Abstract
Helcococci have previously been associated with the colonization of ulcers and infections of the skin and soft tissues. We describe a case of prosthetic joint infection due to a previously undescribed organism that is genetically most closely related to Helcococcus.
CASE REPORT
A 73-year-old Caucasian woman presented with gradually increasing swelling of the right knee and pain over a 2-month period. The patient had a history of degenerative joint disease and multiple bilateral total knee and hip arthroplasties, with the removal of her right patella and a right total knee replacement 5 years prior to presentation. She had not received any intraarticular glucocorticoid injections.
On physical examination, the patient was afebrile, and other vital signs were stable. An examination of the right knee revealed a massive effusion with overlying erythema and a well-healed, curved longitudinal incision with no sinus formation or drainage. The knee was also warm and diffusely tender to palpation, with painful range of motion.
Initial laboratory studies revealed an elevated erythrocyte sedimentation rate (33 mm/h) and C-reactive protein (52.9 mg/liter). A Gram stain of fluid from the right knee demonstrated many white blood cells and no organisms. Knee fluid inoculated into Bactec anaerobic and aerobic blood cultures grew gram-variable cocci after 5 days of incubation at 35°C. No antibiotics were given initially because of the lack of systemic symptoms and to improve intraoperative culture yield during curative surgical debridement. One month later, the patient underwent arthrotomy with debridement (including synovectomy and irrigation) and removal of the suspected infected right total knee arthroplasty hardware. Intraoperatively, two large abscess cavities were noted in the lateral and medial gutter extending to the medial femoral condyle. After cultures of the purulent material were obtained, 4 g of vancomycin and 4 g of tobramycin antibiotic-impregnated articulating cement spacers were placed in the right knee. Gram staining of these intraoperative knee fluid collections showed 3+ white blood cells but no organisms. This fluid was inoculated onto blood, MacConkey agar plates, chocolate agar plates, brain heart infusion (BHI) broth, and brucella agar anaerobically. These cultures grew 3+ gram-variable cocci anaerobically (brucella agar) in 3 to 4 days and, more slowly, grew aerobically (blood agar) incubated at 35°C in 5% CO2.
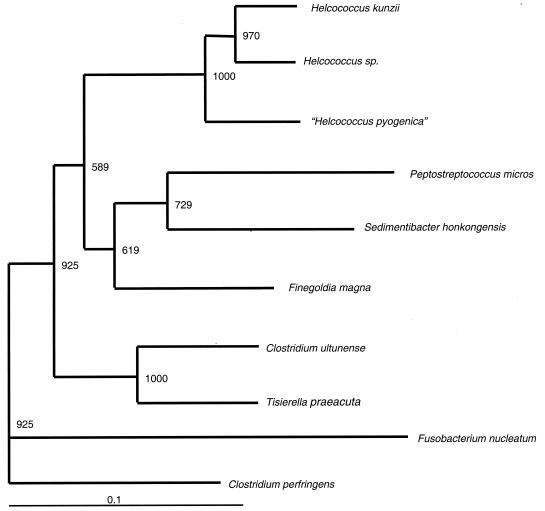
Template DNA extracted from both isolates was amplified by PCR with primers P1 (5′-AGAGTTTGATCCTGGCTCAG-3′) and P3 (5′-TTACCGCGGCTGCTGGCA-3′), and the first ∼500 bp of the 16S rRNA gene from both isolates was sequenced as previously described (7). Pair-wise alignments of the 16S rRNA gene showed that each of the two isolates (at initial presentation and intraoperative cultures) was genetically identical to the other and most closely resembled Helcococcus sp. (93% sequence similarity). A phylogenetic tree was constructed by using PHYLIP (version 3.573; J. Q. Felsenstein, Department of Genetics, University of Washington, Seattle, Wash. [http://evolution.genetics.washington.edu/phylip.html]). The branching order of the neighbor-joining dendrograms was evaluated with 1,000 bootstrap analyses by using the SEQBOOT program in the PHYLIP software package. Our phylogenetic analysis demonstrated that the organism fell within the family Peptostreptococcaceae, most closely related to the Helcococcus genus (Fig. 1). These findings suggest that the isolates most likely represent a previously undescribed species that we have named “Helcococcus pyogenica.”
FIG. 1.
Phylogenetic analysis at the 16S DNA locus of our clinical isolate compared to other members of the Peptostreptococcaceae family, with Clostridium perfringens as the outgroup. The GenBank sequence accession numbers used for analysis are as follows: for Peptostreptococcus micros, U60326; for Helcococcus kunzii, X69837; for Helcococcus sp., Y1629; for “Helcococcus pyogenica,” AY559040; for Finefoldia magna, AY169413; for Sedimentibacter hongkongensis, AF43316; for Clostridium ultunense, Z69293; for Tisierella praeacuta, X80833; for Clostridium perfringens, AB075767; and for Fusobacterium nucleatum, AJ006964.
The isolate was catalase negative, pyrase negative, leucine aminopeptidase (LAP) positive, and esculin hydrolysis negative and grew better anaerobically than aerobically as slowly growing, pinpoint grayish white colonies in 2 to 4 days (nonhemolytic) (Table 1). Antimicrobial susceptibility was determined by using E test methodology (AB Biodisk, Solna, Sweden) according to the instructions of the manufacturer, and Staphylococcus aureus (ATCC 29213) was used as a control organism for the strips; of note, the vancomycin MIC for the control was 0.5 μg/ml and was within quality control limits (0.5 to 2.0 μg/ml). Chocolate agar plates were inoculated with a suspension of the organism standardized to a 0.5 McFarland standard. E test strips were placed, and the plates were incubated at 35°C in 5% CO2 and read at 72 h. E test results of the isolate were as follows: penicillin MIC, 0.032 μg/ml; vancomycin MIC, 8.0 μg/ml; levofloxacin MIC, 0.25 μg/ml; ciprofloxacin MIC, 0.25 μg/ml; and linezolid MIC, 1.0 μg/ml. The vancomycin MIC of 8.0 μg/ml was confirmed by repeat testing with E test.
TABLE 1.
Differentiating features of unusual gram-positive coccia
| Genus | CAT | PYR | LAP | Vancomycin | Gas from glucose | Esculin hydrolysis | Growth in 6.5% NaCl |
|---|---|---|---|---|---|---|---|
| Leuconostoc | − | − | − | R | + | V | V |
| Globicatella | − | + | − | S | − | + | + |
| Pediococcus | − | − | + | R | − | V | V |
| Gemella | − | + | V | S | − | − | − |
| A. viridans | − | + | − | S | − | V | + |
| Helcococcus | − | + | − | S | − | + | V |
| Our organism isolates | − | − | + | MIC = 8.0 μg/ml | − | − | − |
For details, see reference 8. +, positive; −, negative; CAT, catalase; PYR, pyrrolidonylarylamidase; V, variable; R, resistant; S, susceptible.
Since the patient reported developing a rash when she received penicillin in the past, she was initially treated with 750 mg of levofloxacin orally each day but was eventually switched to 400 mg of moxifloxacin orally each day and 600 mg of rifampin orally each day to complete an 8-week course of systemic antibiotics as treatment for prosthetic knee joint infection. Follow-up levels of erythrocyte sedimentation rate and C-reactive protein at 4 weeks of treatment had normalized to 20 mm/h and 14.6 mg/liter, respectively. At the completion of her antibiotic course, the patient was functioning well clinically and underwent the second stage of her right knee surgery with a revision total knee arthroplasty without complications.
We describe a case of prosthetic joint infection with a slowly growing and novel gram-variable coccus. Several lines of evidence suggest that “Helcococcus pyogenica” isolated from this patient was pathogenic and was responsible for severe prosthetic joint infection in this patient: (i) it was isolated on two separate occasions from a sterile site, (ii) definite evidence of infection was present (presence of a large abscess at the time of surgery), (iii) there was an absence of other pathogens, and (iv) the infection responded to antibiotic treatment. The organism shared many phenotypic characteristics with other “unusual gram-positive cocci.” For example, it was gram variable, as it decolorized easily like Gemella hemolysans, and had a colonial morphology similar to Aerococcus viridans and Helcococcus kunzii (from the Greek word “helkos,” meaning wound) (1-6, 9). Attempts to identify this organism by phenotypic methods were unsuccessful, as several important biochemical differences were noted between the various fastidious cocci and our patient's isolate (Table 1). As shown in Table 1, this isolate most closely resembled Pediococcus phenotypically, given that it was catalase negative, pyrase negative, and LAP positive, and it did not hydrolyze esculin. In addition, it demonstrated relative vancomycin resistance (vancomycin MIC, 8.0 μg/ml) like Pediococcus. However, molecular analysis that utilized sequencing of the 16S rRNA gene showed that the organism was most closely related to the Helcococcus genus, sharing only 93% homology with previously published Helcococcus spp.; this result suggests that this organism represents a new species that we have named “Helcococcus pyogenica.”
In summary, we describe a prosthetic joint infection due to a slow-growing and previously undescribed organism whose genetic sequence had not been described in the National Center for Biotechnology Information database and is most closely related genetically to Helcococcus sp. (“Helcococcus pyogenica”; GenBank accession number AY559040). This organism phenotypically resembles several members of the unusual gram-positive cocci and demonstrates relative resistance to vancomycin like Pediococcus. Further characterization of the organism at the biochemical and molecular levels needs to be performed. Finally, the spectrum of disease, natural reservoir, and optimal treatment of this previously undescribed bacterium remain to be defined.
Nucleotide sequence accession number.
The partial gene sequence of the isolate has been deposited in the GenBank sequence database under the accession number AY559040.
REFERENCES
- 1.Caliendo, A. M., C. D. Jordan, and K. L. Ruoff. 1995. Helcococcus, a new genus of catalase-negative, gram-positive cocci isolated from clinical specimens. J. Clin. Microbiol. 33:1638-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chagla, A. H., A. A. Borczyk, R. R. Facklam, and M. Lovgren. 1998. Breast abscess associated with Helcococcus kunzii. J. Clin. Microbiol. 36:2377-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins, M. D., R. R. Facklam, U. M. Rodrigues, and K. L. Ruoff. 1993. Phylogenetic analysis of some Aerococcus-like organisms from clinical sources: description of Helcococcus kunzii gen. nov., sp. nov. Int. J. Syst. Bacteriol. 43:425-429. [DOI] [PubMed] [Google Scholar]
- 4.Collins, M. D., E. Falsen, G. Foster, L. R. Monasterio, L. Dominguez, and J. F. Fernandez-Garazabal. 1999. Helcococcus ovis sp. nov., a gram-positive organism from sheep. Int. J. Syst. Bacteriol. 49:1429-1432. [DOI] [PubMed] [Google Scholar]
- 5.Haas, J., S. L. Jernick, R. J. Scardina, J. Teruya, A. M. Caliendo, and K. L. Ruoff. 1997. Colonization of skin by Helcococcus kunzii. J. Clin. Microbiol. 35:2759-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peel, M. M., J. M. Davis, K. J. Griffin, and D. L. Freedman. 1997. Helcococcus kunzii as sole isolate from an infected sebaceous cyst. J. Clin. Microbiol. 35:328-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pottumarthy, S., A. P. Limaye, J. L. Prentice, Y. B. Houze, S. R. Swanzy, and B. T. Cookson. 2003. Nocardia veterana, a new emerging pathogen. J. Clin. Microbiol. 41:1705-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruoff, K. L. 1999. Leuconostoc, Pediococcus, Stomatococcus, and miscellaneous gram-positive cocci that grow aerobically, p. 306-315. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 9.Stavri, H., T. J. Beveridge, D. Moyles, A. Athamna, and R. J. Doyle. 2002. Hemagglutinin of unusual specificity from Helcococcus kunzii. Arch. Microbiol. 177:197-199. [DOI] [PubMed] [Google Scholar]