Abstract
From December 2002 to June 2003, 14 cultures of Salmonella enterica serotype Babelsberg and 6 cultures of serotype Enteritidis, isolated in France from internationally adopted children, were identified at the French National Reference Center for Salmonella. All serotype Babelsberg isolates were related, as determined by pulsed-field gel electrophoresis, and all serotype Enteritidis strains displayed the same phage type. All serotype Enteritidis and seven serotype Babelsberg isolates produced an SHV-12-like extended-spectrum β-lactamase as determined by sequencing of PCR products and by isoelectrofocusing. Some serotype Enteritidis isolates exhibited additional antimicrobial resistance (aminoglycosides, tetracycline, chloramphenicol, sulfonamides, and trimethoprim). Our investigation indicated that these Salmonella isolates were certainly acquired in the same orphanage in Bamako, Mali, before the children were adopted by French families. An inappropriate use of ceftriaxone was probably the cause of the emergence of such strains. There is an urgent need to determine the origin of the contamination and to introduce adequate antibiotic protocols into this orphanage to prevent further transmission and dissemination. Screening for infections and follow-up, adapted to the origin of the internationally adopted children, should be recommended.
Resistance to the extended-spectrum cephalosporins among members of the family Enterobacteriaceae has become a growing worldwide problem since the first description of a plasmid-mediated extended-spectrum β-lactamase (ESBL) in Klebsiella pneumoniae 20 years ago, shortly after the introduction of the new oxyimino-cephalosporins (11). Among nontyphoidal Salmonella enterica strains, one of the most important food-borne enteric pathogens worldwide, ESBL-producing strains are extremely rare. However, the occurrence of such strains in the different serotypes of this organism has been increasing in recent years. This trend is of particular concern because the extended-spectrum cephalosporins are the antibiotics of choice for children. ESBLs found in Salmonella are most often derivatives of the TEM or SHV families, although some other unrelated enzyme groups including PER and CTX-M have also been found (1, 4, 6, 9, 12, 15, 16, 20, 22, 23, 24).
We report here the first isolation of SHV-12 ESBL-producing S. enterica isolates in France. From December 2002 to June 2003, 14 clinical isolates of S. enterica serotype Babelsberg (48:z4,z23:−) were received for serotyping at the French National Reference Center for Salmonella. Because this serotype is very rare, these isolates attracted our interest. They were recovered from stool specimens from 13 infants (in most cases infants younger than 1 year) and 1 adult. The investigation revealed a common origin. The infants all originated from Mali and had resided in the same orphanage in Bamako before being adopted by French families. The adult was the French adoptive mother of one of the infants. Epidemiological characterization of these isolates was done by pulsed-field gel electrophoresis (PFGE), and the β-lactamases were characterized by isoelectric focusing (IEF) and molecular methods. Five isolates of multiresistant S. enterica serotype Enteritidis detected in adopted infants originating from the same orphanage were also studied.
MATERIALS AND METHODS
Bacterial strains.
The 20 Salmonella clinical isolates used in this study are listed in Table 1. All the isolates were submitted for serotyping to the French National Reference Center for Salmonella (NRC-Salm) from December 2002 to June 2003, except for isolate 3, which was isolated in 2001. The isolates were serotyped on the basis of somatic O and phase 1 and 2 flagellar antigens by agglutination tests with antisera (Bio-Rad, Marnes la Coquette, France, and WHO Collaborative Centre for Reference and Research on Salmonella, Institut Pasteur, Paris, France), as specified by the White-Kauffman-Le Minor scheme (19).
TABLE 1.
Characteristics of the Salmonella isolates under study
| Isolate | Serotype | Source | Sex/age (mo) | Illness | Antimicrobial resistance patterna | pI(s) | bla gene(s) | Size of class I integron and inserted gene cassette |
|---|---|---|---|---|---|---|---|---|
| 1 | Enteritidis | Stools | M/16 | No | ESBL, To, G, Su, T, C, Tp | 5.4, 8.2 | blaTEM, blaSHV | 750 bp |
| 2 | Enteritidis | Stools | M/4 | Yes | ESBL, To, G, Su, T, C, Tp | 5.4, 8.2 | blaTEM-1, blaSHV-12 | 750 bp |
| 3 | Enteritidis | Blood | M/2 | Death | ESBL, Su, Tp | 5.4, 8.2 | blaTEM, blaSHV-12 | 750 bp dfrA15 |
| 4c | Enteritidis | Blood | M/2 | Yes | ESBL, To, G, Su, T, C, Tp | 5.4, 8.2 | blaTEM, blaSHV | 750 bp |
| 5 | Enteritidis | Blood | M/2 | Yes | ESBL, To, G, Su, T, C, Tp | 5.4, 8.2 | blaTEM, blaSHV | 750 bp dfrA15 |
| 6 | Enteritidis | Stools | F/8 | No | ESBL, Su, Tp | 5.4, 8.2 | blaTEM, blaSHV | 750 bp |
| 7 | Babelsberg | Stools | F/<12 | No | ESBL | 5.4, 8.2 | blaTEM, blaSHV | NDb |
| 8 | Babelsberg | Stools | M/<12 | Yes | Ap | 5.4 | blaTEM | ND |
| 9 | Babelsberg | Stools | F/8 | Yes | Ap | 5.4 | blaTEM | ND |
| 10 | Babelsberg | Stools | F/3 | Yes | ESBL | 5.4, 8.2 | blaTEM, blaSHV-12 | ND |
| 11 | Babelsberg | Stools | F/<12 | No | ESBL | 5.4, 8.2 | blaTEM, blaSHV-12 | ND |
| 12 | Babelsberg | Stools | M/<12 | No | Ap | 5.4 | blaTEM | ND |
| 13 | Babelsberg | Stools | F/adult | No | Ap | 5.4 | blaTEM | ND |
| 14 | Babelsberg | Stools | M/2 | No | ESBL | 5.4, 8.2 | blaTEM, blaSHV | ND |
| 15c | Babelsberg | Stools | M/2 | Yes | ESBL | 5.4, 8.2 | blaTEM, blaSHV | ND |
| 16 | Babelsberg | Stools | F/<12 | ?d | Ap | 5.4 | blaTEM | ND |
| 17 | Babelsberg | Stools | F/2 | ? | ESBL | 5.4, 8.2 | blaTEM, blaSHV | ND |
| 18 | Babelsberg | Stools | F/3 | Yes | Ap | 5.4 | blaTEM | ND |
| 19 | Babelsberg | Stools | M/5 | Yes | Ap | 5.4 | blaTEM | ND |
| 20 | Babelsberg | Stools | M/3 | No | ESBL | 5.4, 8.2 | blaTEM, blaSHV | ND |
Abbreviations: Ap, ampicillin; To, tobramycin; G, gentamicin; Tp, trimethoprim; T, tetracycline; Su, sulfonamides; C, chloramphenicol.
ND, none detected.
Same case.
?, not known.
Escherichia coli ATCC 25922 was used as a susceptible control in the disk diffusion method and in MIC determinations. E. coli C1a (nalA) was used for conjugation experiments. E. coli DH5α-T1-R (Invitrogen, Groningen, The Netherlands) was used for heat shock transformation experiments. The reference strain of S. enterica serotype Babelsberg was kindly provided by M. Y. Popoff (WHO Collaborative Centre for Reference and Research on Salmonella). Three S. enterica serotype Babelsberg strains from the NRC-Salm collection (isolated in 1978, 1984, and 1986) were used as controls in the PFGE experiment.
Phage typing.
Phage typing of S. enterica serotype Enteritidis isolates was done by a standard method (25). Phage suspensions were kindly provided by the Health Protection Agency, Colindale, United Kingdom.
PFGE.
Strains of S. enterica serotype Babelsberg were grown on Trypto-Casein soy agar (Bio-Rad) for 48 h at room temperature. After a purity check, several colonies were resuspended in cell suspension buffer (100 mM Tris, 100 mM EDTA [pH 8]) at an optical density at 610 nm of 1.35 to 1.4. A 120-μl volume of the cell suspension containing 10 μg of lysozyme (Sigma-Aldrich Chemie, Steinheim, Germany) was mixed with an equal volume of 1.6% Incert agarose (BMA, Rockland, Maine) and allowed to solidify in 100-μl molds. Plugs were incubated in 1 ml of lysis buffer A (6 mM Tris [pH 7.6], 1 M NaCl, 0.1 M EDTA, 0.5% Brij 58, 0.2% deoxycholate, 0.5% Sarkosyl) with 1 mg of lysozyme and 20 μg of DNase-free RNase. After 2 h at 37°C, the lysis buffer was removed and the plugs were washed with 1 ml of TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA [pH 8.0]) and then incubated for 20 h at 50°C with 1 ml of lysis buffer B (0.25 M EDTA, 20 mM EGTA [pH 9.0]) supplemented with 500 μg of proteinase K and 1% Sarkosyl. The plugs were washed three times with 1.5 ml of TE buffer and three more times with distilled water and then digested with 30 U of XbaI (Roche Diagnostics, Mannheim, Germany) overnight at 37°C. Fragments of DNA were separated by PFGE in a 1% agarose gel (Seakem Gold; BMA) in 0.5× TBE buffer (0.045 M Tris [pH 8], 0.045 M boric acid, 0.01 M EDTA [pH 8.0]), using a CHEF-DR III (Bio-Rad). The running conditions were 6 V/cm at 12°C for 20 h, with pulse times ramped from 2.2 to 63.8 s. Lambda Ladder PFG marker (New England BioLabs, Beverly, Mass.) was used as the molecular size marker.
The gel was stained in 1 μg of ethidium bromide per ml, and the gel image was electronically captured using a video camera interfaced to a microcomputer (ImageMaster VDS; Amersham-Pharmacia Biotech, Freiburg, Germany).
Antimicrobial susceptibility testing.
All isolates were screened for resistance to 32 antimicrobials on Mueller-Hinton (MH) agar by the disk diffusion method following the guidelines of French National Antibiogram Committee (communiqué 2003, www.sfm.asso.fr/nouv/general.php ?pa=2). The following antimicrobials (Bio-Rad) were tested : amoxicillin, amoxicillin-clavulanic acid, ticarcillin, ticarcillin-clavulanic acid, piperacillin, piperacillin-tazobactam, cephalothin, cefamandole, cefoperazone, cefoxitin, ceftriaxone, ceftazidime, cefepime, aztreonam, moxalactam, imipenem, streptomycin, spectinomycin, kanamycin, tobramycin, netilmicin, gentamicin, amikacin, isepamicin, nalidixic acid, pefloxacin, ciprofloxacin, sulfonamides, trimethoprim, chloramphenicol, and tetracycline.
The MICs of the β-lactams were determined by E test (AB Biodisk, Solna, Sweden). The ESBL phenotype was detected by using the ESBL detection E-test strips and the double disk diffusion test.
Preparation of crude extracts of β-lactamase and IEF.
Crude extracts of β-lactamases were obtained by sonication. IEF was performed with a PhastSystem apparatus (Amersham-Pharmacia Biotech) using polyacrylamide gels containing ampholytes with a pH range of 3 to 9 (PhastGel IEF 3-9; Amersham-Pharmacia Biotech). β-Lactamase activity was revealed by staining the gel with 0.5 mg of the chromogenic β-lactam nitrocefin (Oxoid, Basingstoke, England) per ml. Crude extracts of the TEM-1 (isoelectric point [pI] 5.4), TEM-2 (pI 5.6), SHV-1 (5.7), PSE-2 (pI 6.1), SHV-1 (pI 7.6), and SHV-12 (pI 8.2) isolates were used as controls.
PCR amplification of β-lactamase genes and class I integrons, and sequence analysis.
Total DNA was extracted using the InstaGene matrix kit (Bio-Rad) as recommended by the manufacturer. Primers TEM-F (5′-ATAAAATTCTTGAAGACGAAA-3′) and TEM-R (5′-GACAGTTACCAATGCTTAATC-3′) were used to amplify a 1,080-bp fragment of the blaTEM gene (14). Primers SHV-F (5′-TTATCTCCCTGTTAGCCACC-3′) and SHV-R (5′-GATTTGCTGATTTCGCTCGG-3′) were used to amplify a 795-bp internal fragment of the blaSHV gene (3). All amplifications were performed on 50-μl samples containing DNA (2.5 μl), primers (50 pmol each), deoxynucleoside triphosphate (200 μM), Taq DNA polymerase (1.25 U of Ampli Taq Gold; Roche Diagnostics) and its buffer, MgCl 2 (2 mM), and dimethyl sulfoxide (10%). The cycling conditions included 10 min of denaturation at 94°C (1 cycle); 30 s of denaturation at 94°C, 30 s of annealing at 50°C, and 1 min of polymerization at 72°C (35 cycles); followed by 10 min of extension at 72°C.
For amplification of class I integrons, primers 5′-CS (5′-GGCATCCAAGCAGCAAGC-3′) and 3′-CS (5′-AAGCAGACTTGACCTGAT-3′) were used (13). The PCR mix was identical to that described above. The cycling conditions included 10 min of denaturation at 94°C (1 cycle); 30 s of denaturation at 94°C, 30 s of annealing at 55°C, and 2 min of polymerization at 72°C (30 cycles); followed by 10 min of extension at 72°C.
The purified PCR fragments were sequenced on both strands by Genome Express (Meylan, France) using an ABI 100 DNA sequencer (Applied Biosystems, Foster City, Calif.). The nucleotide sequence was analyzed with Lasergene software (Dnastar, Madison, Wis.). The BLASTN program of NCBI (http://www.ncbi.nlm.nih.gov) was used for databases searches.
Resistance transfer determination.
A resistance transfer experiment was carried out with four ESBL-producing isolates and two TEM-producing isolates on liquid and solid media (8). E. coli C1a resistant to nalidixic acid was used as the recipient strain. Transconjugants were selected on Mueller-Hinton agar supplemented with ceftazidime (2 mg/liter) or amoxicillin (100 mg/liter) and nalidixic acid (68 mg/liter). Transformation of plasmid DNA from Salmonella isolates was performed using heat shock transformation with DH5α-T1-R competent E. coli. Transformants were selected on MH agar containing ceftazidime (2 mg/liter) or amoxicillin (100 mg/liter).
Plasmid analysis.
Plasmid DNA was purified from bacterial cells by an alkaline lysis procedure using the plasmid mini kit or the plasmid midi kit (Qiagen, Courtaboeuf, France) and subjected to agarose gel electrophoresis (0.8% agarose). Molecular sizes of plasmids were determined using Taxotron software (Institut Pasteur, Paris, France) by reference to the following plasmids of known sizes (RP4, 54 kb; and pIP173, 126 kb) mixed with a supercoiled DNA ladder (Invitrogen). For Southern hybridization, plasmid DNA was transferred to a nylon membrane (Hybond N+; Amersham-Pharmacia Biotech) and hybridized with PCR-generated probes for blaSHV (795 bp) and blaTEM (1,080 bp) as labeled and detected using the ECL random-prime labeling and detection systems, version II, and Hyperfilm ECL (Amersham Biosciences, Freiburg, Germany).
RESULTS
Molecular and phage typing.
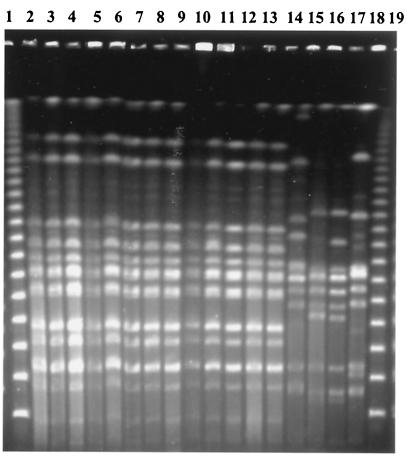
The clonal relatedness of the 14 serotype Babelsberg isolates was assessed by PFGE analysis of XbaI-digested chromosomal DNA (Fig. 1). The same PFGE pattern was identified in all 14 isolates under study. Three serotype Babelsberg strains from our collection and the reference strain displayed different restriction patterns. The same phage type, PT 4, was identified among the serotype Enteritidis isolates.
FIG. 1.
PFGE of XbaI-digested genomic DNA from 17 S. enterica serotype Babelsberg isolates. Lanes: 1 and 19, bacteriophage lambda DNA concatemers (New England Biolabs); 2 to 14, isolates 7, 10, 11, 14, 15, 17, 20, 8, 9, 12, 16, 18, and 19, respectively (isolate 13 is not shown here); 15 to 17, NRC-Salm collection strains; 18, reference strain.
Characterization of β-lactamase genes and class I integrons.
The pattern of antimicrobial resistance of serotype Babelsberg isolates was consistent with the expression of ampicillin resistance (Apr) in seven cases and ESBL production in seven other cases (Table 1). All serovar Enteritidis isolates produced a β-lactamase consistent with an ESBL, and, in contrast to serotype Babelsberg, there were additional resistances depending on the isolates (tobramycin, gentamicin, sulfonamides, trimethoprim, chloramphenicol, and tetracycline) (Table 1). ESBL detection E-test strips and double disk diffusion test confirmed the presence of ESBL. The β-lactam MICs of for representative clinical isolates and their DH5α-T1-R transformants are listed in Table 2.
TABLE 2.
MICs of β-lactams for S. enterica serotype Enteridis isolate 1 (ESBL), serotype Babelsberg isolate 10 (ESBL), serotype Babelsberg isolate 12 (ApR), their respectives DH5α E. coli transformants, and E. coli DH5α
| β-Lactam(s)a | MIC (mg/liter) for:
|
||||||
|---|---|---|---|---|---|---|---|
| Enteritidis isolate 1 | E. coli Tf 1-1b | Babelsberg isolate 10 | E. coli Tf 10-1 | Babelsberg isolate 12 | E. coli Tf 12-1 | E. coli DH5α | |
| Ampicillin | >256 | >256 | >256 | >256 | >256 | >256 | 2 |
| Amoxicillin-CLA | 16 | 4 | 16 | 16 | 16 | 16 | 2 |
| Ticarcillin | >256 | >256 | >256 | >256 | >256 | >256 | 2 |
| Ticarcillin-CLA | >256 | 4 | >256 | 128 | 64 | 32 | 1 |
| Piperacillin | >256 | 32 | >256 | >256 | >256 | 256 | 0.5 |
| Piperacillin-TZB | >256 | 0.5 | >256 | 64 | 4 | 1 | 0.5 |
| Cefoxitin | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Ceftazidime | >256 | 16 | >256 | >256 | 0.5 | 0.125 | 0.125 |
| Ceftazidime-CLA | 0.5 | 0.125 | 0.25 | 0.125 | NT | NT | NT |
| Ceftriaxone | 32 | 1 | 32 | 8 | 0.06 | 0.03 | 0.03 |
| Cefotaxime-CLA | 0.125 | 0.03 | 0.06 | 0.03 | NT | NT | NT |
| Cefepime | 4 | 0.25 | 4 | 2 | 0.125 | 0.06 | 0.03 |
| Aztreonam | 256 | 8 | >256 | >256 | 0.06 | 0.03 | 0.03 |
| Imipenem | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 |
Abbreviations: CLA, clavulanic acid; TZB, tazobactam.
Tf, transformant.
NT, not tested.
The ESBL-producing isolates exhibited a high level of resistance to penicillins, aztreonam, and ceftazidime (MIC ≥ 256 mg/liter) and a lower level of resistance to ceftriaxone (MIC = 32 mg/liter). PCR analysis with the blaSHV primers showed the presence of a 795-bp fragment only in the seven serotype Babelsberg ESBL-producing isolates and in all the serotype Enteridis isolates. The sequence from both strands of PCR products from four isolates (isolates 2, 3, 10, and 11) was 100% homologous to the sequence of the respective segment of the blaSHV-12 gene (GenBank accession number AY293070). IEF was compatible with the DNA analysis in showing a β-lactamase with pI of 8.2 (known for SHV-12) in all the ESBL-producing Salmonella isolates.
PCR analysis identified the 1,080-bp fragment of the blaTEM gene in all 21 isolates. Sequencing of the entire nucleotide sequence from both strands in isolate 2 identified blaTEM-1. It was consistent with the production of a β-lactamase with a pI of 5.4 found in all the isolates.
All the serotype Enteritidis isolates but none of the serotype Babelsberg contained class I integrons (Table 1). A PCR product of approximately 750 bp was obtained from each isolate of serotype Enteritidis when the 5′-CS and 3′-CS primers were used. The sequence of both strands of PCR products from isolates 3 and 5 was 100% homologous to the entire sequence of the dfrA15 (dhfrXV) gene cassette encoding resistance to trimethoprim (GenBank accession number Z83311).
Resistance transfer and plasmid analysis.
Conjugation studies were performed to determine whether ampicillin or ESBL resistance could be transferred from serotypes Babelsberg and Enteritidis to E. coli. Despite multiple attempts with different isolates (isolates 1, 3, 9, 10, and 14) in liquid or on solid media, resistance could not be transferred by conjugation to E. coli. Only β-lactam resistances (ampicillin and ESBL) were transferred by heat shock bacterial transformation followed by selection with amoxicillin or ceftazidime and nalidixic acid.
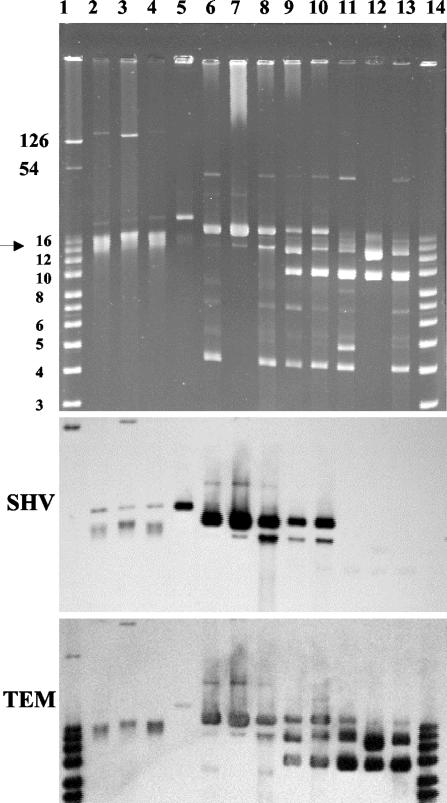
ESBL-producing Salmonella serotype Enteritidis isolates contained two plasmids, a large one (>125 kb) and a small one (ca. 21 kb) (Fig. 2). Hybridization experiments revealed that the blaSHV gene was located on the 21-kb plasmid. In isolate 6, blaSHV was located on both plasmids. After transfer of plasmid DNA from isolate 1 to E. coli, a single plasmid of approximatively 21 kb was found in transformant 1-1. Hybridization was strongly positive with the blaSHV probe. The transformant did not express aminoglycoside or other associated resistances but expressed only pI 8.2 ESBL. It exhibited a lower resistance to β-lactams than did the parental strain as indicated by the MIC determination (Table 2), thus indicating that blaSHV-12 was the only antimicrobial resistance gene located on the 21-kb plasmid. The other genes might be carried by the large plasmid.
FIG. 2.
Plasmid content analysis of selected S. enterica serotype Enteritidis and Babelsberg isolates (top) and Southern hybridization with DNA probes corresponding to blaSHV-type (SHV) and blaTEM-type (TEM) genes (middle and bottom). The chromosome position is indicated by arrowheads. Lanes: 1, supercoiled DNA ladder (Invitrogen) plus RP4 and pIP173 plasmids which served as molecular size markers (band sizes in kilobase pairs); 2, serotype Enteritidis ESBL isolate 5; 3, serotype Enteritidis ESBL isolate 6; 4, serotype Enteritidis ESBL isolate 1; 5, E. coli transformant 1-1; 6, serotype Babelsberg ESBL isolate 10; 7, E. coli transformant 10-1; 8, serotype Babelsberg ESBL isolate 11; 9, serotype Babelsberg ESBL isolate 7; 10, serotype Babelsberg ESBL isolate 14; 11, serotype Babelsberg ApR isolate 9; 12, E. coli transformant 9-1; 13, serotype Babelsberg ApR isolate 8; 14, supercoiled DNA ladder (Invitrogen).
A common 4-kb plasmid was found in all the serotype Babelsberg isolates. ESBL-producing isolates contained a common 19-kb plasmid and, depending on the isolate, up to four additional plasmids. Transfer of a plasmid from isolate 10 resulted in E. coli DH5α transformant 10-1, which contained a single 19-kb plasmid. This plasmid hybridized with the blaSHV and blaTEM probes. The transformant expressed pI 8.2 ESBL and pI 5.4 β-lactamase as indicated by IEF analysis, thus indicating that both genes were carried by the 19-kb plasmid. E. coli transformant 10-1 exhibited a similar level of β-lactam resistance to that of the parental strain (Table 2).
Apr producing serotype Babelsberg isolates contained up to five plasmids ranging from 4 to 45 kb. A 10-kb plasmid was common to all the Apr serotype Babelsberg isolates and E. coli DH5α transformant 12-1. This plasmid hybridized with the blaTEM probe. It was also present in two tested ESBL-producing serotype Babelsberg isolates (isolates 7 and 14).
DISCUSSION
SHV-12 ESBL was identified in 1997 in a K. pneumoniae strain isolated in Switzerland (17). This ESBL differs from SHV-5 by a single leucine-to-glutamine substitution at position 35 in the SHV-5 protein (numbering in accordance with the scheme in reference 2). Since 1998, SHV-12-encoding plasmids have been found in K. pneumoniae isolates from hospitals in Taiwan, Korea, and Italy (10, 18, 26). The first report of SHV-12 in the genus Salmonella, published in 2001, described five strains (four from humans and one from food) of serotype Keurmassar (35:c:1,2) isolated in 2000 in Dakar, Senegal (6). A second report published in 2002, identified five strains of serotype Enteritidis, harboring at least two types of SHV-12-encoding plasmids, in Italy from 1994 to 1997 (24). Two hypotheses have been suggested to explain the emergence of ESBL-producing Salmonella strains. The first involves nosocomial acquisition by exchange of mobile genetic elements, such as plasmids and transposons, between enteric bacteria frequently encountered in hospitals and selected by traces of antimicrobial agents (oxyimino-cephalosporins) used in humans. There have been several reports of outbreaks of ESBL-producing Salmonella infection in nosocomial environment in both developed and developing countries (9, 15, 16, 20, 23). The second hypothesis is the transmission via the food chain (6). Cardinale et al. (6) described the dual emergence of SHV-12-producing serotype Keurmassar in humans and in a poultry product in Senegal, a country close to Mali. Extensive use of antimicrobial agents as feed additives for farm animals (especially in the poultry industry) could be an essential factor for the emergence of these ESBL-producing Salmonella strains (6). SHV-12-producing E. coli strains have been isolated in healthy chickens in Spain (5).
In our study, the contamination certainly occurred in the orphanage in Bamako because (i) all the infants were adopted by unrelated families living in different regions of France; (ii) serotype Babelsberg is extremely rare in France, with only 1 isolate among 98,692 Salmonella strains reported to the NRC-Salm during the period from 1999 to 2001; (iii) the Babelsberg isolates were clonally related as assessed by PFGE (which was discriminatory for this serotype), and the Enteritidis isolates displayed the same phage type; and (iv) SHV-12 has been identified in Mali (F. X. Weill, unpublished observation) and in Senegal (6). A preliminary visit was made to the orphanage in September 2003. We observed an inappropriate use of ceftriaxone (bad indications, too short a duration of treatment, etc.) in infants, which could explained the emergence of the ESBL-producing strains. There is an urgent need to conduct a more detailed investigation in this orphanage to determine the origin of the contamination and to introduce adequate antibiotic and hygiene protocols in order to prevent further transmission and dissemination. Even if there were more carriage than illness, some infections, especially with the multiresistant Enteritidis serotype, were very severe (one infant infected with isolate 3 died from the infection). The SHV-12 resistance pattern, associated with other resistance markers in serotype Enteritidis, drastically reduced the therapeutic options for these severe infections in infants (the use of fluoroquinolones in children is not approved in France).
The present study also documents an uncommon way of dissemination of ESBL-producing Salmonella through internationally adopted children. Several reports from U.S. authors indicated that children adopted from abroad are at increased risk of infections acquired in their country of origin (7, 21). In these studies, the adopted children rarely had infections with Salmonella (less than 2%) compared to tuberculosis or hepatitis B, but none of the children were adopted from Africa.
Concerning the risk for transmission of infection from these infants to their adoptive families and close contacts, just one case of serotype Babelsberg carriage in an adoptive mother was identified in our study. It seems very important that the adoptive families be aware of the spectrum of possible infections in children and, in particular, the potential of ESBL-producing Salmonella infection. On arrival to France or any country of residence of the adoptive families, screening for infections and follow-up adapted to the origin of the adoptees should be recommended.
Acknowledgments
We thank all the corresponding laboratories of the French Salmonella network and, in particular, F. Geffroy (Laboratoire de Microbiologie, Centre Hospitalier de Cornouaille, Quimper, France), G. Chambreuil (Laboratoire, Centre Hospitalier de La Roche-sur-Yon, France), and P. Plessis (Laboratoire, Centre Hospitalier de Morlaix, France) for providing strains.
REFERENCES
- 1.AitMhand, R., A. Soukri, N. Moustaoui, H. Amarouch, N. ElMdaghri, D. Sirot, and M. Benbachir. 2002. Plasmid-mediated TEM-3 extended-spectrum β-lactamase production in Salmonella typhimurium in Casablanca. J. Antimicrob. Chemother. 49:169-172. [DOI] [PubMed] [Google Scholar]
- 2.Ambler, R. P., A. F. Coulson, J. M. Frere, J. M. Ghuysen, B. Joris, M. Forsman, R. C. Levesque, G. Tiraby, and S. G. Waley. 1991. A standard numbering scheme for the class A β-lactamases. Biochem. J. 276:269-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arlet, G., M. Rouveau, and A. Philippon. 1997. Substitution of alanine for aspartate at position 179 in the SHV-6 extended-spectrum β-lactamase. FEMS Microbiol. Lett. 152:163-167. [DOI] [PubMed] [Google Scholar]
- 4.Bauernfeind, A., I. Stemplinger, R. Jungwirth, P. Mangold, S. Amann, E. Akalin, O. Ang, C. Bal, and J. M. Casellas. 1996. Characterization of β-lactamase gene blaPER-2, which encodes an extended-spectrum class A β-lactamase. Antimicrob. Agents Chemother. 40:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briñas, L., M. A. Moreno, M. Zarazaga, C. Porrero, Y. Sáenz, M. García, L. Dominguez, and C. Torres. 2003. Detection of CMY-2, CTX-M-14, and SHV-12 β-lactamases in Escherichia coli fecal-sample isolates from healthy chichens. Antimicrob. Agents Chemother. 47:2056-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardinale, E., P. Colbachini, J. D. Perrier-Gros-Claude, A. Gassama, and A. Aidara-Kane. 200. Dual emergence in food and humans of a novel multiresistant serotype of Salmonella in Senegal: Salmonella enterica subsp. enterica serotype 35:c:12.. J. Clin. Microbiol. 39:2373-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, L. H., E. D. Barnett, and M. E. Wilson. 2003. Preventing infectious diseases during and after international adoption. Ann. Intern. Med. 139:371-378. [DOI] [PubMed] [Google Scholar]
- 8.Courvalin, P., F. Goldstein, A. Philippon, and J. Sirot (ed.). 1985. Conjugaison chez les bacilles à gram négatif, p. 267-269. In L'antibiogramme. Mpc-vidéom, Paris, France.
- 9.Hammami, A., G. Arlet, S. Ben Redjeb, F. Grimont, A. Ben Hassen, A. Rekik, and A. Philippon. 1991. Nosocomial outbreak of acute gastroenteritis in a neonatal intensive care unit in Tunisia caused by multiply drug resistant Salmonella wien producing SHV-2 β-lactamase. Eur. J. Clin. Microbiol. Infect. Dis. 10:641-646. [DOI] [PubMed] [Google Scholar]
- 10.Kim, J., Y. Kwon, H. Pai, J. W. Kim, and D. T. Cho. 1998. Survey of Klebsiella pneumoniae strains producing extended-spectrum β-lactamases: prevalence of SHV-12 and SHV-2a in Korea. J. Clin. Microbiol. 36:1446-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knothe, H., P. Shah, V. Krcmery, M. Antal, and S. Mitsuhashi. 1983. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 11:315-317. [DOI] [PubMed] [Google Scholar]
- 12.Lee, K., D. Yong, J. H. Yum, H. H. Kim, and Y. Chong. 2003. Diversity of TEM-52 extended-spectrum β-lactamase-producing non-typhoidal Salmonella isolates in Korea. J. Antimicrob. Chemother. 52:493-496. [DOI] [PubMed] [Google Scholar]
- 13.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mabilat, C., S. Goussard, W. Sougakoff, R. C. Spencer, and P. Courvalin. 1990. Direct sequencing of the amplified structural gene and promoter for the extended-broad-spectrum β-lactamase TEM-9 (RHH-1) of Klebsiella pneumoniae. Plasmid 23:27-34. [DOI] [PubMed] [Google Scholar]
- 15.Makanera, A., G. Arlet, V. Gautier, and M. Manai. 2003. Molecular epidemiology and characterization of plasmid-encoded β-lactamases produced by Tunisian clinical isolates of Salmonella enterica serotype Mbandaka resistant to broad-spectrum cephalosporins. J. Clin. Microbiol. 41:2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morosini, M. I., J. Blazquez, M. C. Negri, R. Canton, E. Loza, and F. Baquero. 1996. Characterization of a nosocomial outbreak involving an epidemic plasmid encoding for TEM-27 in Salmonella enterica subspecies enterica serotype Othmarschen. J. Infect. Dis. 174:1015-1020. [DOI] [PubMed] [Google Scholar]
- 17.Nuesch-Inderbinen, M. T., F. H. Kayser, and H. Hachler. 1997. Survey and molecular genetics of SHV β-lactamases in Enterobacteriaceae in Switzerland: two novel enzymes, SHV-11 and SHV-12. Antimicrob. Agents Chemother. 41:943-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perilli, M., E. Dell'Amico, B. Segatore, M. R. de Massis, C. Bianchi, F. Luzzaro, G. M. Rossolini, A. Toniolo, G. Nicoletti, and G. Amicosante. 2002. Molecular characterization of extended-spectrum β-lactamases produced by nosocomial isolates of Enterobacteriaceae from an Italian nationwide survey. J. Clin. Microbiol. 40:611-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popoff, M. Y. 2001. Antigenic formulas of the Salmonella serovars, 8th ed. WHO Collaborating Center for Reference and Research on Salmonella. Institut Pasteur, Paris, France.
- 20.Revathi, G., K. P. Shannon, P. D. Stapleton, B. K. Jain, and G. L. French. 1998. An outbreak of extended-spectrum, β-lactamase-producing Salmonella senftenberg in a burns ward. J. Hosp. Infect. 40:295-302. [DOI] [PubMed] [Google Scholar]
- 21.Saiman, L., J. Aronson, J. Zhou, C. Gomez-Duarte, P. San Gabriel, M. Alonso, S. Maloney, and J. Schulte. 2001. Prevalence of infectious diseases among internationally adopted children. Pediatrics 108:608-612. [DOI] [PubMed] [Google Scholar]
- 22.Tzouvelekis, L. S., E. Tzelepi, P. T. Tassios, and N. J. Legakis. 2000. CTX-M-type β-lactamase: an emerging group of extended-spectrum enzymes. Int. J. Antimicrob. Agents 14:137-142. [DOI] [PubMed] [Google Scholar]
- 23.Vahaboglu, H., S. Dodanli, C. Eroglu, R. Ozturk, G. Soyletir, I. Yildirim, and V. Avkan. 1996. Characterizations of multiple-antibiotic-resistant Salmonella typhimurium strains: molecular epidemiology of PER-1 producing isolates and evidence for nosocomial plasmid exchange by a clone. J. Clin. Microbiol. 34:2942-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villa, M., C. D. Mammina, V. Miriagou, L. S. Tzouvelekis, P. T. Tassios, A. Nastasi, and A. Carattoli. 2002. Multidrug and broad-spectrum cephalosporin resistance among Salmonella enterica serotype Enteritidis clinical isolates in Southern Italy. J. Clin. Microbiol. 40:2662-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward, L. R., J. D. H. De Sa, and B. Rowe. 1987. A phagetyping scheme for Salmonella enteritidis. Epidemiol. Infect. 99:291-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan, J. J., S. M. Wu, S. H. Tsai, J. J. Wu, and I. J. Su. 2000. Prevalence of SHV-12 among clinical isolates of Klebsiella pneumoniae producing extended-spectrum β-lactamases and identification of a novel AmpC enzyme (CMY-8) in Southern Taiwan. Antimicrob. Agents Chemother. 44:1438-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]