide-based vaccines. The recent interest in the protein-based pneumococcal vaccines has led to efforts toward development of an in vitro assay for noncapsular antigens that can help in predicting and quantitating the protective activity of antibodies against protein vaccine candidates. Various investigators have tried to correlate anti-protein antibody titers, surface binding (by a whole-cell enzyme-linked immunosorbent assay [ELISA]), and a surface killing assay with in vivo protection (9–12). The notion of in vitro antibody-mediated complement deposition as a possible surrogate for predicting in vivo protection has been proposed by Goulart et al. and Ochs et al. (13, 14). However, these investigators did not validate it with in vivo protection experiments. Availability of a robust in vitro assay would help in minimizing the use of animal models for testing protein vaccine candidates.
PspA is a polymorphic, surface-associated choline-binding protein (15). PspA has a predominantly α-helical coiled coil structure (16, 17). It is present in essentially all clinical isolates studied to date and is being pursued as a promising vaccine candidate (18). Based on the amino acid sequence, PspA has been classified into three families and six clades. Family 1 includes clades 1 and 2, family 2 includes clades 3, 4, and 5, and family 3 includes clade 6 (19). Studies have shown that 94 to 99% of the pneumococcal isolates analyzed belong to PspA families 1 and 2 (20, 21).
The complement-mediated clearance of pneumococci is an important component of the host defense mechanism (22). A PspA-deficient strain is cleared faster than wild-type pneumococci, and an anti-PspA antibody facilitates complement-dependent phagocytosis of S. pneumoniae (23). Ren and coworkers have demonstrated that anti-PspA antibodies enhance complement activation and deposition on pneumococcal surface and thus help in clearance (24).
Active immunization with PspA in animal models has proven to be protective against invasive disease and nasopharyngeal carriage (25). Mice immunized with DNA vaccine expressing the extracellular domain of PspA were protected against an intraperitoneal challenge with a pneumococcal strain bearing PspA from the same clade (26). The B cell epitopes recognized by protective monoclonal antibodies (MAbs) have been mapped to the 192- to 260-amino acid region (27). Daniels et al. recently demonstrated that the proline-rich region of PspA contains surface-accessible epitopes that are protective in both active and passive mouse protection experiments (28). PspA has been shown to elicit high antibody titers in humans, and human anti-PspA sera can protect mice against pneumococcal challenge when transferred passively (18, 29).
There is evidence to suggest that not all anti-PspA antibodies are protective. The goals of the present study were to identify anti-PspAhkR36A MAbs that recognize conserved cross-protective B cell epitopes, and since the in vitro surrogate of protection is not well established for noncapsular antigens, we evaluated the surface binding, complement deposition, and bactericidal activity of anti-PspAhkR36A MAbs as potential in vitro correlates of protection. We found that all of the 18 anti-PspAhkR36A MAbs recognized family 1 PspAs and did not bind PspAs representing families 2 and 3. We identified 4 anti-PspAhkR36A MAbs (P1E11, M4F4, P2A4, and P2B5) that augmented complement deposition on pneumococci, exhibited bactericidal activity, and conferred protection against PspA clade 1- and 2-bearing S. pneumoniae strains in passive mouse protection experiments. Further, our data with strain WU2 suggested that anti-PspAhkR36A MAb-dependent complement deposition on pneumococci strongly correlated with in vivo protection. We observed that all of the protective anti-PspAhkR36A MAbs exhibited bactericidal activity; however, not all of the anti-PspAhkR36A MAbs that showed bactericidal activity conferred in vivo protection.
MATERIALS AND METHODS
Mice.
Six- to 8-week-old BALB/c (female) and CBA/N (male/female) inbred strains of mice were obtained from the Small Animal Facility of the National Institute of Immunology. Animals were rested and handled in accordance with the institutional animal ethics committee guidelines. Blood samples from healthy donors were taken with the approval of and following the guidelines of the institutional human ethics committee. Experiments involving recombinant DNA and handling of S. pneumoniae were carried out in accordance with the institutional biosafety committee guidelines.
Pneumococcal strains, plasmids, and culture conditions.
The pneumococcal strains and plasmids used in this study are listed in Table S1 in the supplemental material. Pneumococcal strains were maintained in Todd-Hewitt broth supplemented with 0.5% yeast extract (THY) or on a plate with tryptic soy agar (TSA) supplemented with 5% (vol/vol) sheep blood at 37°C in the presence of 5% CO2. The mid-logarithmic-phase pneumococcal cultures were stored with 15% (vol/vol) glycerol or 17% fetal calf serum, aliquoted, and stored at −70°C as described previously (30).
Escherichia coli cells were maintained in Luria-Bertani broth or on a Luria-Bertani agar plate with antibiotic(s) wherever required.
Molecular cloning, overexpression, and purification of recombinant PspA.
The subfragments encoding the N-terminal (surface exposed) region of PspA for clade 3 (PspATIGR4) and clade 5 (PspAATCC 6303) were amplified by PCR using genomic DNA from the pneumococcal strains TIGR4 and ATCC 6303, respectively, following the cloning strategy and PCR conditions described by Rohatgi et al. (30). The corresponding plasmid constructs for PspA clade 1 (pUAB069, strain L82016 [PspAL82016]), clade 4 (pUAB100, strain JCP#56 [PspAJCP#56]), and clade 6 (pUAB104, strain BG9300 [PspABG9300]) were kindly provided by Susan Hollingshead, University of Alabama, USA. The plasmid construct for PspA clade 2 (R36A [PspAR36A]) was published previously from our laboratory (30). For expression purposes, pQE-30 Xa- and pET-20b-based constructs were transformed into E. coli expression strains SG13009 and BL-21(DE3), respectively. Recombinant PspA was purified using nickle-nitriloacetic acid (Ni-NTA) affinity chromatography (Sigma-Aldrich, USA). The purity of the protein preparation was found to be >95% by SDS-PAGE and was of the expected molecular size.
Enzyme-linked immunosorbent assay.
The reactivities of the 18 IgG anti-PspAhkR36A MAbs with recombinant PspAs representing the six clades of PspA were analyzed by an ELISA. Briefly, 96-well polystyrene microtiter plates (Greiner Bio-One, Germany) were coated overnight at 4°C with recombinant PspA (50 μl of 2 μg/ml per well) in 100 mM carbonate-bicarbonate buffer (pH 9.5). The plates were washed with phosphate-buffered saline (PBS) containing 0.05% Tween 20 (PBST) and blocked with PBS containing 2% bovine serum albumin (BSA) at 37°C for 1 h. After washing with PBST, the plates were incubated with the culture supernatant from the 18 anti-PspAhkR36A hybridomas (in duplicate) at 37°C for 1 h. The plates were washed with PBST and incubated with horseradish peroxidase-conjugated goat anti-mouse Ig antibody (diluted 1 in 2,500; Becton-Dickinson Bioscience, USA) followed by incubation at 37°C for 1 h. The color was developed using 3,3′,5,5′-tetramethylbenzidine-H2O2 as the substrate, and absorbance was recorded at 450 nm.
Generation of polyclonal sera against recombinant PspAR36A.
Six- to 8-week-old female BALB/c mice were immunized subcutaneously with 25 μg recombinant PspAR36A emulsified with Imject alum (1:1 [wt/wt]) (Pierce, USA). On days 14 and 28, mice were given a booster injection with the same amount of antigen emulsified as described above. The control mice received only Imject alum in PBS. One week after the second booster, hyperimmune serum was isolated after mice were bled retro-orbitally, and an ELISA was performed to determine the PspA-specific antibody titer.
Surface staining with anti-PspAhkR36A MAbs and anti-PspAR36A hyperimmune serum.
The surface binding of anti-PspAhkR36A MAbs with S. pneumoniae strains that express clade 1 PspA (BG8838) and clade 2 PspA (WU2 and D39) was analyzed using flow cytometry as described previously (31). Briefly, mid-logarithmic-phase (optical density at 600 nm [OD600] of 0.4) pneumococci (107 CFU) were washed with PBS and incubated with 200 μl of culture supernatant from the hybridomas at room temperature for 1 h. After washing with PBS, pneumococci were incubated with fluorescein isothiocyanate (FITC)-conjugated F(ab′)2 fragment goat anti-mouse IgG plus IgM(H+L) antibody (diluted 1 in 200) (Jackson ImmunoResearch Laboratory, USA) followed by incubation at room temperature for 1 h. After washing, pneumococci were fixed with 2% paraformaldehyde (PFA) for 10 min at 4°C, and surface staining was analyzed by flow cytometry (FACSCalibur; Becton-Dickinson Bioscience).
The surface binding of the anti-PspAR36A polyclonal sera (diluted 1 in 200) was analyzed with strains BG8838 (clade 1), D39 (clade 2), TIGR4 (clade 3), EF5668 (clade 4), ATCC 6303 (clade 5), and BG6803 (clade 6) using flow cytometry as described above. Geometric mean fluorescence intensity (GMFI) values equal to or greater than twice the value obtained with the preimmune control were considered significant.
Purification of anti-PspAhkR36A MAbs.
Hybridomas secreting anti-PspAhkR36A MAbs were generated and cultured as previously described (30). Ascitic fluid was generated for the anti-PspAhkR36A MAb-secreting hybridomas. Briefly, BALB/c mice (3 mice per hybridoma) were injected intraperitoneally with 0.5 ml incomplete Freund's adjuvant (Sigma-Aldrich). Three days later, 5 × 106 hybridoma cells were injected intraperitoneally. After 7 to 10 days, ascitic fluid from the peritoneal cavity was centrifuged at 500 × g for 10 min at 4°C, and the supernatant was collected and stored at −70°C. The MAbs were purified from ascitic fluid using protein G-Sepharose beads (GE Healthcare, USA) as described earlier (32). The protein concentration of the purified MAb was estimated using a micro BCA protein assay kit (Thermo-Scientific, USA).
Passive mouse protection assay.
Groups of eight 6- to 8-week-old CBA/N mice were injected intraperitoneally with purified anti-PspAhkR36A MAb or a matched isotype control. The amounts of MAb administered for the high- and low-dose experiments were 5 and 1.25 mg/kg body weight, respectively. An hour later, mice were challenged intravenously with 107 CFU (100 times the 50% lethal dose [LD50]) of strain BG8838 and 103 CFU (100 times the LD50) of WU2 (27). The survival of mice was monitored every 12 h for the first 10 days and every 24 h for the next 11 days.
Blood bactericidal assay.
The anti-PspAhkR36A MAbs were analyzed by a blood bactericidal assay as described previously (33). Briefly, human peripheral blood was collected using recombinant hirudin from yeast (100 U/ml blood) as an anticoagulant. Pneumococci (500 CFU in 10 μl) were incubated with 235 μl of blood in the presence of either purified anti-PspAhkR36A MAb or the corresponding isotype control (5 μl of 1 mg/ml). Samples were incubated at 37°C with rotation for 2 h for D39 and 3 h for BG8838. The surviving bacteria were enumerated by plating serial dilutions (in duplicate) on TSA plates, and the mean values obtained were used to calculate bactericidal activity as described below. The data are presented as percent killing, which was arrived at using the formula ([colony count with appropriate isotype control MAb − colony count with anti-PspAhkR36A MAb] divided by colony count with the appropriate isotype control MAb) × 100.
Complement deposition assay.
The complement deposition assay was performed as described by Ochs et al. with some modifications (14). Briefly, mid-logarithmic-phase pneumococci (107 CFU) were washed with PBS and incubated with either an anti-PspAhkR36A MAb or a matched isotype control (100 μl of 20 μg/ml) for 1 h at room temperature. Treated pneumococci were incubated in 200 μl of normal human serum pooled from three healthy donors (diluted to 10% in Hanks' balanced salt solution with Ca2+ and Mg2+ ions) (Biological Industries, Israel) for 30 min at 37°C. The mixture was incubated on ice for 10 min and washed with PBS, and the incubation was continued with PBS-1% BSA containing mouse anti-human C3 MAbs (diluted 1 in 50) (Abcam, USA) for 45 min on ice. The bound anti-C3 antibody was detected using an FITC-conjugate F(ab′)2 fragment goat anti-mouse IgG plus IgM(H+L) antibody (diluted 1 in 200). Pneumococci were fixed with 2% PFA for 10 min on ice and analyzed by flow cytometry.
Statistical analysis.
Statistical analysis was done using GraphPad Prism version 6 software (GraphPad Software Inc., USA). The passive mouse protection data were compared using the log rank test. We used a parametric Pearson's linear regression correlation analysis to evaluate a possible correlation between the anti-PspAhkR36A MAb-dependent deposition of complement on pneumococci and passive mouse protection. A P value of <0.05 was considered statistically significant.
RESULTS
Cross-reactivity and surface staining of anti-PspAhkR36A MAbs is restricted to PspA family 1.
We previously reported that the relative avidity of the primary IgG anti-PspA polyclonal antibody was higher than that of recombinant PspAR36A in the sera of mice immunized with heat-killed R36A (30). In this study, we analyzed 18 IgG anti-PspAhkR36A MAbs for the extent of cross-reactivity with recombinant PspA belonging to the 6 PspA clades (i.e., PspAL82016, PspAR36A, PspATIGR4, PspAJCP#56, PspAATCC 6303, and PspABG9300) by an ELISA (Table 1). All of the 18 MAbs reacted with PspAs representing clades 1 and 2 (family 1). None of the MAbs bound PspAs representing families 2 (clades 3, 4, and 5) and 3 (clade 6), suggesting that the anti-PspAhkR36A MAbs were family 1 specific.
TABLE 1.
ELISA-based analysis for reactivity of anti-PspAhkR36A MAbs with recombinant PspAs representing the 6 PspA cladesa
| Hybridoma | Fold change for pneumococcal strain (PspA family/clade): |
|||||
|---|---|---|---|---|---|---|
| L82016 (1/1)b | R36A (1/2)b | TIGR4 (2/3) | JCP#56 (2/4) | ATCC 6303 (2/5) | BG9300 (3/6) | |
| B3D12 | 16.5 | 17.2 | 0.8 | 0.8 | 0.8 | 1.2 |
| B3H8 | 1.8 | 2.1 | 1.0 | 0.9 | 1.0 | 0.8 |
| L5C8 | 17.1 | 16.7 | 0.8 | 0.8 | 0.8 | 0.8 |
| L5F10 | 19.5 | 26.9 | 0.8 | 0.7 | 0.9 | 1.2 |
| M4F4 | 30.0 | 32.5 | 1.1 | 1.2 | 1.6 | 1.2 |
| P1E11 | 29.2 | 31.5 | 1.4 | 1.4 | 1.7 | 1.6 |
| D1A5 | 10.6 | 11.9 | 0.9 | 0.8 | 0.7 | 0.9 |
| K1B12 | 15.9 | 16.5 | 0.7 | 0.7 | 0.6 | 0.9 |
| M6B2 | 13.5 | 14.5 | 1.0 | 1.4 | 1.3 | 1.9 |
| P2F9 | 9.8 | 13.4 | 0.9 | 1.0 | 0.9 | 1.0 |
| P2A4 | 21.6 | 19.9 | 1.0 | 1.5 | 0.9 | 1.0 |
| P2B5 | 22.0 | 24.0 | 1.0 | 1.2 | 1.2 | 1.2 |
| J4C1 | 28.2 | 29.3 | 1.0 | 0.8 | 0.7 | 0.8 |
| P2C2 | 16.2 | 17.0 | 0.6 | 0.6 | 0.6 | 0.6 |
| A1D9 | 2.0 | 2.7 | 0.6 | 0.7 | 0.7 | 0.6 |
| C4B4 | 18.1 | 18.1 | 0.8 | 0.6 | 0.6 | 0.7 |
| F4B6 | 24.7 | 26.3 | 1.0 | 0.9 | 0.8 | 1.1 |
| D3H6 | 4.0 | 5.7 | 0.7 | 0.7 | 0.7 | 1.1 |
The culture supernatants from the 18 anti-PspAhkR36A MAb-secreting hybridomas were tested for reactivity with recombinant PspAs representing the 6 clades of PspA (extracellular domain with or without the proline-rich region) by an ELISA. The numerical values represent fold change in absorbance relative to the control.
These columns show fold change values that were greater than twice the value obtained with the control and were considered significant.
Next, we assessed whether these 18 anti-PspAhkR36A MAbs bind to the surface of pneumococci. For this purpose, we tested strains BG8838 (clade 1) and WU2 and D39 (clade 2). Of the 18 MAbs, BG8838 and WU2 were recognized by 9 and 10 MAbs, respectively (Table 2). As expected, all 18 anti-PspA MAbs stained D39. Nine MAbs bound one clade 1 and two clade 2 PspA-expressing strains. Eight (i.e., B3D12, B3H8, L5C8, L5F10, M4F4, P1E11, P2A4, and P2B5) of these were selected at random for further analysis.
TABLE 2.
Surface binding of anti-PspAhkR36A MAbs with strains expressing family 1 PspAa
| MAb | GMFI (fold change) for: |
||
|---|---|---|---|
| BG8838 clade 1 | WU2 clade 2 | D39 clade 2 | |
| IgG1 IC | 4.77 (1.0) | 3.75 (1.0) | 3.67 (1.0) |
| B3D12 | 13.10 (2.8) | 5.18 (1.4) | 64.80 (17.7) |
| B3H8 | 14.70 (3.1) | 5.49 (1.5) | 8.75 (2.4) |
| L5C8 | 18.70 (3.9) | 3.82 (1.0) | 137.00 (37.3) |
| L5F10 | 18.90 (4.0) | 7.22 (1.9) | 66.20 (18.1) |
| M4F4 | 59.50 (12.5) | 11.80 (3.2) | 140.00 (38.2) |
| P1E11 | 81.80 (17.2) | 14.20 (3.8) | 146.00 (39.8) |
| D1A5 | 4.04 (0.8) | 107.00 (28.5) | 52.90 (14.4) |
| K1B12 | 3.86 (0.8) | 9.39 (2.5) | 69.80 (19.0) |
| M6B2 | 5.64 (1.2) | 19.20 (5.1) | 174.00 (47.4) |
| P2F9 | 4.80 (1.0) | 4.12 (1.1) | 14.30 (3.9) |
| IgG2a IC | 4.82 (1.0) | 2.82 (1.0) | 4.68 (1.0) |
| P2A4 | 23.60 (4.9) | 11.10 (3.9) | 59.70 (12.8) |
| P2B5 | 17.60 (3.7) | 16.10 (5.7) | 33.90 (7.3) |
| J4C1 | 3.88 (0.8) | 3.27 (1.2) | 60.80 (13.0) |
| P2C2 | 3.52 (0.7) | 8.93 (3.2) | 34.80 (7.4) |
| IgG2b IC | 4.16 (1.0) | 2.28 (1.0) | 3.21 (1.0) |
| A1D9 | 4.35 (1.1) | 16.3 (7.2) | 47.40 (14.8) |
| C4B4 | 10.80 (2.6) | 22.10 (9.7) | 45.00 (14.1) |
| F4B6 | 3.70 (0.89) | 56.7 (24.9) | 41.00 (12.8) |
| IgG3 IC | 4.26 (1.0) | 2.82 (1.0) | 3.21 (1.0) |
| D3H6 | 4.41 (1.0) | 4.32 (1.5) | 25.80 (8.1) |
Surface binding of 18 anti-PspAhkR36A MAbs with strains BG8838 (clade 1) and WU2 and D39 (clade 2) was analyzed by flow cytometry. Strain D39 was included in the analysis as the anti-PspA MAbs were raised against R36A, an unencapsulated derivative of D39. Pneumococci were incubated with 200 μl of culture supernatant from the 18 anti-PspAhkR36A MAb-secreting hybridomas followed by staining with the appropriate FITC-conjugated secondary antibody. Matched isotype control MAbs were included, and samples were analyzed using a flow cytometer. The surface binding is expressed as geometric mean fluorescence intensity (GMFI), and the fold change relative to the corresponding isotype control MAb (assigned 1.0) is given in parentheses. A ≥2-fold increase in the surface staining relative to that of the corresponding isotype control was considered significant. IgG1 IC, IgG2a IC, IgG2b IC, and IgG3 IC represent isotype control MAbs for the isotypes IgG1, IgG2a, IgG2b, and IgG3, respectively.
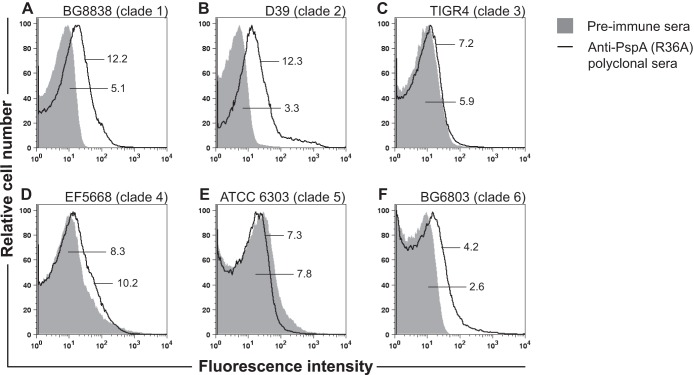
To confirm our observation that the anti-PspA antibody response elicited against heat-killed R36A was family 1 specific, the anti-PspAR36A hyperimmune serum was used to test surface binding with strains expressing PspAs belonging to families 1, 2, and 3 by flow cytometry (Fig. 1). The data demonstrate that anti-PspAR36A hyperimmune serum binds to family 1 PspA-expressing strains BG8838 (clade 1) and D39 (clade 2). No binding was observed with PspA family 2-bearing strains TIGR4 (clade 3), EF5668 (clade 4), and ATCC 6303 (clade 5) (Fig. 1). Similarly, there was no surface staining with the PspA family 3-expressing strain BG6803 (clade 6). Thus, our flow cytometry data demonstrate that the binding of anti-PspAR36A hyperimmune serum to pneumococcal strains was family 1 specific, and this corroborates the data obtained with anti-PspAhkR36A MAbs (Table 2).
FIG 1.
Surface binding of anti-PspAR36A polyclonal sera with pneumococcal strains representing the 6 clades of PspA. Surface binding with strains BG8838 (A), D39 (B), TIGR4 (C), EF5668 (D), ATCC 6303 (E), and BG6803 (F) was analyzed by flow cytometry using anti-PspAR36A polyclonal and preimmune sera. Preimmune serum (solid gray histogram) was used as the negative control. The GMFI values for the anti-PspAR36A polyclonal and preimmune sera are shown. GMFI values of ≥2 times the value obtained with the preimmune sera were considered significant.
In vivo protective efficacy of anti-PspAhkR36A MAbs.
The relative protective activities of anti-PspAhkR36A MAbs B3D12, B3H8, L5C8, L5F10, M4F4, P1E11, P2A4, and P2B5 were assessed by a passive mouse protection assay using BG8838 (clade 1) and WU2 (clade 2) as the challenge strains. CBA/N mice were given either purified anti-PspAhkR36A or the corresponding isotype control MAb (high dose; 5 mg/kg body weight) intraperitoneally. One hour later, mice were challenged intravenously with either BG8838 or WU2. The mouse survival data with BG8838 as the challenge strain (Fig. 2A, C, and E) suggested that anti-PspAhkR36A MAbs M4F4, P1E11, and L5C8 provided 87.5, 75, and 50% protection, respectively, compared to that for the corresponding isotype control (12.5%) (Fig. 2A). The MAbs L5F10, B3H8, and B3D12 provided 75, 62.5 and 37.5% protection, respectively, when challenged with BG8838 (Fig. 2C). All mice in the set that received the IgG1 isotype control MAb died within 7 days. The MAbs P2A4 and P2B5 protected 100 and 62.5% of the mice, respectively, when challenged with BG8838, compared to 12.5% in the case of the isotype control (Fig. 2E). In experiments where mice were challenged with WU2 (Fig. 2B, D, and F), M4F4 and P1E11 provided 100% protection. L5C8 failed to confer any protection, like the IgG1 isotype control MAb (Fig. 2B). L5F10 conferred 62.5% protection and B3H8 and B3D12 provided ≤25% protection when mice were challenged with WU2 (Fig. 2D). All of the mice in the corresponding control group died within 3.5 days. P2A4 provided 100% and P2B5 provided 87.5% protection when mice were challenged with WU2, whereas the corresponding IgG2a isotype control MAb protected only 12.5% of the mice (Fig. 2F).
FIG 2.
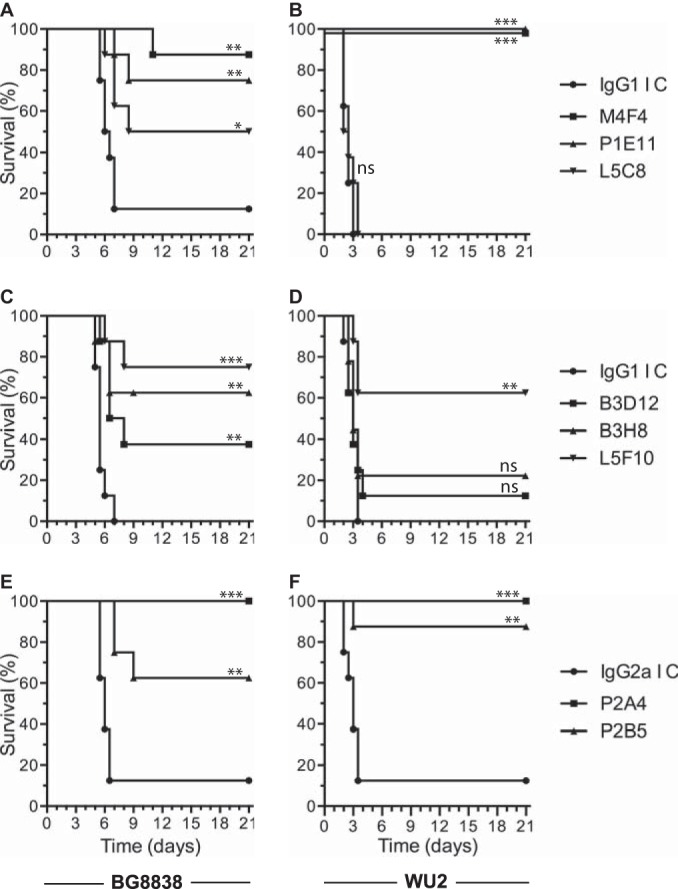
Relative efficacy of anti-PspAhkR36A MAbs to protect mice against intravenous challenge. CBA/N mice were injected with purified anti-PspAhkR36A MAb M4F4, P1E11, or L5C8 (A and B), B3D12, B3H8, or L5F10 (C and D) or P2A4 or P2B5 (E and F) intraperitoneally at 5 mg/kg body weight (high dose). The corresponding isotype control MAb (IgG1 IC or IgG2a IC) was included in each set as the negative control. One hour later, mice were challenged with 107 CFU of BG8838 (A, C, and E) or 103 CFU of WU2 (B, D, and F), and mouse survival was recorded. The data for the group given anti-PspAhkR36A MAb were compared with those for the respective isotype control MAb using the log rank test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, statistically not significant.
On the basis of results of the passive mouse protection experiments described above, MAbs M4F4, P1E11, L5F10, P2A4, and P2B5 were selected as they conferred >50% protection against the 2 challenge strains tested. We repeated the experiments with a lower dose (1.25 mg/kg body weight) of the purified anti-PspAhkR36A MAbs and the corresponding isotype control. The mouse survival data following challenge with BG8838 suggested that P1E11, M4F4, and L5F10 provided 75, 62.5, and 37.5% protection, respectively, whereas all mice in the control group died within 8 days (Fig. 3A), while P2A4 and P2B5 provided 62.5 and 50% protection, respectively, compared to the IgG2a isotype control for which 12.5% survival was observed (Fig. 3C). The mouse survival data with WU2 as the challenge strain suggested that P1E11, M4F4, and L5F10 provided 87.5, 75, and 37.5% protection, respectively (Fig. 3B), while P2A4 and P2B5 provided 100 and 75% protection, respectively (Fig. 3D). In both of the experiments, all control mice died within 3.5 days. The consolidated data from the passive protection experiments demonstrate that 4 anti-PspAhkR36A MAbs (M4F4, P1E11, P2A4, and P2B5) confer ≥50% protection against intravenous challenge with PspA clade 1- and 2-expressing strains.
FIG 3.
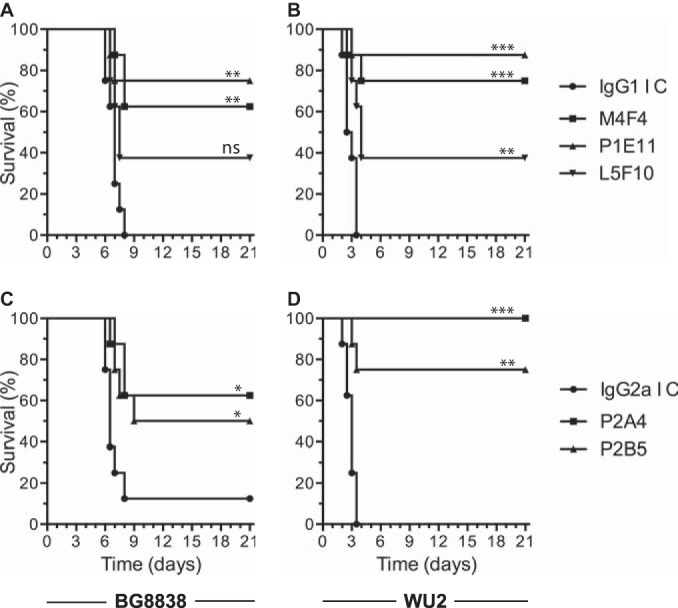
Anti-PspAhkR36A MAbs protect mice against pneumococcal infection even when given at a lower dose. CBA/N mice were injected intraperitoneally with 1.25 mg/kg body weight (low dose) of either anti-PspAhkR36A MAb M4F4, P1E11, or L5F10 (A and B) or P2A4 or P2B5 (C and D). The control group was given the respective isotype control MAb (IgG1 IC or IgG2a IC). Mice were challenged with BG8838 (A and C) or WU2 (B and D) 1 h later, and mouse survival was recorded. For other details, refer to the legend to Fig. 2.
Correlation between in vivo protection and in vitro complement deposition and bactericidal activity.
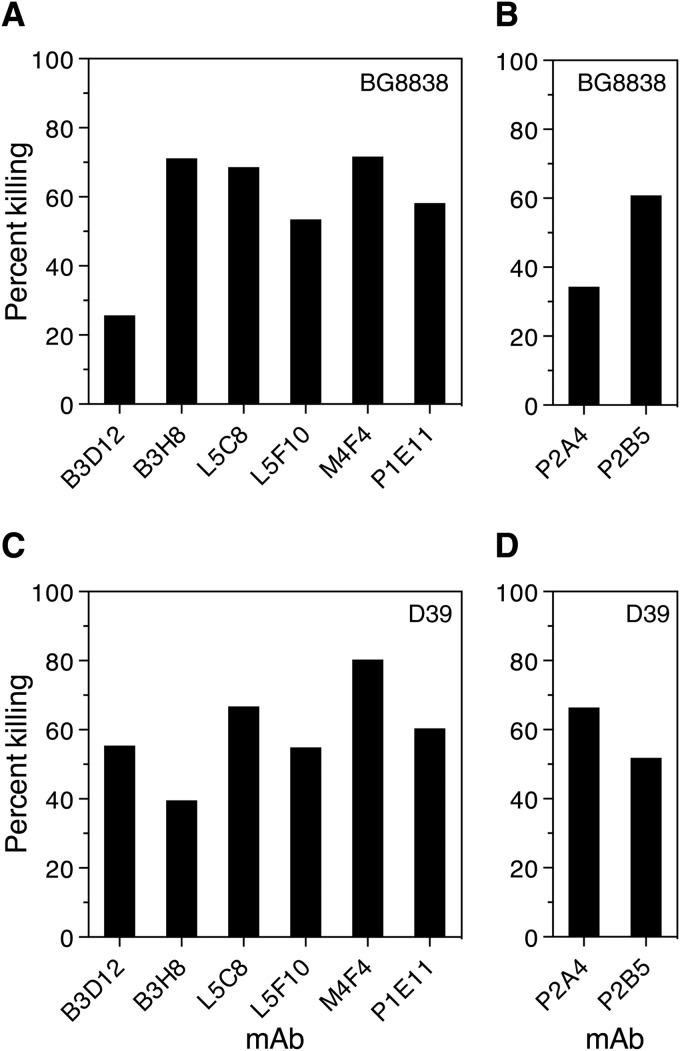
We were interested in determining the possible correlation between in vivo protection and in vitro bactericidal activity and the complement deposition capability of anti-PspAhkR36A MAbs. We employed a blood bactericidal assay to evaluate the ability of the 8 anti-PspAhkR36A MAbs (i.e., B3D12, B3H8, L5C8, L5F10, M4F4, P1E11, P2A4, and P2B5) to kill the PspA family 1-expressing strains BG8838 (clade 1) and D39 (clade 2). Pneumococci were incubated with blood from healthy donors in the presence of either purified anti-PspAhkR36A or a matched isotype control MAb, and the surviving pneumococci were enumerated by plating. The results suggest that all 8 anti-PspAhkR36A MAbs exhibited significant bactericidal activity (ranging from 25.7 to 80.3% compared with that of the matched isotype control) against BG8838 and D39, although the extent of bactericidal activity varied from one MAb to the other (Fig. 4).
FIG 4.
Anti-PspAhkR36A MAbs promote pneumococcal killing by human blood in vitro. S. pneumoniae strains BG8838 (A and B) and D39 (C and D) were incubated with blood from healthy donors and purified anti-PspAhkR36A or a matched isotype control MAb. Matched IgG1 and IgG2a isotype control MAbs were used as the comparator in panels A and C and B and D, respectively (not shown). The contents were rotated for (3 h for BG8838 and 2 h for D39) at 37°C, and the surviving pneumococci were enumerated by plating. The identity of the anti-PspAhkR36A MAb and the percent killing are plotted on the x and y axes, respectively. The percent bacterial killing was calculated as described in Materials and Methods. The assay was performed at least four times, and data from a single representative experiment are shown.
We next tested whether these MAbs can enhance C3 deposition on the surface of clade 1 (BG8838)-expressing and clade 2 (WU2 and D39)-expressing strains (Table 3). Monoclonal antibodies M4F4, P1E11, P2A4, and P2B5 showed significant enhancement in complement deposition on pneumococci compared to that of the corresponding isotype control for the 3 strains analyzed. Seven of the 8 MAbs augmented complement deposition on D39 with B3H8 being the exception. B3H8, however, enhanced deposition of complement on WU2. It was observed that the extent of the enhancement in C3 deposition depended on the anti-PspAhkR36A MAb and the target strain.
TABLE 3.
Flow cytometry-based analysis of complement C3 deposition on the surface of pneumococci in the presence of anti-PspAhkR36A MAbsa
| Strains (clade) | GMFI (fold change) for: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Negative controlb | Positive controlc | IgG1 isotype control | B3D12d | B3H8d | L5C8d | L5F10d | M4F4d | P1E11d | IgG2a isotype control | P2A4e | P2B5e | |
| BG8838 (1) | 4.5 | 16.6 | 15.3 | 29.6 | 30.3 | 24.0 | 14.4 | 71.7f | 233.0f | 12.3 | 39.4f | 106.0f |
| WU2 (2) | 3.0 | 3.4 | 3.2 | 3.8 | 20.5f | 3.4 | 5.2 | 482.0f | 442.0f | 3.6 | 320.0f | 391.0f |
| D39 (2) | 5.5 | 7.1 | 7.0 | 418.0f | 12.9 | 516.0f | 511.0f | 568.0f | 523.0f | 6.1 | 429.0f | 384.0f |
The PspA family 1-expressing strains BG8838 (clade 1) and WU2 and D39 (clade 2) were incubated with either anti-PspAhkR36A or a matched isotype control MAb. The bound complement C3 was detected by flow cytometry using anti-human C3 antibody followed by the appropriate FITC-conjugated secondary antibody.
Pneumococci were incubated with anti-human complement C3 antibody followed by the FITC-conjugated secondary antibody.
Pneumococci were incubated with 10% pooled normal human serum in Hanks' balanced salt solution followed by anti-human C3 antibody and the FITC-conjugated secondary antibody.
These anti-PspAhkR36A MAbs are of the IgG1 isotype.
These anti-PspAhkR36A MAbs are of the IgG2a isotype.
GMFI values greater than or equal to twice the value obtained with the corresponding isotype control were considered significant.
We wanted to find out which aspect or feature of the antibody response correlated with in vivo protection. The surface staining, complement deposition, bactericidal activity, and mouse protection data for MAbs B3D12, B3H8, L5C8, L5F10, M4F4, P1E11, P2A4, and P2B5 are summarized in Table 4. The trend from the data appears to be that the higher the extent of complement deposition, the higher the bactericidal activity and in vivo protection. This is well illustrated by MAbs M4F4, P1E11, P2A4, and P2B5. While all the protective MAbs showed bactericidal activity, not all MAbs that exhibited bactericidal activity showed passive protection. For example, MAbs B3D12, B3H8, and L5C8 exhibited significant bactericidal activity but showed no to poor ability to confer protection when given passively to mice.
TABLE 4.
In vivo protective efficacies of anti-PspAhkR36A MAbs correlate with the extent of complement depositiona
| Experiment | Strain | Result for anti-PspAhkR36A MAb: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| B3D12 | B3H8 | L5C8 | L5F10 | M4F4b | P1E11b | P2A4b | P2B5b | ||
| Surface staining (x)c | BG8838 | 2.8 | 3.1 | 3.9 | 4.0 | 12.5 | 17.2 | 4.9 | 3.7 |
| WU2 | 1.4 | 1.5 | 1.0 | 1.9 | 3.2 | 3.8 | 3.9 | 5.7 | |
| D39 | 17.7 | 2.4 | 37.3 | 18.1 | 38.2 | 39.8 | 12.8 | 7.3 | |
| Complement deposition (x)c | BG8838 | 1.9 | 2.0 | 1.6 | 0.9 | 4.7 | 15.2 | 3.2 | 8.6 |
| WU2 | 1.2 | 6.4 | 1.1 | 1.6 | 150.6 | 138.1 | 88.9 | 108.6 | |
| D39 | 59.7 | 1.8 | 73.7 | 73.0 | 81.1 | 74.7 | 70.3 | 62.9 | |
| Bactericidal activity (%)d | BG8838 | 25.7 | 71.2 | 68.6 | 53.5 | 71.7 | 58.2 | 34.3 | 60.8 |
| D39 | 55.4 | 39.6 | 66.8 | 54.8 | 80.3 | 60.4 | 66.5 | 51.9 | |
| Mouse passive protection assay (%)e | |||||||||
| 5 mg/kg | BG8838 | 37.5 | 62.5 | 50.0 | 75.0 | 87.5 | 75.0 | 100.0 | 62.5 |
| WU2 | 12.5 | 22.5 | 0.0 | 62.5 | 100.0 | 100.0 | 100.0 | 87.5 | |
| 1.25 mg/kg | BG8838 | NDf | ND | ND | 37.5 | 62.5 | 75.0 | 62.5 | 50.0 |
| WU2 | ND | ND | ND | 37.5 | 75.0 | 87.5 | 100.0 | 75.0 | |
BG8838 expresses clade 1 PspA, while WU2 and D39 express clade 2 PspA.
In these columns, the 4 anti-PspAhkR36A MAbs were the most protective.
Surface staining and complement deposition are expressed as fold change (x) in the GMFI value relative to the corresponding isotype control.
Bactericidal activity of the anti-PspAhkR36A MAb is represented as percent killing. A higher value indicates higher bactericidal activity. The assay was not performed with strain WU2.
Mouse protection is expressed as percent survival. A higher value indicates higher protective efficacy. This experiment was not done with strain D39.
ND, not determined.
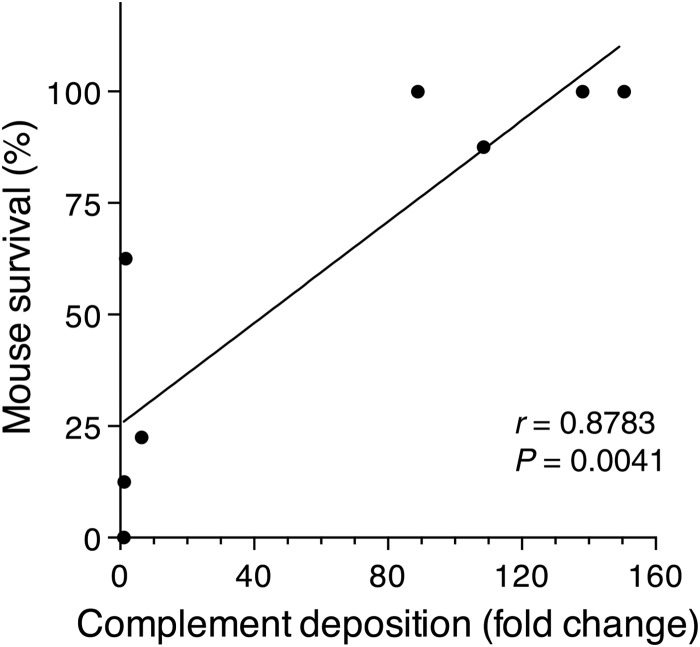
To determine whether in vitro MAb-dependent deposition of complement correlated with passive mouse protection, a parametric Pearson's linear regression correlation analysis was performed. The analysis indicated that there was a positive correlation between in vitro complement deposition and the passive protection for strains WU2 (Fig. 5) and BG8838 (data not shown). The correlation was highly statistically significant for strain WU2 (r = 0.8783, P = 0.0041). Our data suggest that the antibody-dependent deposition of complement on the pneumococcal surface can be a potential in vitro correlate of protection for strains like WU2.
FIG 5.
Analysis of 8 anti-PspAR36A MAbs to assess a possible correlation between MAb-dependent complement deposition and their ability to passively protect mice from an otherwise lethal challenge with strain WU2. Each dot represents an anti-PspAR36A MAb. The correlation between in vitro complement deposition and passive protection was highly significant by a parametric Pearson's linear regression correlation analysis (r = 0.8783, P = 0.0041).
DISCUSSION
PspA is a likely candidate for a protein-based vaccine against pneumococcal infections; however, its serological variability might restrict the coverage of a PspA-based vaccine. For this reason, gaining insight into the nature of the variability of PspAs has been the subject of several studies that are directed at development of a protein-based pneumococcal vaccine. Studies aimed to investigate the level of cross-reactivity among PspAs in mice indicate that antibodies generated against PspA show higher cross-reactivity with the strains expressing PspA of the same family than the strains that bear PspA of a different PspA family. We screened our panel to identify the anti-PspAhkR36A MAbs that exhibited the maximum reactivity across PspA clades. Our ELISA and flow cytometry-based surface staining data revealed that all of the 18 anti-PspA MAbs (raised against the clade 2-expressing strain R36A) recognized family 1 PspA and not PspAs representing families 2 and 3. This is consistent with the observation that mice immunized with DNA vaccine expressing the extracellular domain of PspA were protected from strains bearing PspA of the same clade when tested in an intraperitoneal challenge mouse model. Briles and coworkers, however, reported that human anti-PspA antibodies, when administered to mice, conferred protection against strains expressing PspA belonging to families 1 and 2 (29).
Antibody-dependent complement-mediated phagocytosis is a well-established mechanism of pneumococcal clearance (22). Antibodies directed at pneumococci help in clearance by augmenting opsonization. Antibodies to PspA enhance complement deposition on the pneumococcal surface, thereby contributing to their protective effect (24). A previous report suggested that polyclonal sera against PspA from families 1 and 2 help in enhancing complement deposition (34). Our anti-PspAhkR36A MAbs M4F4, P1E11, P2A4, and P2B5 augmented complement deposition on the 3 PspA family 1-expressing strains analyzed (Table 3). The observed variation in the degree to which various anti-PspAhkR36A MAbs augmented complement deposition across strains may have to do with the chemical nature of the capsule on the target strain, the thickness of the capsule, and the relative accessibility of the pneumococcal surface.
There is evidence to suggest that not all antibodies to PspA are protective indirectly, implying that not all PspA epitopes elicit protective antibody responses. In our previous work, we had observed that P1E11 and P2A4 compete for binding with a PspAR36A subfragment corresponding to a 193- to 286-amino-acid stretch (PspAR36A 193–286), indicating that these two MAbs recognize the same or an overlapping epitope (30). Thus, M4F4, P1E11, P2A4, and P2B5 recognize at least 3 topologically distinct epitopes. The epitopes recognized by P1E11, P2A4, and P2B5 were localized to the PspAR36A 193–286 subfragment. The epitope recognized by M4F4 was mapped to the subfragment PspAR36A 98–192 (30). Our data are consistent with those reported by Roche et al., who localized the cross protection-eliciting region of PspA to the N-terminal 115 amino acid residues and ∼104 C-terminal residues of the extracellular domain from strain EF3296 (12). Knowledge of the epitopes recognized by anti-PspAhkR36A MAbs that do and do not elicit protective responses might be put to use to engineer a PspA vaccine that maximizes the proportion of protective antibodies in the antibodies generated. Fine mapping of the conserved B cell epitopes recognized by the protective anti-PspAhkR36A MAbs M4F4, P1E11, P2A4, and P2B5 can help in development of a superior PspA-based vaccine.
Our data indicate that complement deposition on pneumococci can be a used as a surrogate for the in vivo protection for strains like WU2 (Fig. 5). Roche and coworkers had demonstrated that the antibody titer and surface staining do not correlate with in vivo protection and thus are not useful as a surrogate for protection (12). Cohen et al. reported that a whole-cell ELISA is an inadequate predictor of in vivo protection (11). While the modified surface killing assay for PspA developed by Genschmer et al. is likely to be significant (9), complement deposition as an in vitro correlate of protection is easier to perform and amenable to automation. The complement deposition assay might be potentially useful in quantitating the relative protective efficacy of antibodies against novel protein vaccine antigens (or their subfragments) to confer protection in vivo.
ACKNOWLEDGMENTS
This work was supported in part by the intramural research program of the National Institute of Immunology and by grant BT/PR5037/MED/15/77/2012 from the Department of Biotechnology (DBT), India.
Naeem Khan was the recipient of a Senior Research Fellowship from DBT.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/CVI.00001-14.
REFERENCES
- 1.Lynch JP III, Zhanel GG. 2010. Streptococcus pneumoniae: epidemiology and risk factors, evolution of antimicrobial resistance, and impact of vaccines. Curr Opin Pulm Med 16:217–225. doi: 10.1097/MCP.0b013e3283385653. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. 2007. Pneumococcal conjugate vaccine for childhood immunization—WHO position paper. Wkly Epidemiol Rec 82:93–104. [PubMed] [Google Scholar]
- 3.O'Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, Lee E, Mulholland K, Levine OS, Cherian T, Hib and Pneumococcal Global Burden of Disease Study Team . 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 4.Vila-Corcoles A, Ochoa-Gondar O, Guzman JA, Rodriguez-Blanco T, Salsench E, Fuentes CM. 2010. Effectiveness of the 23-valent polysaccharide pneumococcal vaccine against invasive pneumococcal disease in people 60 years or older. BMC Infect Dis 10:73. doi: 10.1186/1471-2334-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyaji EN, Oliveira ML, Carvalho E, Ho PL. 2013. Serotype-independent pneumococcal vaccines. Cell Mol Life Sci 70:3303–3326. doi: 10.1007/s00018-012-1234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moffitt KL, Malley R. 2011. Next generation pneumococcal vaccines. Curr Opin Immunol 23:407–413. doi: 10.1016/j.coi.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldman C, Anderson R. 2014. Review: current and new generation pneumococcal vaccines. J Infect 69:309–325. doi: 10.1016/j.jinf.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell AM, Mitchell TJ. 2010. Streptococcus pneumoniae: virulence factors and variation. Clin Microbiol Infect 16:411–418. doi: 10.1111/j.1469-0691.2010.03183.x. [DOI] [PubMed] [Google Scholar]
- 9.Genschmer KR, Accavitti-Loper MA, Briles DE. 2013. A modified surface killing assay (MSKA) as a functional in vitro assay for identifying protective antibodies against pneumococcal surface protein A (PspA). Vaccine 32:39–47. doi: 10.1016/j.vaccine.2013.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniels CC, Kim KH, Burton RL, Mirza S, Walker M, King J, Hale Y, Coan P, Rhee DK, Nahm MH, Briles DE. 2013. Modified opsonization, phagocytosis, and killing assays to measure potentially protective antibodies against pneumococcal surface protein A. Clin Vaccine Immunol 20:1549–1558. doi: 10.1128/CVI.00371-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen JM, Wilson R, Shah P, Baxendale HE, Brown JS. 2013. Lack of cross-protection against invasive pneumonia caused by heterologous strains following murine Streptococcus pneumoniae nasopharyngeal colonisation despite whole-cell ELISAs showing significant cross-reactive IgG. Vaccine 31:2328–2332. doi: 10.1016/j.vaccine.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 12.Roche H, Hakansson A, Hollingshead SK, Briles DE. 2003. Regions of PspA/EF3296 best able to elicit protection against Streptococcus pneumoniae in a murine infection model. Infect Immun 71:1033–1041. doi: 10.1128/IAI.71.3.1033-1041.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goulart C, Darrieux M, Rodriguez D, Pimenta FC, Brandileone MC, de Andrade AL, Leite LC. 2011. Selection of family 1 PspA molecules capable of inducing broad-ranging cross-reactivity by complement deposition and opsonophagocytosis by murine peritoneal cells. Vaccine 29:1634–1642. doi: 10.1016/j.vaccine.2010.12.074. [DOI] [PubMed] [Google Scholar]
- 14.Ochs MM, Bartlett W, Briles DE, Hicks B, Jurkuvenas A, Lau P, Ren B, Millar A. 2008. Vaccine-induced human antibodies to PspA augment complement C3 deposition on Streptococcus pneumoniae. Microb Pathog 44:204–214. doi: 10.1016/j.micpath.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crain MJ, Waltman WD II, Turner JS, Yother J, Talkington DF, McDaniel LS, Gray BM, Briles DE. 1990. Pneumococcal surface protein A (PspA) is serologically highly variable and is expressed by all clinically important capsular serotypes of Streptococcus pneumoniae. Infect Immun 58:3293–3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yother J, Briles DE. 1992. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequence analysis. J Bacteriol 174:601–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jedrzejas MJ, Lamani E, Becker RS. 2001. Characterization of selected strains of pneumococcal surface protein A. J Biol Chem 276:33121–33128. doi: 10.1074/jbc.M103304200. [DOI] [PubMed] [Google Scholar]
- 18.Nabors GS, Braun PA, Herrmann DJ, Heise ML, Pyle DJ, Gravenstein S, Schilling M, Ferguson LM, Hollingshead SK, Briles DE, Becker RS. 2000. Immunization of healthy adults with a single recombinant pneumococcal surface protein A (PspA) variant stimulates broadly cross-reactive antibodies to heterologous PspA molecules. Vaccine 18:1743–1754. doi: 10.1016/S0264-410X(99)00530-7. [DOI] [PubMed] [Google Scholar]
- 19.Hollingshead SK, Becker R, Briles DE. 2000. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect Immun 68:5889–5900. doi: 10.1128/IAI.68.10.5889-5900.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hotomi M, Togawa A, Kono M, Ikeda Y, Takei S, Hollingshead SK, Briles DE, Suzuki K, Yamanaka N. 2013. PspA family distribution, antimicrobial resistance and serotype of Streptococcus pneumoniae isolated from upper respiratory tract infections in Japan. PLoS One 8:e58124. doi: 10.1371/journal.pone.0058124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollingshead SK, Baril L, Ferro S, King J, Coan P, Briles DE. 2006. Pneumococcal surface protein A (PspA) family distribution among clinical isolates from adults over 50 years of age collected in seven countries. J Med Microbiol 55:215–221. doi: 10.1099/jmm.0.46268-0. [DOI] [PubMed] [Google Scholar]
- 22.Brown EJ, Hosea SW, Frank MM. 1983. The role of antibody and complement in the reticuloendothelial clearance of pneumococci from the bloodstream. Rev Infect Dis 5(Suppl 4):S797–S805. doi: 10.1093/clinids/5.Supplement_4.S797. [DOI] [PubMed] [Google Scholar]
- 23.Ren B, Li J, Genschmer K, Hollingshead SK, Briles DE. 2012. The absence of PspA or presence of antibody to PspA facilitates the complement-dependent phagocytosis of pneumococci in vitro. Clin Vaccine Immunol 19:1574–1582. doi: 10.1128/CVI.00393-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren B, Szalai AJ, Hollingshead SK, Briles DE. 2004. Effects of PspA and antibodies to PspA on activation and deposition of complement on the pneumococcal surface. Infect Immun 72:114–122. doi: 10.1128/IAI.72.1.114-122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu HY, Nahm MH, Guo Y, Russell MW, Briles DE. 1997. Intranasal immunization of mice with PspA (pneumococcal surface protein A) can prevent intranasal carriage, pulmonary infection, and sepsis with Streptococcus pneumoniae. J Infect Dis 175:839–846. doi: 10.1086/513980. [DOI] [PubMed] [Google Scholar]
- 26.Ferreira DM, Miyaji EN, Oliveira ML, Darrieux M, Areas AP, Ho PL, Leite LC. 2006. DNA vaccines expressing pneumococcal surface protein A (PspA) elicit protection levels comparable to recombinant protein. J Med Microbiol 55:375–378. doi: 10.1099/jmm.0.46217-0. [DOI] [PubMed] [Google Scholar]
- 27.McDaniel LS, Ralph BA, McDaniel DO, Briles DE. 1994. Localization of protection-eliciting epitopes on PspA of Streptococcus pneumoniae between amino acid residues 192 and 260. Microb Pathog 17:323–337. doi: 10.1006/mpat.1994.1078. [DOI] [PubMed] [Google Scholar]
- 28.Daniels CC, Coan P, King J, Hale J, Benton KA, Briles DE, Hollingshead SK. 2010. The proline-rich region of pneumococcal surface proteins A and C contains surface-accessible epitopes common to all pneumococci and elicits antibody-mediated protection against sepsis. Infect Immun 78:2163–2172. doi: 10.1128/IAI.01199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Briles DE, Hollingshead SK, King J, Swift A, Braun PA, Park MK, Ferguson LM, Nahm MH, Nabors GS. 2000. Immunization of humans with recombinant pneumococcal surface protein A (rPspA) elicits antibodies that passively protect mice from fatal infection with Streptococcus pneumoniae bearing heterologous PspA. J Infect Dis 182:1694–1701. doi: 10.1086/317602. [DOI] [PubMed] [Google Scholar]
- 30.Rohatgi S, Dutta D, Tahir S, Sehgal D. 2009. Molecular dissection of antibody responses against pneumococcal surface protein A: evidence for diverse DH-less heavy chain gene usage and avidity maturation. J Immunol 182:5570–5585. doi: 10.4049/jimmunol.0803254. [DOI] [PubMed] [Google Scholar]
- 31.Daniels CC, Briles TC, Mirza S, Hakansson AP, Briles DE. 2006. Capsule does not block antibody binding to PspA, a surface virulence protein of Streptococcus pneumoniae. Microb Pathog 40:228–233. doi: 10.1016/j.micpath.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 32.Manivel V, Sahoo NC, Salunke DM, Rao KV. 2000. Maturation of an antibody response is governed by modulations in flexibility of the antigen-combining site. Immunity 13:611–620. doi: 10.1016/S1074-7613(00)00061-3. [DOI] [PubMed] [Google Scholar]
- 33.Briles DE, Forman C, Horowitz JC, Volanakis JE, Benjamin WH Jr, McDaniel LS, Eldridge J, Brooks J. 1989. Antipneumococcal effects of C-reactive protein and monoclonal antibodies to pneumococcal cell wall and capsular antigens. Infect Immun 57:1457–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren B, Szalai AJ, Thomas O, Hollingshead SK, Briles DE. 2003. Both family 1 and family 2 PspA proteins can inhibit complement deposition and confer virulence to a capsular serotype 3 strain of Streptococcus pneumoniae. Infect Immun 71:75–85. doi: 10.1128/IAI.71.1.75-85.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]